Abstract
Myosin is the chemomechanical energy transducer in striated heart muscle. The myosin cross-bridge applies impulsive force to actin while consuming ATP chemical energy to propel myosin thick filaments relative to actin thin filaments in the fiber. Transduction begins with ATP hydrolysis in the cross-bridge driving rotary movement of a lever-arm converting torque into linear displacement. Myosin regulatory light chain (RLC) binds to the lever-arm and modifies its ability to translate actin. Gene sequencing implicated several RLC mutations in heart disease and three of them are investigated here using photoactivatable GFP tagged RLC (RLC-PAGFP) exchanged into permeabilized papillary muscle fibers. Single lever-arm probe orientation is detected in the crowded environment of the muscle fiber by using the RLC-PAGFP with dipole orientation deduced from the 3-spatial dimension fluorescence emission pattern of the single molecule. Symmetry and selection rules locate dipoles in their half sarcomere, identify those at minimum free-energy, and specify active dipole contraction intermediates. Experiments were performed in a microfluidic chamber designed for isometric contraction, total internal reflection fluorescence detection, and 2-photon excitation second harmonic generation to evaluate sarcomere length. The RLC-PAGFP reports apparently discretized lever-arm orientation intermediates in active isometric fibers that on average produce the stall force. Disease linked mutants introduced to RLC move intermediate occupancy further down the free-energy gradient implying lever-arms rotate more to reach stall force because mutant RLC increases lever-arm shear strain. Lower free-energy intermediate occupancy involves less energy conversion efficiency in the fiber relating a specific myosin function modification to the disease implicated mutant.
INTRODUCTION
Myosin is an energy transducer converting ATP free-energy into mechanical work. It has a globular head or subfragment 1 (S1) and a linear tail domain for myosin assembly into thick filaments that interdigitate with actin thin filaments in striated muscle. Myosin’s function is to power the relative sliding of thick and thin filaments during contraction. S1 contains the actin binding site and a lever-arm whose rotary movement while myosin is strongly actin bound cyclically generates tension to move the load as shown in Figure 1. The lever-arm converts torque generated in the motor into the linear displacement characteristic to contraction and undergoes shear strain due to the load resisting translation. Strain under load is shared among the lever-arm and the myosin light chains, essential and regulatory, that bind to the lever-arm. In vitro evidence indicates human cardiac regulatory light chain (HCRLC) probably stabilizes lever-arm structure (1) and that HCRLC mutants implicated in disease lower velocity, force, and strain sensitivity (2) suggesting they alter lever-arm processing of shear stress.
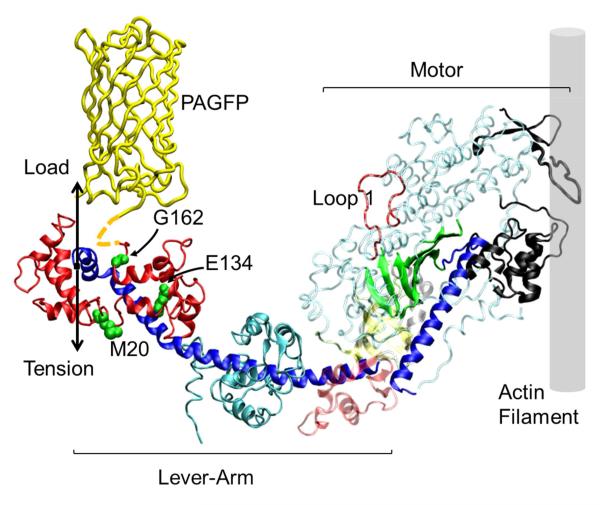
Figure 1.
The human β-cardiac myosin S1 motor binds the actin filament and generates tension with the lever-arm swing. In a muscle fiber the lever-arm links to the myosin filament where Load and Tension vectors are shown. The regulatory light chain (HCRLC in bold red) indicates positions 20, 134, and 162 with mutations implicated in HCM. The PAGFP is linked to the light chain (dashed yellow linker near G162) and reports lever-arm orientation. The ELC is shown in cyan. In the motor, black regions are the actin binding site, the blue α-helix is the switch 2 helix and the green β-sheet is the active site with entrance at Loop 1.
Photoactivatable green fluorescent protein (PAGFP) linked to the HCRLC C-terminus (HCRLC-PAGFP) is expressed in bacteria and exchanged with >50% efficiency with the endogenous RLC in permeabilized porcine cardiac papillary muscle fibers. Upon photoactivation, a sparse population of tagged myosins fluoresce that we detect individually despite the high myosin concentration in the sarcomere. Single molecules emit in a pattern uniquely representing the PAGFP dipole moment orientation as it tracks lever-arm orientation (3). Previously, we observed lever-arm orientation from single myosins in relaxation, rigor, and isometric contraction (4). The active isometric cross-bridge had apparently quantized lever-arm orientations suggesting the presence of sub-states in its force generation cycle.
Single molecule detection (in principal) samples each state of a system addressing problems inherent to an ensemble average approach although practical limitations persist (5). Here we measured PAGFP orientation on myosin in the muscle in rigor, active isometric, and relaxed conditions representing different free-energy and actin attachment states. In a basic contraction mechanism, the myosin states transit from one to another producing force as the actin attached myosin rotates a lever-arm performing the power-stroke. Direct single molecule power-stroke transition detection is beyond current capabilities. Instead we use dispersion in the active isometric fiber to represent cross-bridge states at the high free-energy side of the transition and the unique low free-energy rigor state to represent the low free-energy side. We do not image the same single molecule across the three physiological states for two reasons. First, because the fiber moves from its relaxed or rigor position while the activated fiber becomes isometric and again as the active isometric fiber is returned to a resting state. The movement makes recovery of the single molecule of interest uncertain and impractical. Second, because rigor tension imposition from relaxed or active states produces rigor dispersion in which an unknown subset of rigor cross-bridges reach their unique free-energy minimum. To manage these contingencies we compute the set of likely transitions linking different single molecules in disperse active isometric states to their unique minimum free-energy rigor configuration. Compatible single molecules conform to a symmetry rule associating myosin molecules in the same half-sarcomere and a selection rule associating the disperse active states with their lowest free-energy rigor-state partner. Overall this approach is practical and avoids the rigor dispersion ambiguity. Results are from identical permeabilized fibers except for the introduction of HCRLC point mutations.
Here we report findings on the cardiac papillary muscle fiber system perturbed by three hypertrophic cardiomyopathy (HCM) linked mutations of HCRLC (gene MYL2) (6). Mutants M20L, E134A, and G162R span the length of the regulatory light chain. They survey HCRLC for sensitivity to mutation with M20 near the phosphorylation site at Ser15 (7), E134 a charged residue probably participating in HCRLC/lever-arm binding (8), and G162 at the C-terminus adjacent to the myosin short α-helix at the end of, and orthogonal to, the lever-arm long α-helix. Measurements with exchanged HCRLC-PAGFP show disease linked mutants invert intermediate state occupation favoring lower free-energy in the active cycle when compared to WT. Lower free-energy intermediate occupancy causes lower energy conversion efficiency in the fiber. These findings demonstrate a myosin functional modification characterized at the single molecule level, and in the context of the crowded muscle fiber, linking the HCM phenotype to lower efficiency for ATP free-energy conversion to mechanical work.
METHODS
Preparation of HCRLC
cDNA of wild type MYL2 was a gift from Dr. D. Szczesna-Cordary, University of Miami. The HCRLC-GFP was constructed as previously described (9). Mutagenesis of GFP to PAGFP was also previously described (10). HCRLC mutants M20L, E134A, and G162R were made in the HCRLC-PAGFP pET3d plasmid using the QuikChange site directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Expressed protein was purified and used for fiber exchange as described (4). Protein concentrations used in the fiber exchange were 62.5-108.1 μM.
Solutions
Fiber solutions are based on the protocol from Szczesna-Cordary et al. (11) and were prepared exactly as described previously (4). Rigor, relax, and active solutions were adjusted to 150 mM ionic strength by varying potassium propionate (KPr) concentration and using the program FIBER PCA.xls to estimate total concentrations of species (12). Relax and active solutions are pCa 8 and 4.
Permeabilized fiber preparation
Fiber preparation, dissection, light chain exchange, and tension measurements were carried out as previously described (4). Fiber bundles used in tension or microscope experiments were 3-10 fibers each (width 50-100 μm) and 2 mm long. After completing tension measurements, fibers were severed from the tension transducer and prepared for microscope experiments or protein extraction and SDS/PAGE as described (4).
SDS-PAGE from fiber extracted proteins
Exchanged fibers were tension tested and prepared as described above then proteins were extracted and subjected to SDS-PAGE analysis to measure light chain extraction and reconstitution efficiency as described previously (4). The summary of SDS-PAGE results from tagged HCRLC (WT-PAGFP, M20L-PAGFP, E134A-PAGFP, and G162R-PAGFP) indicate fractional endogenous light chain removal of (0.53±0.03, 0.34±0.20, 0.58±0.16, and 0.60±0.06) and full replacement to within error.
Microfluidic chamber for TIRF and SHG microscope measurements
Experiments were performed in microfluidic channel depicted in Figure 2 and constructed using the toner transfer method as described earlier (13). Microfluidic channel construction is described further in the Supporting Information.
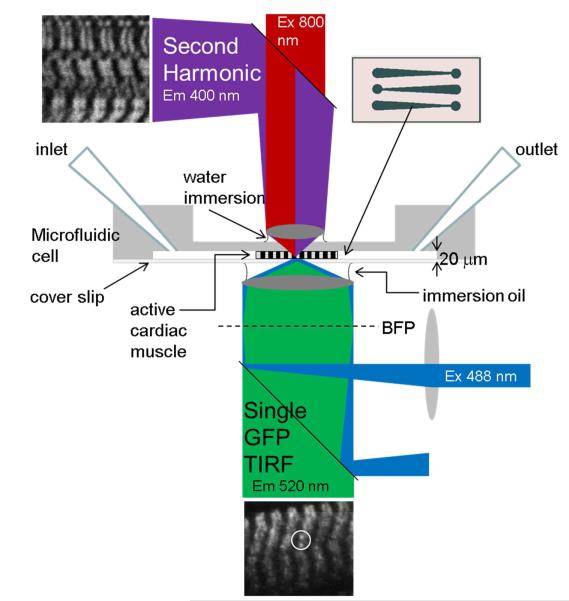
Figure 2.
TIRF observation characterizes single myosin lever-arms in the muscle fiber using exchanged HCRLC-PAGFP. TIRF excitation uses 488 nm laser light focused on the back focal plane (BFP) to excite photoactivated PAGFP that emits with peak intensity at 520 nm. Infrared (800 nm) light focused on the fiber causes myosin SHG to image the periodic myosin structure and monitor sarcomere length. The microfluidic chamber was constructed from a brass master etched into the pattern shown in the insert. Three channels were constructed with depths of 20 (shown) to 40 μm to accommodate different fiber bundle sizes and to apply mild pressure on the contracting fiber to keep it stationary during observation. The channel is also the conduit to exchange solutions. TIRF and SHG experiments are performed sequentially.
Fiber bundles were placed on the polydimethylsiloxane (PDMS) substrate positioned channel side up. Channel width and depth were selected to provide optimal access to solution and minimum compression to hold the activated fiber isometric. The PDMS substrate with mounted fiber was inverted channel side down and placed on a clean glass coverslip. The PDMS made a spontaneous water tight seal and relaxing solution was immediately flowed through the channel. The sample was then ready for microscope experiments.
Connective tissue present in the bundle adhered to the large surface area presented by the microscopically rough PDMS surface allowing a minimum of compression to immobilize the contracting fiber. Fiber length was observed before and after single molecule TIRF measurements to check for length change. Experiments were terminated when the fiber shortened more than ~5% from the starting resting length. Following single molecule experiments using TIRF on the inverted wide field microscope the chamber was immediately taken to the upright scanning 2 photon microscope for measurement of the SHG signal.
Microscopy
Figure 2 shows excitation and emission pathways for the TIRF widefield microscope (Olympus IX71) and the multiphoton excitation (MPE) scanning upright microscope (Olympus FV1000 and BX61) for SHG. Experiments were done sequentially, first the TIRF experiments using a 100X, 1.49 NA oil immersion objective then the SHG experiments using a 25X, 1.05 NA water immersion objective. The water immersion objective has 2 mm working distance to image the fiber through 1 mm of PDMS.
The single molecule TIRF microscope setup using photoactivatable probes is identical to that described previously (3, 4). Experiments involve axial-scanning by image space camera translation. A microscope stage with leadscrew drives and stepper motors (LEP, Hawthorne, NY) translate the EMCCD camera (Hamamatsu C9100-13, Hamamatsu-City, Japan) with submicrometer resolution. The quantitatively analyzed axial-scanned imagery contained 5 frames in a scene covering 1 mm in image space or 100 nm in object space. The 5 frames take 5 seconds to collect with ~40% and 60% of the time interval used to reposition the camera and collect photons. Images span 4.6 seconds of real time because the final camera translation is irrelevant. The porcine left ventricle papillary cardiac muscle fibers used here contains predominantly the β-cardiac myosin heavy chain isoform. The β-cardiac myosin has cycling kinetics 2- to 10-fold slower than the α-cardiac isoform (14, 15). Isometric α-cardiac cross-bridges in permeabilized transgenic mouse myofibrils cycle with a periodicity of ~3 sec (16) suggesting the cross-bridges observed here are approximately static during the 4.6 sec data collection time interval. Two photoactivated single myosins are circled in the representative TIRF image of active isometric permeabilized papillary fiber shown in Figure 2 where 488 nm illumination produces PAGFP emission at ~520 nm.
SHG from striated muscle originates from thick filament myosin rod domains but not S1 (17). SHG images were collected from the permeabilized papillary cardiac muscle fibers used in the single molecule experiments to estimate and compare sarcomere length from relaxed and isometric active fibers. SHG images were Fourier transformed and the spatial frequencies compared as described in Supporting Information. A representative SHG image of a relaxed permeabilized papillary fiber is shown in Figure 2 where 800 nm illumination produces the ~400 nm back scattered SHG image. After SHG imaging, fibers were removed from the microfluidic and placed in activating solution to qualitatively test contractility. Fibers normally shortened to about half the starting length.
Photoselection by photoactivation
Total probe molecules N convert from un-photoactivated (NB) to photoactivated (NA) species by irreversible isomerization, NB ↔NA, as described previously (4). Experiments utilized sparse photoactivation to obtain the highest possible directional ordering in photoactivated probes. Measuring emission dipole orientation of the photoactivated species is the goal accomplished with emission spatial pattern analysis.
Interpretation of single molecule emission patterns
Light collected by the objective forms a real image of the emitter dipole on the CCD detector. The image is a 3-dimensional pattern that devolves into six basis patterns with coefficients that depend algebraically on the dipole orientation. Given the basis patterns, we invert an observed image to deduce the coefficients by using maximum likelihood fitting for Poisson distributed uncertainties. The coefficients establish a one-to-two correspondence between pattern and dipole moment orientations because a particular dipole and its spatial inversion produce the same emission pattern (3, 4). Dipole moment orientation is represented by coordinates (β,α) for β the spherical polar angle and α the azimuth in muscle fiber coordinates where the z-axis is parallel to the fiber symmetry axis, x-axis is in the plane of the coverslip where total internal reflection occurs (TIR interface), and y-axis is normal to the coverslip plane pointing into the aqueous medium. Inversion symmetry implies (β,α) and (π–β,α+π) are equivalent hence the entire solution set of dipole orientations can be shown on the domain defined by 0 ≤ β ≤ π and 0 ≤ α ≤ π. P-polarization illumination, with electric field predominantly oriented normal to the TIR interface and the fiber symmetry axis, is used for photoactivation and emission excitation (18).
Single molecule emission is documented by 5 images in a scene covering lateral and axial dimensions of the pattern. When analyzed, each 5 image scene converts to the emission dipole moment orientation coordinates (β,α). We recorded scenes from fibers in rigor, relaxation, and isometric contraction at room temperature. Each physiological state was characterized by ≥ 48 dipole coordinates (≥ 96 including those related by inversion symmetry) for PAGFP tagged WT, M20L, E134A, and G162R HCRLC exchanged cross-bridges.
Principal angle
We investigate cross-bridge rotation for force production during contraction by assuming the HCRLC-PAGFP rotates as a rigid body and computing principal angle transitions between single lever-arm fixed probe dipoles. Unit vector represents a single dipole orientation computed from coordinates (β,α) using = {Sin[β]Cos[α],Sin[β]Sin[α],Cos[β]}. Let vectors and represent the ith and jth unit dipoles for a set of molecules then the principal angle transition between them is given by,
| (1) |
Sets and are observed from the same fiber physiological state representing orientation dispersion or from different physiological states representing state transition.
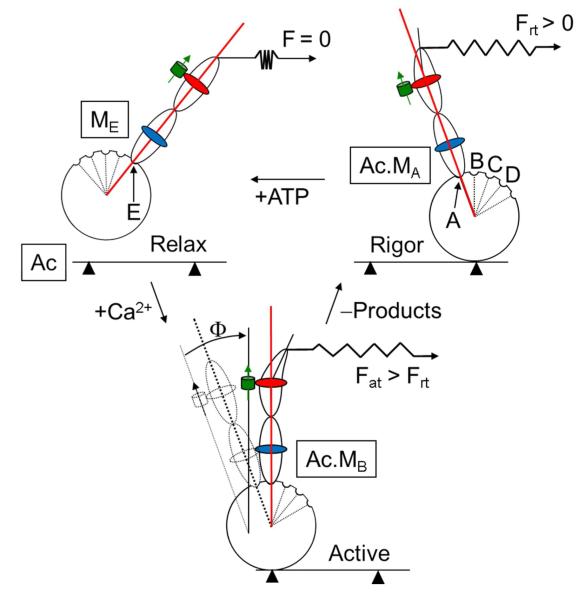
Figure 3 shows a schematic cross-bridge cycle and the meaning of the principal angle state transition, Φ, relating lever-arm positions. A myosin cross-bridge in rigor supports significant rigor tension, Frt, with the lever-arm strained and in the minimum free-energy A-state. ATP binding releases the cross-bridge from actin and re-primes myosin forming the maximum free-energy E-state in relaxation that is weakly actin bound and does not support tension. Addition of Ca2+ initiates contraction causing strong actin binding and the lever-arm rotation in the power stroke. The principal angle, Φ, describing cross-bridge orientation transition between states is depicted in the figure. For simplicity, we represent the tension generating lever-arm swing movement in a plane also containing the PAGFP dipole moment although this is unlikely to happen. Φ amplitude is ≤ the real lever-arm principal swing angle amplitude, Ψ, because just a component of the PAGFP dipole moment lies in the lever-arm swing plane. Thus the Φ’s correspond to a lower bound on Ψ. Free-energy for the lever-arm swing,
| (2) |
as derived previously (4) and for spring constant k ≈ 2 pN/nm in WT skeletal myosin (19), step-size h ≈ 5.2 nm (20), and lever-arm length l ≈ 8.5 nm (21). Eq. 2 implies that tanΦ ≤ h/l or that Φ ≤ 33 deg covers the power stroke. This is the Φ-domain chosen for the Φ-permutation and Φ-density histograms defined and discussed subsequently.
Figure 3.
Simplified cross-bridge cycle in contraction detailing myosin’s discrete sub-states (A-E) and their correspondence to lever-arm orientation. The rigor cross-bridge binds actin (Ac) in the A-state (MA), exerts force Frt, and maintains the PAGFP (green cylinder with arrow) tagged HCRLC (red) in the A-state orientation. ELC is shown in blue. ATP addition relaxes the fiber by binding to myosin and detaching actin. The relaxed cross-bridge assumes the E-state (ME) with the lever-arm re-primed for a new power stroke. Ca2+ addition activates the fiber producing force Fat. In isometric contraction single lever-arms assume one of their intermediates (A-E) depicted here as MB which is a high force producing intermediate having a characteristic orientation indicated by Φ.
Cross-bridge dispersion
Rigor cross-bridges are formed from a single turn-over of ATP when they perform a final power stroke then release product. Rising tension in the fiber implies that cross-bridges releasing ATP hydrolysis products do not all reach their free-energy minimum configuration contributing to rigor cross-bridge orientation dispersion (only the free-energy minimum configuration is indicated in Figure 3). Rising tension in the active isometric fiber implies that unsynchronized cross-bridges reach their stall force at various orientation states in the free-energy pathway leading to active cross-bridge orientation dispersion.
The transitions from disperse active cross-bridges to the minimum free-energy rigor cross-bridge are what we wish to identify and characterize. Hence, while active dispersion is an interesting part of the active-rigor principal angle transition, rigor dispersion is not. We identify the minimum free-energy rigor orientation to minimize rigor dispersion in the active-rigor principal angle. The same concept applies to the relax-rigor principal angle. We introduce a selection rule to identify the minimum free-energy rigor cross-bridge dipole orientation and a symmetry rule to sort dipoles into their half sarcomere subsets. The rules promote correct assignment of active dipole intermediates to their minimum free-energy end point.
Dipole orientation selection and symmetry rules
Photoactivation photoselection selects a similarly oriented dipole set from all dipoles in the muscle fiber. Within the photoactivated set, dipole dispersion due to rigor tension produces a set of dipoles related to a unique minimum free-energy conformation. If Φ describes the angular dispersion among single lever-arm fixed probe dipoles in rigor, we specify the set of all ordered pairs of single molecules in rigor with Φ falling into A-E sub-states of rising Φ values (Figure 3). From the ordered pairs of single molecules we identified each in that has a transition to each of the B-E sub-states. This set of dipoles, always denoted , is the minimum free-energy rigor A-state representing the power-stroke end point. This is the selection rule.
Each has an associated projection scalar,
| (3) |
for dipoles qualifying in the selection rule, and arbitrary constants c1 and c2 selected to avoid instances when si,j(v,u) = 0. Pseudo-vector combines vectors related by a principal angle that falls within the A-E sub-states. Its projection onto the x- or z-axes will reverse sign in a y-axis rotation of the fiber through 180 degrees that exchanges the half-sarcomeres. Requiring the sign of si,j(v,u) to be conserved for every qualifying places in a unique half-sarcomere. We find that for is never zero and where we have dropped the first index since sj(v,u) specifies the half-sarcomere for for all . We also require active or relaxed isometric dipoles, or , participating in the A-E configurations transitioning to to have scalar si,j(a,u) or si,j(r,u) with sign equal to the sign of sj(v,u). This is the symmetry rule requiring active-rigor or relaxed-rigor transitioning between dipoles in the same half-sarcomere.
The “Φ-permutations” for active-rigor or relaxed-rigor transitions collect all instances of allowed transitions between dipoles. Active-rigor or relaxed-rigor allowed transitions are identified by ordered pairs or . They amplify common relationships between dipoles in the two physiological states. The “Φ-density” for active-rigor or relaxed-rigor transitions is a probability density. It sorts instances of allowed transition between dipoles. The allowed or dipoles falling in the active or relaxed A-state are removed from the active or relaxed list such that they participate just once. The remaining active or relaxed or dipoles falling in the B-state are identified next then also removed from the list. This process continues in sequence to the E-state.
Repetitive relationships in or will appear in either the Φ-permutation or Φ-density. The Φ-permutation has better statistics because the total number of instances is larger due to re-sampling since or ’s occasionally qualify as allowed with more than one . The Φ-density or dipoles occur just once in their lowest free-energy allowed instance. If or aligns by coincidence with two or more ’s it could be incorrectly assigned in the Φ-density. We cannot rule out this possibility without direct knowledge of the or correspondence by tracking a single molecule through each physiological state but this circumstance is impractical and undesirable for reasons already mentioned. However, incorrect assignment is unlikely given the high selectively of probe photoactivation, the selection rule as applied to , and the symmetry rule agreement between or and .
RESULTS
Fiber tension measurements
Isometric tension follows linearly light chain extraction and reconstitution of untagged and PAGFP tagged WT HCRLC in permeabilized porcine papillary muscle fibers (4, 11). We showed that peak tension declines with endogenous RLC removal and recovers with its replacement. We also showed that extracting conditions (with magnesium chelator CDTA) efficiently removes endogenous light chain while inducing a small fraction of irreversibly actin bound myosin (dead heads). We measured ~20% decline in peak isometric tension by treatment with extracting conditions minus CDTA when there is no endogenous RLC removal. Hence, peak isometric tension recovery of ≥ 80% indicates a full recovery of tension following light chain extraction and reconstitution. Table 1 summarizes isometric tension measurements for reconstituted fibers where 1-Tex/T0 measures the endogenous RLC extracted fraction for Tex the tension after extraction and T0 the tension before extraction. Endogenous RLC extraction efficiency averages to ~50%. The normalized reconstituted tension Trc/T0 represents the exchange efficiency provided the exchanged HCRLC supports full isometric tension. Mutants considered here fully recover tension when exchanged except for E134A and E134A PAGFP that lose ~10 and ~15% of peak tension.
Table 1.
Papillary muscle fiber isometric tension. 1
| HCRLC variant | Extracted fraction (1-Tex/T0) |
Tension recovery (Trc/T0) |
n |
|---|---|---|---|
| WT | 0.53±0.09 | 0.85±0.03 | 5,4 2 |
| M20L | 0.51±0.1 | 0.86±0.05 | 3 |
| E134A | 0.54±0.04 | 0.75±0.01 | 4 |
| G162R | 0.53±0.02 | 0.91±0.03 | 4 |
| WT-PAGFP | 0.48±0.04 | 0.84±0.03 | 10 |
| M20L-PAGFP | 0.50±0.04 | 0.88±0.03 | 6 |
| E134A-PAGFP | 0.52±0.04 | 0.71±0.03 | 8 |
| G162R-PAGFP | 0.48±0.02 | 0.84±0.02 | 11 |
T0 is initial tension, Tex tension after extraction, and Trc tension measured after extraction and reconstitution of the HCRLC variants. Average T0 is 155±20 kN/m2 for the fibers tested. Tension recovery ≥ 80% indicates full recovery (see RESULTS: Fiber tension measurements). Errors are standard error of the mean for n experiments.
One fiber broke during reconstitution.
Figure 4 shows the SDS-PAGE gel for native RLC extraction and reconstitution with (human MYL2 mutant) E134A-PAGFP where Lanes 1 and 2 are control and reconstituted fibers and Lane 3 contains reference purified proteins. Protein molar concentrations listed to the right are normalized relative to essential light chain (ELC). The ELC is not affected by the extraction/reconstitution treatment. Extraction removes 58% of the endogenous RLC and reconstitution is 100% with the tagged human mutant. The fiber tension data for E134A and E134A-PAGFP taken together with the SDS/PAGE data from extracted proteins indicated E134A inhibits peak isometric tension. The other mutants do not affect peak isometric tension. SDS-PAGE from WT and other mutants confirms exchange efficiencies of ~50% and our ability to fully replace extracted endogenous RLC with the recombinant HCRLC or HCRLC-PAGFP.
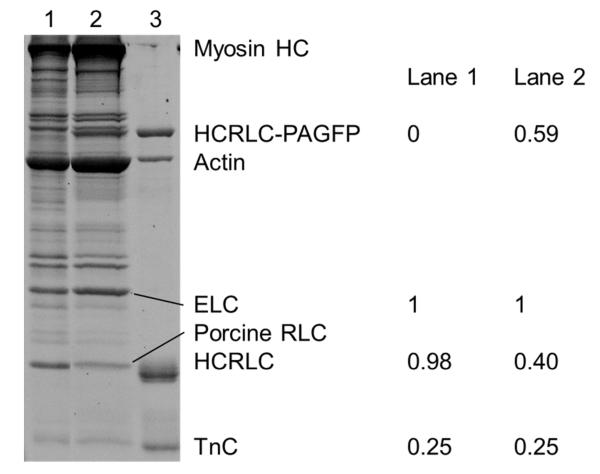
Figure 4.
SDS-PAGE of the fiber protein extract and purified proteins. Lane 1 is the untreated fiber control, lane 2 the E134A-PAGFP exchanged fiber, and lane 3 purified proteins identified on the right of the gel except for ELC and porcine RLC. Quantitative evaluation of the untreated and exchanged fibers was done with ImageJ and using the ELC content as standard. Error is ~10%. Stoichiometric replacement of the porcine RLC is indicated by the sum of Lane 2 bands at HCRLC-PAGFP and porcine RLC equaling 1 to within error. HC is heavy chain and TnC is troponin C.
Single molecule measurements
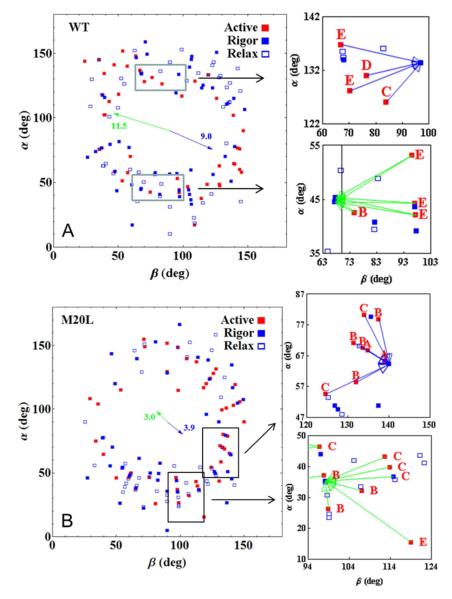
We measure single lever-arm orientation as described in METHODS. Figure 5 shows scatter plots for single dipole coordinates, (β,α), in WT and M20L species measured from fibers in rigor (solid blue square), isometric contraction (solid red square), and relaxation (open blue square). Scatter plots for all species (WT, M20L, E134A, and G162R) are compared in Supporting Information Figure S3. Extremes in the (β,α) domain, and in the central region where (β,α) ≈ (90,90), are unpopulated by cross-bridges in all states. The extremes are excluded by photoselection from p-polarization photoactivation light that favors <β> ≈ <α> ≈ 90 deg. The central depletion zone is from steric/molecular-crowding constraints that depopulate the central zone in favor of orientations that allow strong interaction with actin since favorable regions are roughly equivalent in rigor, active, and relaxation. The steric/molecular-crowding constraints originate from the thick filament (4). Average dipole orientation in each physiological state (active isometric, rigor, or relax) include contributions from both half-sarcomeres and does not vary for WT and mutant species beyond standard deviation. The standard deviations (σ) themselves are similar for all cases but differ with respect to the degree of freedom such that σβ ~ 36 deg and σα ~ 43 deg so that all the scatter plots are ellipsoidal with slightly longer dimension on the α-axis.
Figure 5.
Dipole moment spherical polar coordinates (β,α) scatter plot for fibers in rigor (blue solid square), isometric contraction (red solid square), and relaxation (open blue square) and for the WT and M20L species. Coordinates are defined relative to a lab frame z-axis parallel to the fiber symmetry axis, x-axis in the plane of the coverslip, and y-axis normal to the coverslip plane pointing into the aqueous medium. Right panels show enlargements of the rectangular regions in the scatter plots. They depict the arrow clusters associating various high free-energy active isometric coordinates in sub-states A-E (Figure 3) at the base of the arrow with their unique minimum free-energy rigor A-state at the pointy ends. Blue or green arrows designate a positive or negative projection scalar (eq. 3). Arrow clusters like those in the enlargements populate the entire scatter plot but were left out here for clarity. The full set of clustered arrows for WT and all mutant species is shown in Supporting Information (Figure S3). Blue and green arrows arranged like the hands of a clock are the average orientation of all the same colored clustered arrows (including those not shown here outside of the rectangules). They are referred to in the text as (±) and have amplitudes proportional to the average free-energy separating the isometric active cross-bridge from the minimum free-energy rigor A-state cross-bridge computed using eq. 2.
Selected portions of the scatter plots in Figure 5 are enlarged to show the principal rotation relationship between active and minimum free-energy rigor A-state coordinates identified as described in METHODS. Enlarged portions contain arrow clusters associating several high free-energy active isometric coordinates in sub-states A-E (Figure 3) with a unique minimum free-energy rigor A-state. Blue/green arrows designate the +/− symmetry (eq. 3) indicating the half-sarcomere dipole location. WT active-rigor transition clusters rotate from higher free-energy sub-states using mostly the β-degree of freedom while M20L active-rigor transition clusters rotate from lower free-energy sub-states using more the α-degree of freedom. This observation is quantified by the diverging blue and green arrows arranged like the hands of a clock in the middle of the scatter plots in Figure 5 and denoted here as (±). They are the average orientation of all the same colored clustered arrows (including those not shown here outside of the rectangles) and have amplitudes proportional to the average free-energy separating the isometric active coordinate from the minimum free-energy rigor A-state coordinate computed using eq. 2 in units of pN·nm and with standard error of the mean < 25%. Because Ψ ≥ Φ, eq. 2 estimates a lower bound on the free-energies, however, the relative amplitudes of (±)’s from WT and M20L species are estimated without ambiguity assuming the relationship between Ψ and Φ does not change among WT and mutant species (discussed below). The free-energy depicted by (±) amplitude for the mutant species shown in Figure 5 anticipates their altered relative stiffness (affecting k in eq. 2) as estimated below. (±) orientation is qualitative because of the outliers however the trend between WT and all mutants is consistent. All arrow clusters and (±)’s for the fiber species are shown in Supporting Material Figure S3.
The (±)’s are shorter and rotate clockwise in the mutant compared to WT species. (±) length change indicates the mutant isometric active cross-bridge occupies lower free-energy sub-states compared to WT. (±) rotation indicates the mutant isometric active cross-bridge undergoes a smaller polar but larger azimuthal angular change as active sub-states transition to the minimum free-energy rigor A-state configuration. Placing this in the context of a cross-bridge cycle, force without work production happens quickly as relaxed cross-bridges activate and stretch their elastic element against resisting force. Work production follows as resisting force yields allowing filament sliding and the completion of the cross-bridge cycle. Figures 5 and S3 characterize the work production phase. Mutants have diminished work production ((±) amplitude) accompanying the cross-bridge active-rigor transition. The reduced work production correlates with a reduced polar and increased azimuthal angular change ((±) orientation) suggesting the free-energy liberated by motor domain ATPase is partially dissipated in the mutant by unproductive or counter-productive azimuthal rotation.
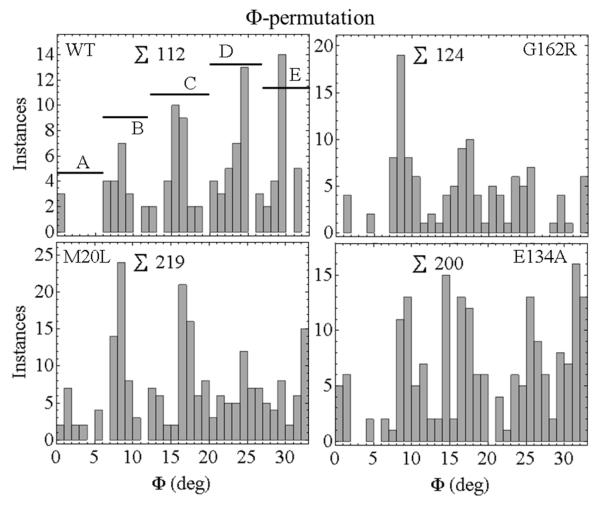
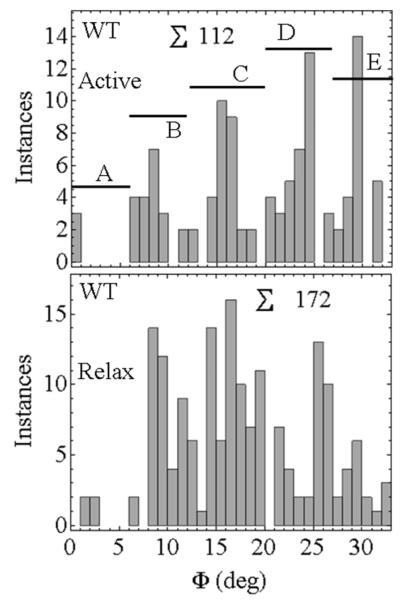
Figure 6 shows the Φ-permutation for active-rigor transitioning. We chose states A, B, C, D, and E corresponding to 0 ≤ Φ < 7, 7 ≤ Φ < 12, 12 ≤ Φ < 20, 20 ≤ Φ < 27, and 27 ≤ Φ < 33. These choices are somewhat arbitrary but they provide the framework for discussion. Discretized Φ-permutations are apparent in each species except E134A. The latter has a relatively featureless Φ-permutation while the other mutants lose their discreteness after the C-state. Figure 7 contrasts Φ-permutations for active rigor and relax-rigor transitions from the WT HCRLC-PAGFP. Φ-permutations are relatively featureless in relax-rigor compared to active-rigor transitioning implying angular discreteness is introduced by actin binding. The lack of discreteness in E134A active-rigor transitioning (Figure 6), its similarity to WT relax-rigor transitioning (Figure 7), and the unique loss of peak isometric tension in E134A (Table 1), together suggest E134A affects actin binding in contraction. Dipole orientation within the discretized packets of the Φ-permutations are constant among WT and mutant species (excluding E134A) implying the relationship between Φ and Ψ does not change among species.
Figure 6.
Φ-permutations for the active-rigor transitions in WT and mutant HCRLC-PAGFP exchanged permeabilized papillary muscle fibers. The histogram is discretized corresponding to A- to E-state intermediates. ∑ indicates total instances.
Figure 7.
Φ-permutations for the active-rigor (Active) and relax-rigor (Relax) transitions in WT HCRLC-PAGFP exchanged permeabilized papillary muscle fibers. ∑ indicates total instances.
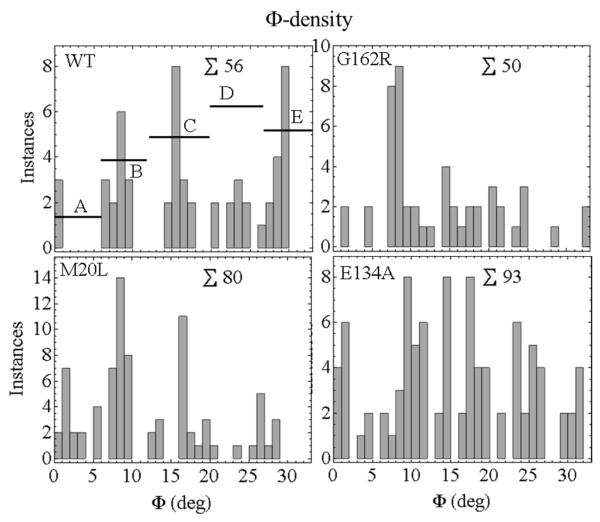
The Φ-density in an active isometric fiber is a subset of the Φ-permutation computed as described in METHODS. The Φ-densities for the WT, G162R, M20L, and E134A HCRLC-PAGFP exchanged fibers are indicated in Figure 8. The occupation probabilities for the A-E sub-states are derived from the Φ-densities by summing instances over the angular sub-state domains then dividing by the total instances to give probability. Occupation probabilities listed in Table 2 show the distinctiveness of the WT and mutant isoforms. They quantitate the trend evident in Figure 8 that Φ-density shifts towards the lower free-energy end of the power-stroke upon introduction of mutant HCRLC-PAGFP. Φ-density loss at the high free-energy end explains the concomitant loss of the discretized orientation feature at the high free-energy end for the mutants other than E134A seen in the Φ-permutations of Figure 6.
Figure 8.
Φ-densities for single myosins in an isometric active fiber. The WT and mutant species maintain different Φ-densities in isometric contraction. The altered intermediate densities reflect lowered lever-arm stiffness due to mutation in HCRLC. The E134A mutant also has lower isometric tension (Table 1). ∑ indicates total instances.
Table 2.
Sub-state occupation probability, maximum isometric tension, and cross-bridge stiffness for HCRLC-PAGFP exchanged permeabilized papillary muscle fibers. 1
| State | WT | M20L | E134A | G162R |
|---|---|---|---|---|
| A- | 0.054 ± 0.003 | 0.22 ± 0.04 | 0.128 ± 0.002 | 0.08 ± 0.03 |
| B- | 0.25 ± 0.07 | 0.35 ± 0.10 | 0.28 ± 0.01 | 0.44 ± 0.07 |
| C- | 0.26 ± 0.05 | 0.28 ± 0.02 | 0.31 ± 0.02 | 0.24 ± 0.02 |
| D- | 0.18 ± 0.02 | 0.11 ± 0.06 | 0.20 ± 0.04 | 0.18 ± 0.06 |
| E- | 0.25 ± 0.01 | 0.05 ± 0.02 | 0.08 ± 0.04 | 0.06 ± 0.01 |
| Fat | 1. | 1.05 ± 0.07 | 0.85± 0.07 | 1.00 ± 0.06 |
| Relative stiffness | 0.44 ± 0.02 | 0.32 ± 0.02 | 0.33 ± 0.01 | 0.35 ± 0.01 |
Errors represent standard error of the mean for n = 3-4. The full data set for single molecule dipole orientations was partitioned into n = 3 or 4 parts then occupation probabilities computed for each part. The n results were averaged and the standard error of the mean computed. Fat is peak isometric tension normalized to the WT fiber value. Relative stiffness is computed using eq. 4 and Fat.
Lever-arm displacement generating tension is measured at the distal end and is proportional to ΦE - Φi where ΦE is for the E-state configuration where the lever-arm exerts no force and subscript i is the ordinate in Figure 8. Isometric force F,
| (4) |
for k the lever-arm spring constant or stiffness already mentioned in eq. 2 and ρi the Φ-density from Figure 8. We normalize F/l to the WT level then use eq. 4 to estimate the relative stiffness k. Stiffness is significantly lower in the mutant isoforms compared to WT. Stiffness ordering is: WT > G162R > E134A ≈ M20L with G162R stiffness intermediate and unlike either WT or the lower stiffness mutants. E134A also has lowered isometric tension that affects stiffness separately from occupation probability.
SHG measurements
In a few cases, and following single molecule experiments using TIRF, the microfluidic chamber containing the fiber was immediately taken to the 2-photon upright scanning microscope for SHG measurements. Representative data is shown in Figure 2 and Supporting Information Figure S2. SHG images imply relaxed and active fibers have a similar average sarcomere spacing of 2.33 μm. Otherwise sarcomere spacing was estimated from TIRF images giving equivalent results. SHG imaging has deep penetration and long depth of focus in contrast to TIRF providing contrasting information from deep vs surface sarcomeres about fiber integrity following contraction/relaxation cycles.
DISCUSSION
Single myosin detection in the muscle fiber lattice involves its isolation in a condensed phase where each myosin occupies a volume <10 zeptoL (10−21 L). Sparse labeling techniques are undesirable in this application because how the probe affects fiber function, in this case isometric contraction, cannot be assessed when significantly less that 50% of the myosin is modified. The HCRLC-PAGFP permits the single molecule detection using sparse photoactivation of PAGFP while our exchange with the endogenous RLC is > 50% (Table 1). We developed the analytical tools to uniquely and precisely deduce PAGFP orientation by exploiting the full information content in the 3-spatial dimension fluorescence emission pattern (3). These tools provided the single dipole coordinates shown in Figures 5 and S3. We found previously that in this system molecular crowding and lattice constraints dominate the energy landscape limiting myosin rotational movement to a domain favoring actin interaction (4).
Disease implicated mutation suggest a genetic origin for illness although the pathway linking the diminished mutant protein function and the illness phenotype has proven to be neither simple nor direct. Nonetheless, a genetic origin to clinically diagnosed HCM appears to select the most severely ill indicating a practical reason to identify the protein function affected by the mutant (6). Furthermore, characterizing myosin functionality and its alteration by disease implicated mutation is the only rational means to elucidate the molecular basis for disease and identify the targets for smarter therapy. Heart disease is linked genetically to several cardiac sarcomere proteins including HCRLC. The HCRLC binds and stabilizes the heavy chain for folding and function (22). Disease-linked RLC mutations R58Q and N47K were shown to perturb strain sensitivity of ADP affinity (2). The A13T mutant triggered a hypertrophic remodeling of the heart in transgenic mice despite low incorporation into the cardiac tissue suggesting its effect is nonlinear (23).
We investigated three HCM linked mutants in HCRLC. Figure 1 shows their locations in the β-cardiac myosin homology modeled structure and Figure 3 outlines the role of HCRLC in contraction. In isometric active fibers, the mutants cause a cross-bridge population inversion to higher force generating sub-states (Figure 8 and Table 2) while macroscopic isometric force is unchanged (M20L and G162R) or diminished (E134A). It implies the lever-arms with bound mutant HCRLC strain more under shear force requiring a larger lever-arm rotation to achieve comparable isometric tension. We quantitated larger lever-arm shear strain as diminished lever-arm stiffness in Table 2. In free-energy terms, the power-stroke modification from WT caused by the mutants is indicated in Figures 5 and S3 with (±) change in direction and amplitude. This alteration has the mutant lever-arm dissipating the free-energy liberated by ATP hydrolysis in the motor domain with azimuthal rotation that does not produce work, at the expense of polar rotation, that does. How these modifications affect cardiac muscle performance is unknown but can be framed in two testable hypotheses. The mutant lever-arm preferentially populates lower free-energy sub-states in isometric contraction. Hypothesis 1 presumes these sub-states are stronger actin binders than the higher free-energy sub-states occupied by the WT lever-arm. The former create internal drag on filament sliding by prolonging actin attachment in a normally low duty ratio motor inhibiting sliding and decreasing myosin cycling rate. In the in vitro motility assay, higher duty ratio enhances myosin’s ability to move actin at low surface density and decreased myosin cycling rate lowers motility velocity. Alternatively, Hypothesis 2 presumes the mutant power-stroke cycle is nearly completed at peak force hence as the filament slides slightly under load, mutant lever-arm myosin rapidly detaches from actin. Limited actin attachment time lowers duty ratio and increases myosin cycling rate. In the in vitro motility assay, lower duty ratio lowers myosin’s ability to move actin at low surface density without altering motility velocity since strong binding is normally just 5% of the unloaded cycle time so lowering it more has little impact (24). In either hypothesis, mutant HCRLC alteration of myosin cycling, not related to the time myosin spends tightly actin bound, will affect both duty cycle and motility velocity in the in vitro motility assay. Faster cycling will increase duty cycle and motility velocity while slower cycling will lower duty ratio and motility velocity. Each case has a motility assay fingerprint.
The enlarged heart muscle HCM phenotype is consistent with the mutant HCRLC lever-arm modification that down-modulates energy conversion efficiency such that mutant myosin converts more ATP for the same work done compared to WT. In this case, heart muscle mass compensation would alleviate the immediate problem which appears to be the course taken in humans. We note also that the effect of myosin inefficiency on fiber contractility is difficult to predict hence two divergent hypotheses are feasible. Lowered energy conversion efficiency up- or down-modulates fiber contractility depending on ATPase and actin binding kinetics rationalizing why there is uncertainty surrounding how HCM mutants affect myosin function. If fiber contractility is the readout, then up- and down-modulation could be consistent with the same underlying malady of inefficient energy conversion. It is not surprising then that linking the diminished mutant protein function and the illness phenotype has proven to be difficult.
Animal (mouse) and computational modeling indicated a role for HCRLC phosphorylation in cardiac contraction regulation (25). The proposed mechanisms involve facilitating the actomyosin interaction by repositioning the myosin head in thick filaments (26) and the inhibition of myosin cycling kinetics due to increased lever-arm stiffness (27). The latter was associated with a larger duty ratio increasing the time myosin spends strongly actin bound thereby slowing the ATPase cycle in in vitro studies. Lower lever-arm stiffness in the M20L, E134A, and G162R mutants can then be reasonably associated with a more rapid myosin cycling kinetics consistent with results from in vitro motility of different HCM implicated mutants of HCRLC (28). This is tested for our mutants by the in vitro motility assay mentioned in Hypotheses 1 and 2. Fortunately, the lever-arm stiffness changes due to our mutations and phosphorylation (25) are reciprocal indicating a therapy for mitigating the effect of M20L, E134A, or G162R is HCRLC phosphorylation.
M20L and E134A contrast with G162R suggesting that mutation position in the HCRLC matters. In the latter, tension generating sub-state populations more modestly reconfigure with A state minimally and about equally populated in both WT and G162R. It appears that M20L or G162R myosin compensate for the effect of mutation and produce maximal isometric force with below normal lever-arm stiffness by radically or modestly reconfiguring sub-state populations. Radical reconfiguration in M20L reinforces the suggestion that light chain modification in the N-terminus near Ser15 affects function. E134A is not able to maintain normal isometric force or normal stiffness. Based on data reported here, actin binding in contraction is compromised by the E134A mutation.
Single molecule studies in tissues present unique technical challenges that we have addressed with innovative device design. Evanescent illumination using TIRF selectively excites chromophores within ~100 nm of the glass coverslip/aqueous interface enhancing signal-to-noise ratio by minimizing background light (29). However, coercing the dissected sample into the evanescent field is challenging because the dissected cardiac muscle fibers also contain significant connective tissue that frequently replaces muscle within the evanescent illumination. In addition, activated fibers often lift off the coverslip as they move slightly before reaching maximum tension. The microfluidic chamber provides an external and controllable constraint to confine part of the fiber to the illuminated volume, keep it immobilized during contraction, and the means to exchange bathing solution with pressurized flow through the chamber. Our device was designed to allow access by an upright microscope where we can observe SHG from the muscle thick filaments to assess sarcomere length and evaluate fiber integrity before and after the rigor/relaxation/contraction cycles in the single molecule experiments.
Single molecule studies from the intact contractile system offer the uniqueness of single molecule measurement in the apposite context of the intact muscle fiber. Proper context may be critical for deciphering an elusive molecular mechanism for HCM providing the best means available to identify the targets for a smarter therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Katalin Ajtai for engaging scientific discussion, and, Tom Meier and Steve Krage for exceptional assistance with obtaining the porcine cardiac tissue. TPB gratefully acknowledges inspiration from Tom Bator.
Funding: This work was supported by NIH grants R01AR049277 and R01HL095572 and by the Mayo Foundation
Footnotes
SUPPORTING INFORMATION Supporting information provides detailed descriptions for construction of the microfluidic device appearing in Figure 2 and for the Fourier analysis of SHG images to measure sarcomere length in fibers. Additional data in the form of dipole moment spherical polar coordinates for the mutant and WT species is also provided there. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Pant K, Watt J, Greenberg M, Jones M, Szczesna-Cordary D, Moore JR. Removal of the cardiac myosin regulatory light chain increases isometric force production. The FASEB Journal. 2009;23:3571–3580. doi: 10.1096/fj.08-126672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg MJ, Kazimierczak K, Szczesna-Cordary D, Moore JR. Cardiomyopathy-linked myosin regulatory light chain mutations disrupt myosin strain-dependent biochemistry. Proc. Nat. Acad. Sci. USA. 2010;107:17403–17408. doi: 10.1073/pnas.1009619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burghardt TP. Single Molecule Fluorescence Image Patterns Linked to Dipole Orientation and Axial Position: Application to Myosin Cross-Bridges in Muscle Fibers. Plos One. 2011;6:e16772. doi: 10.1371/journal.pone.0016772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burghardt TP, Josephson MP, Ajtai K. Single myosin cross-bridge orientation in cardiac papillary muscle detects lever-arm shear strain in transduction. Biochemistry. 2011;50:7809–7821. doi: 10.1021/bi2008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burghardt TP, Ajtai K. Single-molecule fluorescence characterization in native environment. Biophys.Rev. 2010;2:159–167. doi: 10.1007/s12551-010-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivotto I, Girolami F, Ackerman M, Nistri S, Bos JM, Zachara E, Ommen SR, Theis JL, Vaubel RA, Re F, Armentano C, Poggesi C, Torricelli F, Cecchi F. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin. Proc. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 7.Josephson MP, Sikkink LA, Penheiter AR, Burghardt TP, Ajtai K. Smooth muscle myosin light chain kinase efficiently phosphorylates serine 15 of cardiac myosin regulatory light chain. Biochem. Biophys. Res. Commun. 2011;416:367–371. doi: 10.1016/j.bbrc.2011.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni S, Hong F, Haldeman BD, Baker JE, Facemyer KC, Cremo CR. Modification of Interface between Regulatory and Essential Light Chains Hampers Phosphorylation-dependent Activation of Smooth Muscle Myosin. J. Biol. Chem. 2012;287:22068–22079. doi: 10.1074/jbc.M112.343491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burghardt TP, Ajtai K, Chan DK, Halstead MF, Li J, Zheng Y. GFP Tagged Regulatory Light Chain Monitors Single Myosin Lever-Arm Orientation in a Muscle Fiber. Biophys. J. 2007;93:2226–2239. doi: 10.1529/biophysj.107.107433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burghardt TP, Li J, Ajtai K. Single Myosin Lever-Arm Orientation in a Muscle Fiber Detected with Photoactivatable GFP. Biochemistry. 2009;48:754–765. doi: 10.1021/bi8017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczesna-Cordary D, Guzman G, Ng SS, Zhao J. Familial hypertrophic cardiomyopathy-linked alterations in Ca2+ binding of human cardiac myosin regulatory light chain affect cardiac muscle contraction. J. Biol. Chem. 2004;279:3535–3542. doi: 10.1074/jbc.M307092200. [DOI] [PubMed] [Google Scholar]
- 12.Dweck D, Reyes-Alfonso A, Potter JD. Expanding the range of free calcium regulation in biological solutions. Anal.Biochem. 2005;347:303–315. doi: 10.1016/j.ab.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Easley CJ, Benninger RK, Shaver JHHWS, Piston DW. Rapid and inexpensive fabrication of polymeric microfluidic devices via toner transfer masking. Lab on a Chip. 2009;9:1119–1127. doi: 10.1039/b816575k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locher MR, Razumova MV, Stelzer JE, Norman HS, Moss RL. Effects of low-level α-myosin heavy chain expression on contractile kinetics in porcine myocardium. Amer. J. Physiol. - Heart and Circulatory Physiology. 2011;300:H869–H878. doi: 10.1152/ajpheart.00452.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossmanith GH, Hoh JFY, Kwan LJ. Influence of V1 and V3 isomyosins on the mechanical behaviour of rat papillary muscle as studied by pseudo-random binary noise modulated length perturbations. J.Muscle Res.Cell Motil. 1986;7:307–319. doi: 10.1007/BF01753651. [DOI] [PubMed] [Google Scholar]
- 16.Muthu P, Mettikolla P, Calander N, Luchowski R, Gryczynski I, Gryczynski Z, Szczesna-Cordary D, Borejdo J. Single molecule kinetics in the familial hypertrophic cardiomyopathy D166V mutant mouse heart. J. Mol. Cell. Cardiol. 2010;48:989–998. doi: 10.1016/j.yjmcc.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plotnikov SV, Millard AC, Campagnola PJ, Mohler WA. Characterization of the Myosin-Based Source for Second-Harmonic Generation from Muscle Sarcomeres. Biophys. J. 2006;90:693–703. doi: 10.1529/biophysj.105.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burghardt TP, Thompson NL. Evanescent intensity of a focused gaussian light beam undergoing total internal reflection in a prism. Optical Engineering. 1984;23:62–67. [Google Scholar]
- 19.Uyeda TQP, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Nat. Acad. Sci. USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steffen W, Smith D, Simmons R, Sleep J. Mapping the actin filament with myosin. Proc. Nat. Acad. Sci. USA. 2001;98:14949–14954. doi: 10.1073/pnas.261560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayment I, Holden HM. Myosin subfragment-1: structure and function of a molecular motor. Curr. Opin. Struct. Biol. 1993;1993:3944–3952. [Google Scholar]
- 22.Lowey S, Waller GS, Trybus KM. Function of skeletal muscle myosin heavy and light chain isoforms by an in vitro motility assay. J. Biol. Chem. 1993;268:20414–20418. [PubMed] [Google Scholar]
- 23.Kazmierczak K, Muthu P, Huang W, Jones M, Wang Y, Szczesna-Cordary D. Myosin regulatory light chain mutation found in hypertrophic cardiomyopathy patients increases isometric force production in transgenic mice. Biochem.J. 2012;442:95–103. doi: 10.1042/BJ20111145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uyeda TQP, Kron SJ, Spudich JA. Myosin step size: Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 25.Sheikh F, Ouyang K, Campbell SG, Lyon RC, Chuang J, Fitzsimons D, Tangney J, Hidalgo CG, Chung CS, Cheng H, Dalton ND, Gu Y, Kasahara H, Ghassemian M, Omens JH, Peterson KL, Granzier HL, Moss RL, McCulloch AD, Chen J. Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease. J. Clin. Invest. 2012;122:1209–1221. doi: 10.1172/JCI61134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine RJ, Chantler PD, Kensler RW, Woodhead JL. Effects of phosphorylation by myosin light chain kinase on the structure of Limulus thick filaments. J. Cell Biol. 1991;113:563–572. doi: 10.1083/jcb.113.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg MJ, Mealy TR, Watt JD, Jones M, Szczesna-Cordary D, Moore JR. The molecular effects of skeletal muscle myosin regulatory light chain phosphorylation. American Journal of Physiology - Regulatory Integrative & Comparative Physiology. 2009;297:R265–R274. doi: 10.1152/ajpregu.00171.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg MJ, Watt JD, Jones M, Kazmierczak K, Szczesna-Cordary D, Moore JR. Regulatory light chain mutations associated with cardiomyopathy affect myosin mechanics and kinetics. J. Mol. Cell. Cardiol. 2009;46:108–115. doi: 10.1016/j.yjmcc.2008.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stout AL, Axelrod D. Evanescent field excitation of fluorescence by epiillumination microscopy. Applied Optics. 1989;28:5237–5242. doi: 10.1364/AO.28.005237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.