Summary
Cell growth is regulated by coordination of both extracellular nutrients and intracellular metabolite concentrations. AMP activated kinase and mammalian target of rapamycin complex 1 serve as key molecules that sense cellular energy and nutrients levels, respectively. In addition, the dioxygenase family, including prolylhydroxylase, lysine demethylase, and DNA demethylase, has emerged as possible sensors of intracellular metabolic status. The interplay among nutrients, metabolites, gene expression, and protein modification are involved in the coordination of cell growth with extracellular and intracellular conditions.
A fundamental issue in cell biology is how cells coordinate their growth with nutrient availability. Cells have developed exquisite mechanisms to sense nutrient status and adjust their behavior to maintain growth or cope with stress. Extensive studies have revealed that the AMP activated kinase (AMPK) acts as a master energy sensor to modulate cellular activities in response to energy stress, while the target of rapamycin (TOR) regulates cell growth by monitoring levels of amino acids and growth stimulating signals. The dioxygenase family, including prolylhydroxylase, lysine demethylase, and DNA demethylase, has emerged as possible sensors of metabolic status to regulate gene expression and cellular functions. Therefore, nutrients and metabolites actively participate in cellular regulation through a variety of mechanisms.
Mechanisms of nutrient sensing
Energy sensing
Living cells use ATP as the most important direct energy source. Hydrolysis of ATP to ADP and phosphate (or AMP and pyrophosphate) provides energy for most biological processes. The ratio of ATP to ADP and AMP is a barometer of cellular energy status, and is therefore tightly monitored by the cell. In eukaryotic cells, AMP-activated protein kinase (AMPK) serves as a key cellular energy sensor and a master regulator of metabolism to maintain energy homeostasis (Fig. 1) (Carling, 2004; Hardie, 2007). AMPK exists as heterotrimeric complexes consisting of a catalytic α subunit and two regulatory subunits, β and γ. AMPK senses energy levels by direct binding of AMP, ADP or ATP via the adenine nucleotide-binding sites of the γ subunit. Binding of AMP or ADP leads to conformational change of the enzyme and activates AMPK through several mechanisms, including allosteric activation, promoting the phosphorylation of the conserved threonine in the activation loop of AMPK by upstream kinases while at the same time preventing its dephosphorylation of the activation loop (Hardie, 2011). Among the three mechanisms, inhibiting dephosphorylation of the activation loop is most crucial for AMPK activation by AMP or ADP in vivo. AMP-promoted activation was considered for a long time as the key mechanism of AMPK regulation. However, recent studies demonstrated that ADP, with a similar affinity as AMP in binding the γ subunit, is also able to activate AMPK (Oakhill et al., 2011; Xiao et al., 2011). Because intracellular concentrations of ADP are usually much higher than AMP, ADP has emerged as a more relevant physiological activator of AMPK under most stress. However, the AMP-specific ability of AMPK allosteric activation, which can increase AMPK activity approximately 10 fold, makes AMP as an essential regulator under severe energy stress. Thus, AMPK activity is capable of responding to the different levels of energy demands responding appropriately.
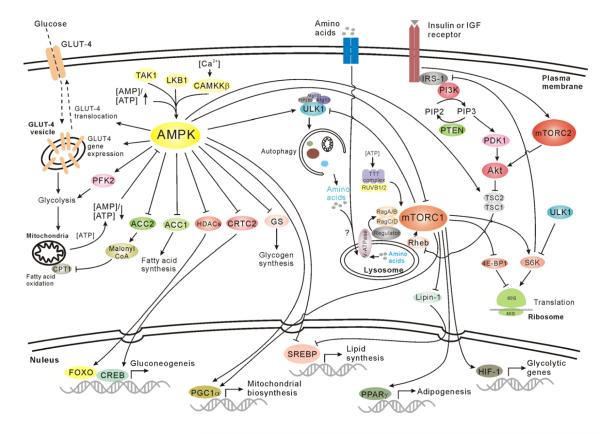
Figure 1.
Nutrient sensing and metabolism pathways. AMP-activated kinase (AMPK) is activated by energy stress and promotes catabolic pathways to produce ATP while switching off ATP-consuming anabolic pathways. In contrast, mammalian target of rapamycin complex 1 (mTORC1) activation under nutrient-sufficiency leads to significant elevation of anabolic processes, such as protein and lipid synthesis. Abbreviations are: GLU4, glucose transporter type 4; CPT1, Carnitine Palmitoyltransferase-1; TAK1, TGFβ-activated kinase 1; LKB1, liver kinase B1 (a key AMPK activator tumor suppressor); CAMKKβ, Calmodulin-dependent protein kinase kinase β; ACC, acetyl CoA carboxylase; PFK2, Phosphofructokinase 2; GS, glycogen synthase; HDACs, histone deacetylases; CRTC2, CREB-regulated transcription co-activator 2; FOXO, forkhead box protein O; CREB, cAMP response element-binding protein; ULK, UNC-51-like kinase; FIP200, 200 kDa FAK family kinase-interacting protein; ATG, autophagy-related; PI3K, phosphoinositide 3-kinase; IRS1, insulin receptor substrate 1; PTEN, phosphatase and tensin homologue; PDK1, 3-phosphoinositide-dependent protein kinase 1; TSC1/2, tuberous sclerosis 1/2; Rheb, Ras homologue enriched in brain; v-ATPase, vacuolar H+-adenosine triphosphatase; 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; S6K, ribosomal S6 kinase; Ragulator, a protein complex responsible for lysosomal recruitment and activation of Rag GTPases; PGC1α, peroxisome proliferator-activated receptor-γ coactivator 1α; PPARγ, peroxisome proliferator-activated receptor-γ; SREBP, sterol regulatory element-binding protein; HIF, Hypoxia-inducible factors. Stimulatory interactions are indicated with ↓ and inhibitory interactions are indicated with ⊥.
Amino acid sensing
Amino acids, the building blocks of proteins, are also essential nutrients for cell growth. Amino acids can also be used for the synthesis of nucleic acid, glucose, and ATP. In all eukaryotes, the target of rapamycin (TOR), a conserved central cell growth modulator and serine/threonine kinase, is tightly regulated by amino acid availability. Mammalian TOR (mTOR) exists in two different complexes, mTORC1 and mTORC2, which are distinguished by unique accessory proteins: Raptor and Rictor, respectively. Furthermore, the two mTOR complexes are differentially regulated and only mTORC1 is sensitive to amino acids (Fig. 1).
Extensive efforts have been taken to elucidate the mechanisms through which mTORC1 senses intracellular amino acids. The Rag GTPases have been shown to be key transducers between amino acids and mTORC1 activation (Fig. 1) (Kim et al., 2008; Sancak et al., 2008). The Rag proteins form heterodimers consisting of RagA or RagB bound to RagC or RagD and are localized on lysosomes. Amino acids activate the heterodimers by promoting GTP loading of RagA/B and GDP loading of RagC/D. The active Rag heterodimers physically interact with mTORC1 through binding to Raptor, thereby recruiting mTORC1 to the surface of lysosomes where it may interact with and be activated by the small GTPase Rheb (Inoki et al., 2003a; Saucedo et al., 2003; Stocker et al., 2003). Rheb is regulated by growth factor signals via the action of the TSC1/TSC2 tumor suppressor proteins. This model explains why mTORC1 activation requires both growth factors and amino acids. Recent studies have demonstrated that the Ragulator complex, which is associated with the lysosome membrane, promotes nucleotide exchange and activation of Rag GTPases (Sancak et al., 2010). The Ragulator complex functions as a guanine nucleotide exchange factor (GEF) for RagA/B (Bar-Peled et al., 2012), although the precise mechanism remains to be uncovered. Interestingly, VAM6 has been reported to function as a GEF for the yeast Rag homologs (Binda et al., 2009); however, the question has been raised as to whether mammalian VAM6 homolog has GEF activity towards Rag GTPases. Recent work further demonstrated that the Ragulator complex is regulated by the lysosomal vacuolar H+-adenosine triphosphatase (v-ATPase), which senses intra-lysosomal amino acid levels by an unknown mechanism (Zoncu et al., 2011). These studies argue that amino acid signaling to mTORC1 begins within the cell instead of at the plasma membrane, and the lysosome appears to be a key player (Fig. 1). Besides the Ragulator complex, other proteins have been implicated in the regulation of Rag GTPases, including leucyl-tRNA synthetase in both yeast and mammals that may serve as intracellular leucine sensor (Bonfils et al., 2012; Han et al., 2012), signaling adaptor protein p62 that promotes the formation of the active Rag heterodimers (Duran et al., 2011) and SH3 domain-binding protein 4 (SH3BP4) that binds to the inactive Rag GTPase complex under nutrient stress and inhibits the formation of active Rag complex (Kim et al., 2012).
GCN2 (general control non-derepressible-2) kinase functions as an amino acid sensor in eukaryotic cells. Under amino acid deficiency, uncharged aminoacyl-tRNA accumulates and binds to the aminoacyl-tRNA synthetase-like domain in GCN2. This leads to the activation of GCN2 kinase (Hinnebusch, 2005). Activated GCN2 is able to phosphorylate eukaryotic translation initiation factor 2-alpha (eIF2α) and reduces general translational initiation. Additionally, activated GCN2 induces the translation of specific mRNAs, such as the transcription factor ATF4, which promotes expression of stress responsive genes (Wek et al., 2006). Therefore, activation of GCN2 and inhibition of mTORC1 similarly act to suppress global protein synthesis in response to amino acid deficiency. GCN2 may also be involved in the regulation of mTOR signaling; although, the mechanisms involved in this is currently not clear (Novoa et al., 2001; Watanabe et al., 2007).
Oxygen sensing
Besides the conventional nutrients, such as amino acids and glucose, oxygen is also critical for cell growth and cellular activity and can be considered as a nutrient. Prolyl hydroxylase domain protein (PHD) carries out one of the best-characterized oxygen sensing mechanisms in cellular regulation (Reviewed by Kaelin and Ratcliffe, 2008). PHD is a member of the dioxygenase family that uses molecular oxygen and α-ketoglutarate (α-KG) to hydroxylate conserved prolyl residues in hypoxia inducible factor α (HIFα), which is the activating subunit of the HIF heterodimeric transcription factor (Fig. 2B). Hydroxylated HIFα is recognized by the von Hippel-Lindau tumor suppressor protein pVHL, which is a substrate recognition subunit of an E3 ubiquitin ligase. Recruitment of hydroxylated HIFα to the pVHL-E3 ubiquitin ligase leads to its ubiquitination and a rapid clearance by the proteasome system. During HIFα hydroxylation, molecular oxygen level is a key determinant in controlling PHD activity (Nakayama et al., 2004). When oxygen levels are high, such as during normoxic conditions, HIFα is rapidly hydroxylated and degraded resulting in low HIFα protein levels under oxygen sufficient conditions. However, when oxygen levels are low, referred to as hypoxia, the activity of PHD is inhibited due to the lack of this essential co-factor in the hydroxylation reaction.
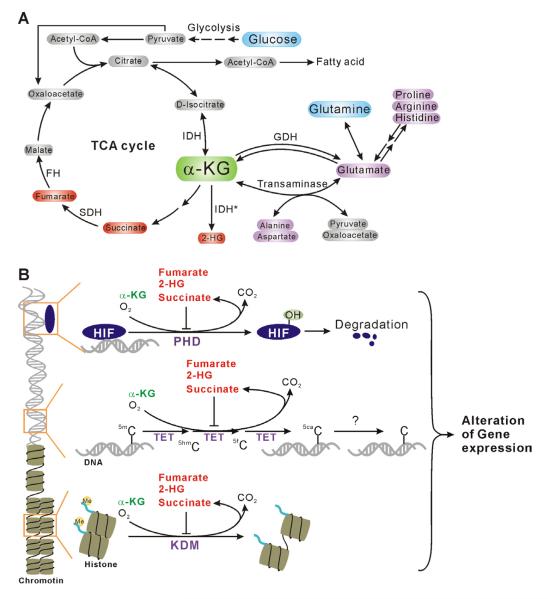
Figure 2.
Dioxygenases in sensing metabolic intermediates and epigenetic regulation. (A) Schematics of α-KG metabolism and TCA cycle. The mutant isocitrate dehydrogenase (IDH) (indicated with a star) may decrease α-KG and generate a new oncometabolite 2-hydroxylglutarate (2-HG). (B) A proposed role of dioxygenases in metabolite sensing and epigenetic modifications. Using α-KG as a key substrate, dioxygenases is involved in HIFα hydroxylation, DNA demethylation, and histone demethylation. All processes are inhibited by normal metabolites, such as succinate and fumarate, as well as oncometabolite 2-HG. α-KG, α-ketoglutarate; GDH, glutamate dehydrogenase; SDH, succinate dehydrogenase; FH, fumarase; TET, ten-eleven translocation; KDM, lysine demethylase; PHD, prolyl hydroxylase domain protein.
Under hypoxic conditions, HIFα stabilization and accumulation leads to the activation of a transcriptional program preparing cells to adapt to the conditions. For example, HIFα induces expression of glycolytic genes, allowing cells to use oxygen-independent glycolysis instead of oxygen-dependent oxidative phosphorylation to produce energy to maintain essential cellular activity (Gordan et al., 2007). In addition, HIFα activation also stimulates expression of genes to promote blood vessel growth, thus increasing oxygen supply to tissues experiencing hypoxia (Majmundar et al., 2010). Notably, HIF1α also inhibits mTORC1 activity during hypoxia by inducing the expression of REDD1 (also known as RTP801 or DDIT4) (Brugarolas et al., 2004; Corradetti et al., 2005; Sofer et al., 2005), which represses mTORC1 by promoting the release of sequestered TSC2 from 14-3-3 proteins (DeYoung et al., 2008). Inhibition of mTORC1 by HIF1α may protect cells from hypoxic stress by reducing ATP-consuming protein synthesis while increasing autophagy (discussed in following paragraphs). Finally, the interplay between HIF1α and MYC, a key transcriptional factor in cell growth, plays important roles in the regulation of cell growth and metabolism during hypoxia (Reviewed by Dang et al., 2008; Gordan et al., 2007).
FIH (factor inhibiting HIF1) is a PHD domain containing dioxygenase and also participates in oxygen sensing through regulation of the HIF transcription factor. FIH hydroxylates HIFα on asparagine residues in an oxygen dependent manner (Hewitson et al., 2002; Lando et al., 2002a). The asparagine hydroxylation does not promote HIFα degradation but rather inhibits the transcriptional activation activity of HIFα, possibly by interfering with co-activators interactions, such as the histone acetyltransferase p300 (Lando et al., 2002b; Mahon et al., 2001). Therefore, the adaptive transcriptional program activated by lack of cellular oxygen is under multifaceted control by dioxygenases, directly linking oxygen availability to HIF1α stability.
Regulation of metabolism by nutrient signaling
Nutrient sensors play an essential role in maintaining cellular homeostasis by regulating downstream metabolic processes. For example, activation of AMPK strongly promotes catabolic pathways to produce ATP while switching off ATP-consuming anabolic pathways. In contrast, mTORC1 activation under nutrient-sufficient conditions leads to significant elevation of anabolic processes, such as protein synthesis, to generate building materials required for cell growth.
Glucose homeostasis
Activation of AMPK by increased energy demands promotes the utilization of glucose to generate ATP through multiple modes of action (Fig. 1). First, AMPK up-regulates glucose uptake by promoting the expression and function of glucose transporters (Hardie, 2011). Second, AMPK promotes glycolysis under anaerobic conditions by phosphorylating and activating 6-phosphofructo-2-kinase (PFK-2), the enzyme responsible for the synthesis of glycolytic activator fructose 2,6-bisphosphate (Marsin et al., 2000). Third, AMPK promotes mitochondrial biogenesis. Fourth, AMPK inhibits gluconeogenesis, an energy consuming glucose synthetic pathway mainly found in the liver (Hardie, 2011). Finally, AMPK is able to phosphorylate and inhibit glycogen synthase (GS) and thus reduce glycogen synthesis (Carling and Hardie, 1989; Jorgensen et al., 2004). Therefore, AMPK modulate glucose and glycogen metabolism through multiple mechanisms.
AMPK can also influence glucose homeostasis by inhibiting mTORC1. It has been well established that activation of mTORC1 promotes glucose uptake and glycolytic flux by increasing both transcription and translation of HIF1α (Duvel et al., 2010; Wouters and Koritzinsky, 2008). However, there seems to be a dilemma that mTORC1 positively regulates HIF1α while itself is inhibited by hypoxia. This may serve as a feedback mechanism. Further studies are needed to elucidate the mechanisms and functional implication of mTOR-dependent regulation of HIF1α. Interestingly, hyperactivation of mTORC1 under nutrient-excess conditions has a negative feedback effect on glucose catabolism. Prolonged activation of S6 kinase 1 (S6K1, a major downstream substrate of mTORC1) caused by mTOR hyperactivation phosphorylates and dampens the insulin receptor substrate (IRS1) (Harrington et al., 2004; Shah and Hunter, 2006; Tremblay et al., 2007), leading to insulin desensitization and inhibition of Akt activity (Fig. 1). Consistently, mice deficient for S6K1 showed elevated insulin sensitivity when fed on high fat diet (Um et al., 2004). In addition, phosphorylation of Grb10 by mTORC1 may also suppress signaling of insulin and other receptor tyrosine kinases (Hsu et al., 2011; Yu et al., 2011). This feedback loop reduces glucose uptake and glycogen synthesis, and increases gluconeogenesis as results of mTORC1 hyperactivation.
Recent studies have revealed new roles of mTORC1 in intracellular energy control. Inhibition of the hyperactivated mTORC1 in TSC1/2 mutant cells is sufficient to protect cell death from glucose deprivation (Choo et al., 2010). Notably, deprivation of both glucose and glutamine, two major carbon sources feeding mitochondrial (tricarboxylic acid) TCA cycle, inhibits mTORC1 activity in a manner independent of AMPK, TSC1/2 or Rag GTPases (Kim et al., 2013). Instead, energetic stress induced by glucose and glutamine starvation inhibits mTORC1 signaling through repressing Tel2-Tti1-Tti2-RUVBL1/2 complex, an AAA+ ATPase-containing complex. Consistently, ATP depletion by mitochondrial inhibition significantly blocks mTORC1 activation and lysosomal localization, which is also independent of AMPK or Rag GTPases (Kim et al., 2013). These studies suggest another potential mechanism of mTORC1 regulation by cellular energy status.
Protein synthesis and autophagy
Protein synthesis is the best-characterized process regulated by mTORC1. mTORC1 plays a key role in translational control by phosphorylating a myriad of translation regulators, including eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) and S6K1 (Ma and Blenis, 2009). Inhibition of the 4E-BP family of translation repressor plays a major role in the translation activation by mTOR (Fig. 1) (Thoreen et al., 2012). The role of mTOR in translational regulation has been extensively reviewed (Ma and Blenis, 2009) and will not be discussed in detail here. Notably, since protein synthesis consumes a large portion of cellular energy, AMPK activation induced by energy stress significantly inhibits protein synthesis, resulting in the AMPK-mTORC1 crosstalk (Fig. 1). AMPK attenuates mTORC1 signaling through phosphorylation and activation of tuberous sclerosis 2 (TSC2) (Inoki et al., 2003b), a negative regulator of mTORC1. AMPK also directly phosphorylates Raptor, which induces 14-3-3 binding to raptor and repression of mTORC1 activity (Gwinn et al., 2008). Besides the indirect effect through mTORC1, AMPK inhibits translation by inhibiting the eukaryotic elongation factor-2 (eEF2) kinase, which phosphorylates and inhibits eEF2 (Browne et al., 2004; Horman et al., 2002).
Under severe energy or nutrient stress conditions, a mere inhibition of biosynthesis is not sufficient and cells have to find strategies to cope with resource shortage for survival. Autophagy is a critical process for cell survival under stress conditions. It enables the recycling of non-essential macromolecules and organelles to provide nutrients and energy to maintain essential activity and biosynthesis of pivotal components (He and Klionsky, 2009; Mizushima and Komatsu, 2011). Autophagy is strongly induced during nutrient deprivation. Based on its central role in nutrient metabolism, TOR functions as a primordial negative regulator of autophagy in organisms from yeast to mammals (Neufeld, 2010). The best-characterized mechanism is through inhibition of the autophagy-initiating Atg1 (homologous to ULK1 in mammals) kinase complex (Fig. 1) (Reviewed by Mizushima et al., 2011), although TORC1 may phosphorylate other components to suppress autophagy. In yeast, active TORC1 disrupts the Atg1 complex by phosphorylating Atg13 whereas, in mammalian cells, mTORC1 phosphorylates ULK1 and blocks its activation under nutrient sufficiency conditions (Kamada et al., 2000; Kim et al., 2011). Recently, mTOR was found to play a positive role in the termination of autophagy upon prolonged starvation and to facilitate the restoration of functional lysosomes in multiple animal species (Yu et al., 2010). Interestingly, autophagy, which is inhibited by mTORC1, may also negatively influence cell growth signaling. Overexpression of the autophagy initiating kinase Atg1/ULK1 represses S6K activity in both Drosophila and mammalian cells (Lee et al., 2007; Scott et al., 2007). These findings suggest an intricate interplay between autophagy and mTORC1.
As expected, AMPK plays a positive role in autophagy induction in response to glucose starvation (Liang et al., 2007; Meley et al., 2006). AMPK may indirectly induce autophagy by inhibiting mTORC1. Indeed, two recent studies uncovered a direct mechanism by AMPK to promote autophagy (Egan et al., 2011; Kim et al., 2011). AMPK phosphorylates and activates the autophagy essential kinase ULK1. As the major energy sensor, AMPK may induce autophagy by regulating additional autophagic machinery downstream of the ULK1 complex.
Mitochondrial biogenesis
As the “energy factory” for the cell, mitochondrial biogenesis in the long term increase the energy generation through oxidative catabolism. Chronic activation of AMPK by treating rodents with either AMPK activator (Narkar et al., 2008; Winder et al., 2000) or drug inducing energy stress (Zong et al., 2002) caused a significant increase in the expression of mitochondrial genes and mitochondrial biogenesis in muscle. Interestingly, although mTOR functions in anabolic pathways that are generally antagonized by AMPK, these two kinases seem to display consistent regulation on mitochondrial biogenesis (Fig. 1), possibly due to similar demand for energy. For example, hyperactivation of mTORC1 increases mitochondrial DNA content and the expression of many oxidative-related genes (Cunningham et al., 2007). In agree with this, Raptor deficiency in skeletal muscle results in a defect in mitochondrial biogenesis and oxidative capacity (Bentzinger et al., 2008). The regulation of mitochondrial biogenesis by AMPK and mTOR seems to converge on the same protein, peroxisome proliferator-activated receptor-γ coactivator 1α (PGC1α), a key nuclear cofactor for mitochondrial biogenesis and oxidative metabolism. AMPK directly phosphorylates PGC1α and promotes the activation of its own transcription (Jager et al., 2007), while mTOR promotes the transcriptional activity of PGC1α by enhancing its interaction with the transcription factor yin-yang 1 (YY1) (Cunningham et al., 2007).
Lipid and nucleic acid metabolism
Another important intracellular energy source, lipid metabolism, is also tightly regulated by AMPK. In fact, one of the best-characterized downstream targets of AMPK is the fatty acid metabolism pathway. AMPK decreases fatty acid synthesis by phosphorylating and inhibiting acetyl CoA carboxylase 1 (ACC1), the key regulatory enzyme in fatty acid synthesis (Fig. 1) (Davies et al., 1992; Munday et al., 1988). In addition, AMPK also down-regulates the expression of enzymes involved in fatty acid synthesis at the transcriptional level, possibly through phosphorylation and inhibition of the lipogenic transcription factor sterol regulatory element-binding protein 1C (SREBP-1C) (Li et al., 2011). Furthermore, AMPK promotes uptake and β-oxidation of fatty acids in mitochondria. This is achieved by AMPK-dependent phosphorylation and inhibition of ACC2, an ACC isoform responsible for synthesis of malonyl CoA, which decreases fatty acid entry into mitochondria by inhibiting the carnitine O-palmitoyltransferase 1 (CPT1) system (Merrill et al., 1997). Consequently, AMPK activation results in inhibition of lipogenesis and activation of fatty acid oxidation.
Lipids are also used as building material for membranes and are in high demand for growing cells. Lipid biosynthesis is coordinated with cell growth and is controlled by mTOR. Inhibition of mTORC1 reduces the expression of SREBP1/2 and prevents SREBP1/2 activation by proteolytic processing, resulting in a significant decrease of lipogenic gene expression (Fig. 1) (Duvel et al., 2010; Porstmann et al., 2008; Wang et al., 2011). A recent study further demonstrated that mTORC1 promotes the function of SREBP through phosphorylating Lipin-1, a phosphatidic acid phosphatase, and preventing its nuclear entry and suppression on SREBP1/2 (Peterson et al., 2011). In addition, mTORC1 may also regulate lipid synthesis by upregulating peroxisome proliferator-activated receptor γ (PPARγ), a key regulator of lipid uptake and adipogenesis (Kim and Chen, 2004; Le Bacquer et al., 2007; Zhang et al., 2009). The ability of mTORC1 to increase lipid synthesis likely contributes to cell growth promotion.
Nucleic acids are another group of biomolecules essential for cell growth. Biosynthesis of nucleotides shares several important characteristics with lipid biosynthesis. For example, both use glucose as a carbon source and consume TCA cycle intermediates. In addition, they also require reductive power in the form of NADPH. Carbon diverted from the glycolytic flux to the pentose phosphate pathway (PPP) is used to generate ribose-5-phosphate (R5P) for de novo nucleotide biosynthesis (Deberardinis et al., 2008). The regulation of nucleotide biosynthesis by metabolic status has emerged as a topic of interest, although the mechanism is still poorly understood. A recent study demonstrated a link between mTOR signaling and nucleotide biosynthesis, in which mTORC1 activation leads to induction of genes encoding the enzymes of the oxidative PPP, possibly mediated by SREBP1 (Duvel et al., 2010). HIF1α is found as another potentially important regulator of nucleotide biosynthesis. Nonhypoxic induction of HIF1α in leukemia cells promotes the flux of glucose carbon through PPP and is critical for leukemia cell growth and survival (Zhao et al., 2010). Together with its function in inducing glycolytic flux, HIF1α seems to play an essential role in directing glucose metabolism for both energy source and building blocks of nucleic acids. It is of note that HIF1α induces nucleotide biosynthesis by promoting the flux of glucose carbon through a nonoxidative arm of PPP (Zhao et al., 2010). In contrast to its role in stimulating glycolytic flux, HIF1α is not the downstream mediator of mTORC1 in promoting nucleotide biosynthesis.
Sensing metabolic intermediates and epigenetic regulation
In addition to extracellular nutrients, the cell has developed intricate mechanisms to sense cellular metabolite concentrations in order to regulate metabolism and cellular functions accordingly. For example, the AMP activated protein kinase (AMPK), as discussed above, is a master cellular energy sensor that regulates a wide range of cellular activities. Moreover, many metabolic enzymes are regulated by allosteric activation or inhibition. Most of the allosteric regulators are often key metabolic intermediates, such as ATP, ADP, NAD, and NADH, to provide feedback inhibition or feed forward activation. For example, AMP activates the glycolytic enzyme phosphofructose kinase (Wegener and Krause, 2002). Conversely, ADP inhibits the activity of PEPCK, a key regulatory enzyme in gluconeogenesis. Therefore, glycolysis and gluconeogenesis are activated and inhibited, respectively, when cellular AMP and ADP levels are elevated under energy stress. This type of regulation usually affects one specific metabolic enzyme at a time and will not be discussed here as a mechanism of global nutrient sensing. In addition, metabolic intermediates can have a global effect by influencing processes involved in broad cellular regulation, particularly through gene expression as discussed below.
Recent studies revealed that the Fe(II)- and α-KG-dependent dioxygenase family may play a broad role in sensing metabolic intermediates to regulate cell growth, or even promote tumorigenesis by altering gene expression., The PHD involved in HIFα hydroxylation is one such example as discussed above (Fig. 2). There is convincing evidence suggesting that PHD may be inhibited by metabolites, such as fumarate and succinate (Koivunen et al., 2007; Selak et al., 2005), which are intermediates of the citric acid cycle. Fumarate and succinate are structurally similar to α-KG and can function as competitive inhibitors to suppress PHD activity when these metabolites are accumulated at high levels, and succinate itself is a byproduct of the hydroxylation reaction (Fig. 2B). This situation is certainly applicable to some type of cancers, which contain mutations in fumarase (FH) or succinate dehydrogenase (SDH) (Koivunen et al., 2007; Selak et al., 2005). Cancer associated mutations (loss of function) of FH or SDH result in abnormal accumulation of succinate or fumarate, respectively, leading to PHD inhibition and HIFα accumulation. Tumors harboring these mutations often have a hypoxic transcription profile. This may provide one mechanism how mutation of these metabolic enzymes influence gene expression and promote tumor growth.
Dioxygenase is a large family of enzymes. There are estimated to be more than 60 α-KG-dependent dioxygenases in humans based on sequence homology of active sites (Rose et al., 2011). The Jumonji C (JmjC) domain histone demethylases, also known as lysine demethylase (KDM), are members of the dixoygenases and their activity depends on molecular oxygen and α-KG (Fig. 2B) (Mosammaparast and Shi, 2010; Tsukada et al., 2006). Moreover, the ten-eleven translocation (TET) tumor suppressor protein, capable of catalyzing the hydroxylation of the 5-methylcytosine leading to demethylation of DNA, is a member of the dioxygenase family (Fig. 2B) (Tahiliani et al., 2009). It has been suggested that activity of both the histone and DNA demethylase may be regulated by metabolic intermediates. For example, high concentration of succinate and fumarate can inhibit the activity of TET2 and KDM4A, a JmjC family histone demethylase (Xiao et al., 2012). These epigenetic regulations will lead to a global alteration of gene expression and eventually influence cellular activities, including metabolism and cell growth. Therefore, the dioxygenase family appears to play a key role in sensing metabolic intermediates to influence epigenetic modifications and gene expression.
Because α-KG is a common metabolic intermediate involved in multiple cellular reactions (Fig. 2A), the concentration of α-KG may reflect cellular metabolic status. The levels of α-KG should be determined by the rates of its production and utilization in the cell. A major source of α-KG is from glutamine, which is first converted to glutamate and then to α-KG by glutamate dehydrogenase (Fig. 2A). Amino acid catabolism of arginine, histidine, and proline produces glutamate that can serve as a source for α-KG. α-KG can also be produced by oxidation of isocitrate in the TCA cycle as well as by transamination of glutamate with other α-ketoacid, such as pyruvate or oxaloacetate. On the other hand, α-KG can be utilized by multiple reactions, including those involved in amino acid biosynthesis, the TCA cycle, and production of isocitrate. The isocitrate produced serves as an important intermediate for acetyl-CoA production and fatty acid synthesis. Furthermore, α-KG is an essential substrate for a large number of dioxygenases as discussed above. The role of α-KG in supporting dioxygenases may serve as a metabolic barometer to globally influence cellular physiology and cell growth.
Altered metabolism is a hallmark of cancer (Vander Heiden et al., 2009). The metabolic changes in cancers may be sensed by the dioxygenases that detect the level of α-KG and its competitive inhibitory metabolites to alter histone and DNA methylation, thus affecting gene expression. Mutations in genes encoding for several metabolic enzymes have been found in human cancer. Eight genes, FH, SDHA, SDHB, SDHC, SDHD, SDHAF2, IDH1 and IDH2, encoding four different metabolic enzymes—FH, SDH, and isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) are mutated, both inherently and somatically, in human cancers (Reviewed by Oermann et al., 2012). A common feature associated with these mutations is the accumulation of metabolites. Fumarate and succinate are elevated in tumors with mutation of FH or SDH. The IDH mutation is particularly notable, for it decreases α-KG and more importantly generates a new oncometabolite, 2-hydroxylglutarate (2-HG) (Fig. 2A) (Dang et al., 2009; Zhao et al., 2009). Interestingly, 2-HG has been shown to inhibit the activity of TET and KDM, suggesting a possible mechanism of 2-HG, hence IDH mutation, in tumorigenesis (Xu et al., 2011). Consistent with TET as a direct target of the oncometabolite 2-HG, mutation of the tumor suppressor TET2, is mutually exclusive with mutation of IDH in leukemia (Figueroa et al., 2010). Moreover, the IDH mutant tumors display a gene expression profile similar to tumors containing the TET2 mutation, supporting both IDH and TET function in the same pathway. Furthermore, JmjC family member KDM has been implicated in human cancer (Rotili and Mai, 2011). Together these observations suggest a critical role for dioxygenase in sensing metabolites, hence the metabolic status, to regulate cell growth.
It is worth noting that fast proliferating embryonic cells display metabolism properties similar to cancer cells, such as elevated glycolysis. Epigenetic modifications and transcription regulation are critically important in cell differentiation and reprogramming. One may speculate that the altered metabolic status may contribute to the maintenance and differentiation of stem cells and progenitor cells because the effect of metabolites on the dioxygenase-dependent epigenetic modification and altered gene expression.
Conclusion
Cell growth has to be coordinated with extracellular nutrients and intracellular metabolite concentrations because they provide both energy and building blocks for synthesis of macro biomolecules. The status of cellular energy and nutrients can be reflected by the activity of AMPK and mTORC1, respectively. AMPK and mTORC1 regulate cell metabolism and growth by phosphorylating a wide spectrum of targets, particularly those involved in protein synthesis, mitochondrial biogenesis, glucose and lipid metabolism, and autophagy. Through AMPK and mTOR, therefore, nutrients modulate cellular metabolic status, which is reflected by the concentrations of key metabolic intermediates, such as α-KG. The dioxygenase family of enzymes monitors α-KG as a substrate and other metabolites as competitive inhibitors to control epigenetic modifications and gene expression. The interplay among nutrients, metabolites, protein phosphorylation, and gene expression are involved in the coordination of cell growth with extracellular and intracellular conditions.
Acknowledgements
The authors wish to thank Drs. Jenna Jewell and Ryan Russell for critical reading of the manuscript. This work is supported by grants from NIH (KLG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D. The AMP-activated protein kinase cascade--a unifying system for energy control. Trends Biochem Sci. 2004;29:18–24. doi: 10.1016/j.tibs.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Carling D, Hardie DG. The substrate and sequence specificity of the AMP-activated protein kinase. Phosphorylation of glycogen synthase and phosphorylase kinase. Biochim Biophys Acta. 1989;1012:81–86. doi: 10.1016/0167-4889(89)90014-1. [DOI] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Carling D, Munday MR, Hardie DG. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur J Biochem. 1992;203:615–623. doi: 10.1111/j.1432-1033.1992.tb16591.x. [DOI] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Horman S, Browne G, Krause U, Patel J, Vertommen D, Bertrand L, Lavoinne A, Hue L, Proud C, Rider M. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr Biol. 2002;12:1419–1423. doi: 10.1016/s0960-9822(02)01077-1. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003a;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003b;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SB, Nielsen JN, Birk JB, Olsen GS, Viollet B, Andreelli F, Schjerling P, Vaulont S, Hardie DG, Hansen BF, et al. The alpha2-5′AMP-activated protein kinase is a site 2 glycogen synthase kinase in skeletal muscle and is responsive to glucose loading. Diabetes. 2004;53:3074–3081. doi: 10.2337/diabetes.53.12.3074. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J. regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, et al. Metabolic Stress Controls mTORC1 Lysosomal Localization and Dimerization by Regulating the TTT-RUVBL1/2 Complex. Mol Cell. 2013 doi: 10.1016/j.molcel.2012.10.003. http://dx.doi.org/10.1016/j.molcel.2012.1010.1003. [DOI] [PMC free article] [PubMed]
- Kim YM, Stone M, Hwang TH, Kim YG, Dunlevy JR, Griffin TJ, Kim DH. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol Cell. 2012;46:833–846. doi: 10.1016/j.molcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J Biol Chem. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002a;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002b;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, Kim JM, Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–365. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsin AS, Bertrand L, Rider MH, Deprez J, Beauloye C, Vincent MF, Van den Berghe G, Carling D, Hue L. Phosphorylation and activation of heart PFK-2 by AMPK has a role in the stimulation of glycolysis during ischaemia. Curr Biol. 2000;10:1247–1255. doi: 10.1016/s0960-9822(00)00742-9. [DOI] [PubMed] [Google Scholar]
- Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- Merrill GF, Kurth EJ, Hardie DG, Winder WW. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Physiol. 1997;273:E1107–1112. doi: 10.1152/ajpendo.1997.273.6.E1107. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Munday MR, Carling D, Hardie DG. Negative interactions between phosphorylation of acetyl-CoA carboxylase by the cyclic AMP-dependent and AMP-activated protein kinases. FEBS Lett. 1988;235:144–148. doi: 10.1016/0014-5793(88)81251-1. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- Oermann EK, Wu J, Guan KL, Xiong Y. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol. 2012;23:370–380. doi: 10.1016/j.semcdb.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem Soc Rev. 2011;40:4364–4397. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- Rotili D, Mai A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Hunter T. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay F, Brule S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, et al. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BT, Ducker GS, Barczak AJ, Barbeau R, Erle DJ, Shokat KM. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc Natl Acad Sci U S A. 2011;108:15201–15206. doi: 10.1073/pnas.1103746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Tambe Y, Inoue H, Isono T, Haneda M, Isobe K, Kobayashi T, Hino O, Okabe H, Chano T. GADD34 inhibits mammalian target of rapamycin signaling via tuberous sclerosis complex and controls cell survival under bioenergetic stress. Int J Mol Med. 2007;19:475–483. [PubMed] [Google Scholar]
- Wegener G, Krause U. Different modes of activating phosphofructokinase, a key regulatory enzyme of glycolysis, in working vertebrate muscle. Biochem Soc Trans. 2002;30:264–270. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Winder WW, Holmes BF, Rubink DS, Jensen EB, Chen M, Holloszy JO. Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol. 2000;88:2219–2226. doi: 10.1152/jappl.2000.88.6.2219. [DOI] [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Mancuso A, Bui TV, Tong X, Gruber JJ, Swider CR, Sanchez PV, Lum JJ, Sayed N, Melo JV, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene. 2010;29:2962–2972. doi: 10.1038/onc.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]