Abstract
Dopa (3,4-dihydroxyphenylalanine) is recognized as a key chemical signature of mussel adhesion and has been adopted into diverse synthetic polymer systems. Dopa’s notorious susceptibility to oxidation, however, poses significant challenges to the practical translation of mussel adhesion. Using a Surface Forces Apparatus to investigate the adhesion of Mfp3 (mussel foot protein 3) slow, a hydrophobic protein variant of the Mfp3 family in the plaque, we have discovered a subtle molecular strategy correlated with hydrophobicity that appears to compensate for Dopa instability. At pH 3, where Dopa is stable, Mfp3 slow like Mfp3 fast adhesion to mica is directly proportional to the mol% of Dopa present in the protein. At pH 5.5 and 7.5, however, loss of adhesion in Mfp3 slow was less than half that occurring in Mfp3 fast, purportedly because Dopa in Mfp3 slow is less prone to oxidation. Indeed, cyclic voltammetry showed that the oxidation potential of Dopa in Mfp3 slow is significantly higher than in Mfp3 fast at pH 7.5. A much greater difference between the two variants was revealed in the interaction energy of two symmetric Mfp3 slow films (Ead = −3 mJ/m2). This energy corresponds to the energy of protein cohesion which is notable for its reversibility and pH-independence. Exploitation of aromatic hydrophobic sequences to protect Dopa against oxidation as well as to mediate hydrophobic and H-bonding interactions between proteins provides new insights for developing effective artificial underwater adhesives.
INTRODUCTION
The California mussel, Mytilus californianus, thrives in the exposed wave-swept habitats along the Pacific coast of North America, secured by its versatile, rapid and permanent adhesion to diverse solid substrata. Given the intense focus on mussel-inspired adhesives in recent years,1–6 investigations of the adhesion mechanism of M. californianus have had a profound impact on the design and production of synthetic adhesive polymers that work in a wet environment as well as spurring new approaches to prevent fouling which is of major economic concern to marine shipping,7 heat exchangers,8 and biomedical implants.9–11
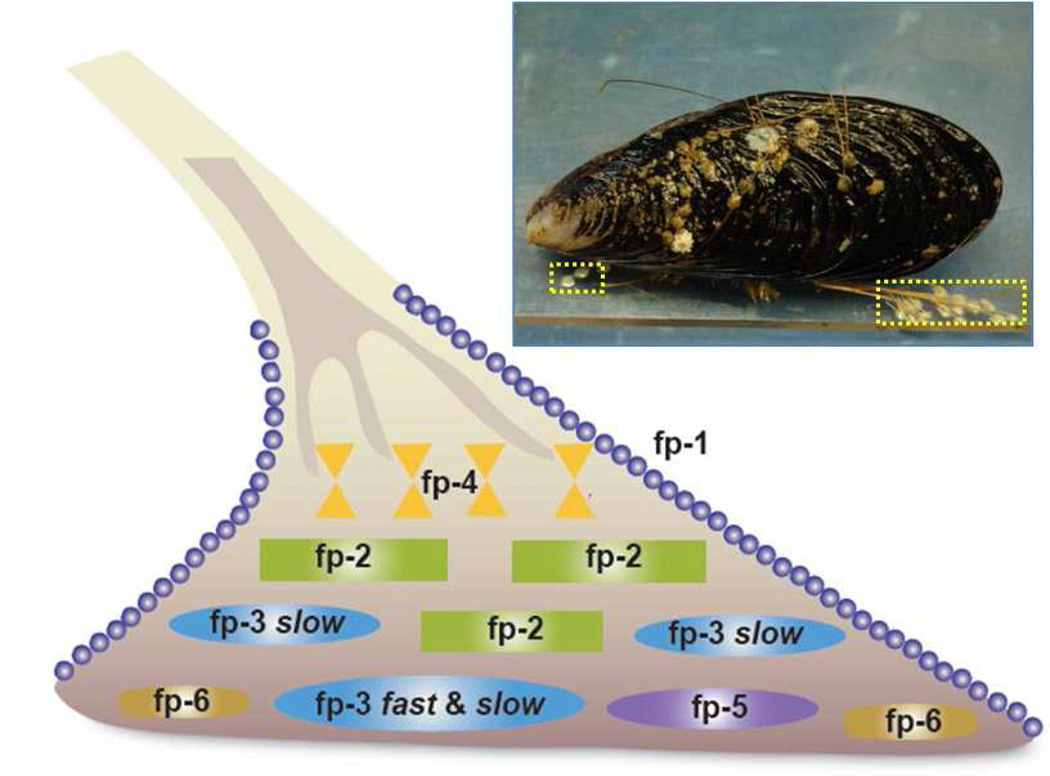
Mussel adhesion is mediated by a holdfast structure known as the byssus, essentially a bundle of leathery threads tipped by flat adhesive plaques that attach to a variety of hard surfaces (Figure 1).12 At least 9 proteins have been characterized from the adhesive plaques of Mytilus species and several of them are well characterized. Dopa is the prominent functionality in most Mfps (mussel foot proteins) that allows them to adhere to various surfaces underwater, as well as contributing to the cohesion within the plaques. Adhesive versatility is facilitated by Dopa’s ability to form hydrogen bonds with hydrophilic surfaces (e.g. mica, hydroxyapatite)13,14 and participate in coordination bonding with metal ions and metal oxides, and undermined by Dopa’s notorious susceptibility to oxidation. 15 For example, following the pH induced autoxidation of Dopa at neutral to basic pH, the adhesion of Dopa-rich Mfp3 fast to a mica surface is almost completely abolished.13,16 Clearly there is little to be gained by adopting a Dopa-like chemistry in synthetic adhesive polymers if autoxidation can’t be controlled. Mussels stringently control Dopa oxidation, and a better understanding of their control mechanisms would help to engineer better biomimetic adhesives. In one known mechanism, antioxidant thiolates in Mfp6 rescue Dopa in Mfp3 and Mfp5 from oxidized Dopaquinones.16
Figure 1.
Byssal plaque proteins of Mytilus. A mussel (M. californianus, inset) is shown attached to a polymer plate. One of its plaques (shown in dotted yellow box) is enlarged as a schematic drawing to illustrate the approximate distribution of known proteins.
In this paper, we show that mussels have another strategy to improve the adhesive performance of Dopa at neutral pH that relies on a subtle and complex interplay of Dopa with local hydropathy as well as changing secondary structure. These important new insights are both observed in the Mfp3 slow group of Mfp3 variants. Dopa-rich Mfp3 has about 30 different variants in the mussel plaque 17, which can be sub-divided to two groups, Mfp3 fast and slow. In previous studies, focus was predominantly on the adhesion properties and performance of Mfp3 fast, which was shown to be able to bridge two mica surfaces, acting like surface glue.13,16,18 The other big family of the Mfp3 variants, Mfp3 slow, is less well understood. The defining differences between Mfp3 fast and Mfp3 slow variants are that their eluting fractions in reverse phase HPLC are well separated as are their mobilities on acetic acid-urea polyacrylamide gels (AU-PAGE).
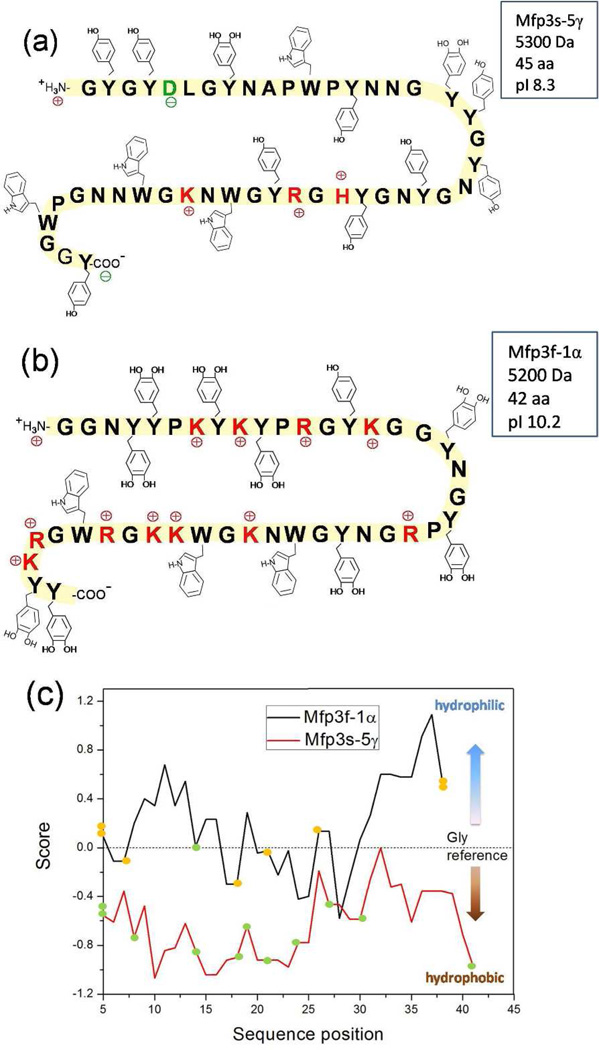
Both Mfp3 fast and slow have low molecular weights with masses between 5~7 KDa. Compositionally (Figure 2), Mfp3 slow has much lower content of basic arginine and lysine; in Mfp3 fast, almost all the tyrosine residues are post-translationally modified to Dopa (17–20 mol %), whereas in Mfp3 slow, approximately half are modified (8–14 mol %). The lower charge density results in Mfp3 slow being a very hydrophobic protein as seen in the hydropathy plot in Figure 2c. As a confirmation of the hydropathy prediction, Mfp3 fast is water soluble while Mfp3 slow is not. Mfp3 slow is the most abundant directly extractable protein from the byssus plaques of M. californianus, with a yield more than twice that of Mfp3 fast. Based on a comparison of the tryptophan (Trp) content of Mfp3 and whole plaque, it is estimated that the proportion of Mfp3 slow in plaque to be at least 16% by weight (Supporting information).
Figure 2.
Sequences of (a) Mcfp3-5γ and (b) Mfp3-1α, with (c) their Hopp and Woods hydropathy (ExPaSy tools) plots using hydropathy values from Nozaki and Tanford19 for standard amino acids and Dopa with window size 9 (Orange spots denote Dopa; green spots denote Tyr). The Dopa residues in the Mfp3-5γ sequence are unassigned and could occur wherever Tyr is present.
In this work, the adhesion of Mfp3 slow to mica, the self-interactions between Mfp3 slow layers, and the interactions (Supporting information) between Mfp3 slow and other proteins (Mfp3 fast and Mfp2) were tested using a surface forces apparatus (SFA). The SFA has proven to be a suitable and powerful technique for studying protein adhesion.17 Cyclic voltammetry was used to compare the redox potential of Dopa in Mfp3 fast and slow. Attenuated total reflectance Fourier transform infrared spectrometry (ATR-FTIR) as well as circular dichorism (CD) were used to study the secondary structure of Mfp3 slow at the same three pHs used for SFA experiments.
EXPERIMENTAL SECTION
Mfp Purification from Mussel Feet
Mussels were collected locally from rocks and jetties around Campus Point and Goleta Pier in Santa Barbara, CA and immediately transferred to shallow holding tanks with circulating raw seawater at 15 °C. About 30 mussels were tethered to each 18 × 25-cm acrylic or glass plate (thickness, 0.5 cm), from which byssal plaques were harvested daily by scraping using a clean single edge razor blade, rinsed with MilliQ water, and stored at −80 °C. About 1000 accumulated plaques were thawed and homogenized in a small volume (5 ml/200 plaques) of 5% acetic acid (v/v) containing 8 M urea on ice using a small hand-held tissue grinder (Kontes, Vineland, NJ). The homogenate was centrifuged for 30 min at 20,000 × g and 4 °C. The soluble acetic acid/urea plaque extracts were subjected to reverse phase HPLC using a 260 × 7-mm RP-300 Aquapore (Applied Biosciences Inc., Foster City, CA) column, eluted with a linear gradient of aqueous acetonitrile. Eluant was monitored continuously at 230 and 280 nm, and 1-ml fractions containing Mfp3 slow were pooled and freeze-dried, injected into Shodex-803 column (5 µm, 8 × 300 mm), which was equilibrated and eluted with 5 % acetic acid in 0.1 % trifluoroacetic acid. Eluant was monitored at 280 nm. Sample purity was assessed by acid urea-PAGE, amino acid analysis, and MALDI time-of-flight mass spectrometry. Fractions with pure Mfp3 slow were diluted with buffer for further studies.
Force Versus Distance Profiles Measurement by the Surface Forces Apparatus
The interactions between the Mfp3 slow films and between a Mfp3 slow film with the film of a different Mfp were measured by a surface forces apparatus (SFA). The SFA technique has been used for many years to measure varies interactions between surfaces in vapors and liquids including van der Waals forces, electrostatic interactions, hydrophobic interactions, and different ligand-acceptor specific interactions.20–22 The details of this technique have been described elsewhere12. Briefly, the SFA can measure the normal (attractive or repulsive) force F between two curved surfaces in cross cylinder geometry as a function of surface separation distance D. The force F(D) measured by the SFA can be converted into the interaction energy between two flat surfaces by applying Derjaguin approximation: E = F(D)/2πR, where R is the radius of the surfaces.
The protein was deposited by placing 20 µl of Mfp3 slow solution (20 µg/ml) together with 100 µl of pH 3 buffer onto on one or both mica surfaces. After 30 min absorption followed by rinsing with buffer with the desired pH, the two surfaces were mounted in the SFA chamber with a droplet of buffer (~100 µl) sandwiched in between for measurement. All experiments were performed at a controlled room temperature (22 °C). To adjust the pH of protein solution droplets on mica surfaces, the following stock buffers were used for rinsing: 0.1 M sodium acetate (EM Science, Gibbstown, NJ) and 0.25 M potassium nitrate titrated by acetic acid (pH 5.5); 0.016 M potassium phosphate monobasic (Mallinckrodt, Hazelwood, MO), 0.084 M potassium phosphate dibasic (EMD Chemicals, Gibbstown, NJ) 0.25 M potassium nitrate (pH 7.5). Milli-Q water (Millipore, Bedford, MA) was used for all the glassware cleaning and solution preparation.
Cyclic Voltammetry
Cyclic voltammetry (CV) was performed using a CHI 500/A electrochemical workstation from CH Instruments Inc. These analyses were carried out using a three-electrode cell, a Pt wire as the counter electrode, a Ag/AgCl reference electrode calibrated with a 5 mM solution of Fc/Fc+ in 0.1 M TBAP/acetonitrile (ACN) as electrolyte solution and a glassy carbon working electrode. For the pH 2 buffer treatment, 0.1mg/ml Mfp3 fast and slow were dissolved in 5% acetic acid. For pH 7.5, concentrated Mfp3 fast and slow dissolved in 5% acetic acid solution were deposited onto the working electrode first, and after being dried at ambient temperature, the electrode was put into 0.1 M phosphate buffer for measurement.
ATR-FTIR Spectroscopy
IR spectra were collected using a Nicolet Magna 850 infrared spectrometer at buffering pH conditions of 3, 5.5 and 7.5, respectively, prepared with deuterium oxide (final protein concentration 0.5 mg/ml). Data were collected using an attenuated total reflectance (ATR) device equipped with a ZnSe crystal. A total of 128 scans were taken at a resolution or 4 cm−1. Fourier self-deconvolution was applied to increase spectral resolution in the amide I′ region (1600–1700 cm−1) as described by Byler et al 23 Origin software (Origin Lab Corporation, Northampton, MA) was used to quantify structural components using a least squares curve-fitting assuming Gaussian peak shapes with peak centers determined from second derivative spectra.
Circular dichroism spectroscopy
Circular dichroism (CD) measurements were performed on an OLIS RSM circular dichroism spectro polarimeter. The cell was maintained at 25 °C using a Quantum temperature controller. Far-UV (250–190 nm) scans were performed in a micro cell (with a path length of 0.05 cm) that required only 300 µl of solution. The computer-averaged trace of 10 scans was employed in all calculations. Signal due to solvent was subtracted. 0.3 mg/ml of Mfp3 fast was measured in acetic acid buffer at pH 3.
RESULTS and ANALYSES
The adhesive properties of Mfp3 slow on mica
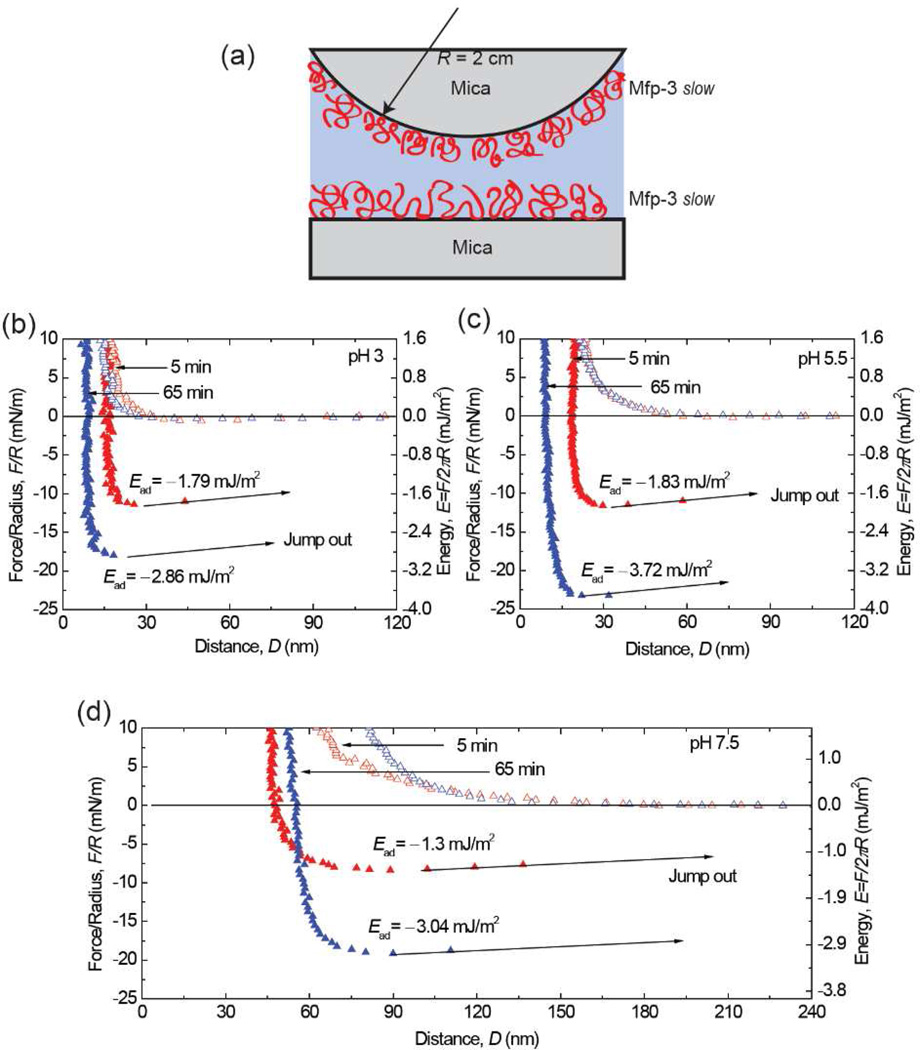
The adhesive properties of Mfp3 slow on mica surfaces were investigated by SFA at three different pHs (Figure 3a). A single normal-force measurement by SFA has nano-Newton, nN (force) and Å, 0.1 nm (distance) resolution and typically requires only 1 µg of protein. Mica is atomically smooth and biologically relevant (cf. clay-based rocks) as a support/substrate surface onto which various Mfps can be adsorbed and their interactions examined at appropriate solution conditions (salinities, pH, temperatures, etc.).16
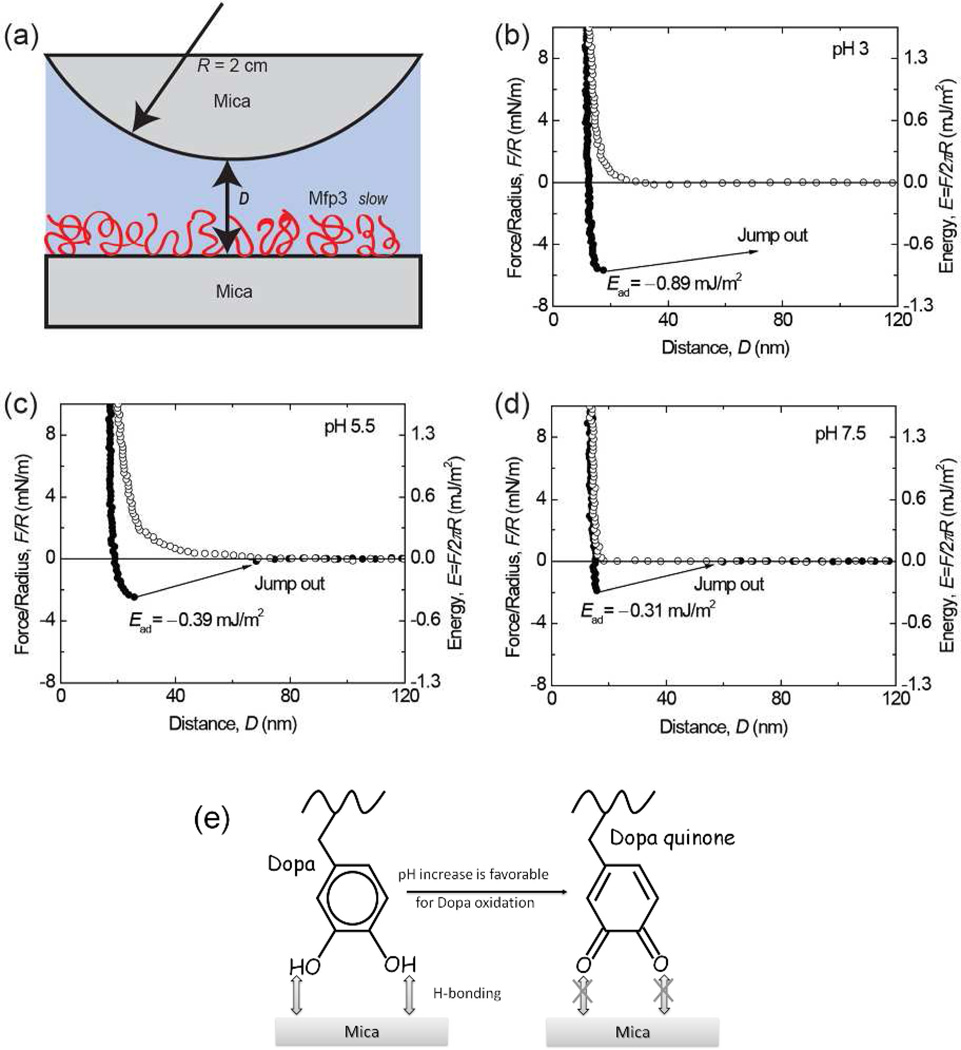
Figure 3.
Interaction between mica and Mfp3 slow adsorbed to mica at pH 3 (b), 5.5 (c), 7.5 (d). The y axis on the left gives the measured force, F/R (normalized by the radius of the surface), whereas the y axis on the right gives the corresponding adhesion energy (E) per unit area between two flat surfaces, defined by E=F/2πR. Approach (unfilled symbols); separation (filled symbols). Separation followed a brief (~3–5 min) contact. (e) The interaction between Dopa and mica is H-bonding, which decreases as pH increases due to Dopa oxidation.
The interaction between one Mfp3 slow coated and another bare mica surface was measured as a function of surface separation distance at pH 3 (Figure 3b). During the approach, the first observable interaction was the “steric” repulsion caused by the entropic confinement of proteins. The surfaces were further compressed until the distance between two surfaces reached a constant value (the “hard wall”). After a short holding period (~3 min), the surfaces were separated and an adhesion energy of Ead~−0.95 mJ/m2 when the surfaces “jumped” apart. When the same experiment was repeated at pH 5.5 and 7.5, the adhesion energy decreased to −0.42 and −0.32 mJ/m2 respectively (Figure 3c and 3d).
These results showed that, similar to the fast Mfp3 variants, an Mfp3 slow film is also capable of adhering well to a bare mica surface (asymmetric configuration). Given that the Dopa-rich Mfp-to-mica interaction is attributed to hydrogen bonds (H-bonds) between the bidentate OH groups of the catecholic side groups and the O atoms in the mica polysiloxane lattice 13,16 and that on average Mfp3 slow has half as much Dopa as Mfp3 fast, it is reasonable to expect weaker adhesion for Mfp3 slow to mica (Ead~−0.95 mJ/m2) at pH 3 than for Mfp3 fast (Ead~−2 mJ/m2). The trend of decreased adhesion with increasing pH (Figure 3e) also parallels what was previously observed for Mfp3 fast.13 However, at pH 7.5, the adhesion of Mfp3 slow is still significant, being about 1/3 of that measured at pH 3, whereas for Mfp3 fast no adhesion was measurable at pH 7.5. This discrepancy suggests that Dopa residues in the two protein variants (Mfp3 fast and slow) may be differentially prone to oxidation at the same pH. To verify that Dopa is essential for Dopa/mica bonding, Dopa of Mfp3 slow in the asymmetric configuration was periodate oxidized. Following periodate addition, the adhesion was abolished (Figure S1), which suggests Mfp3 slow with oxidized Dopa cannot adhere to mica.
Cyclic Voltammetry on Mfp3 fast and slow
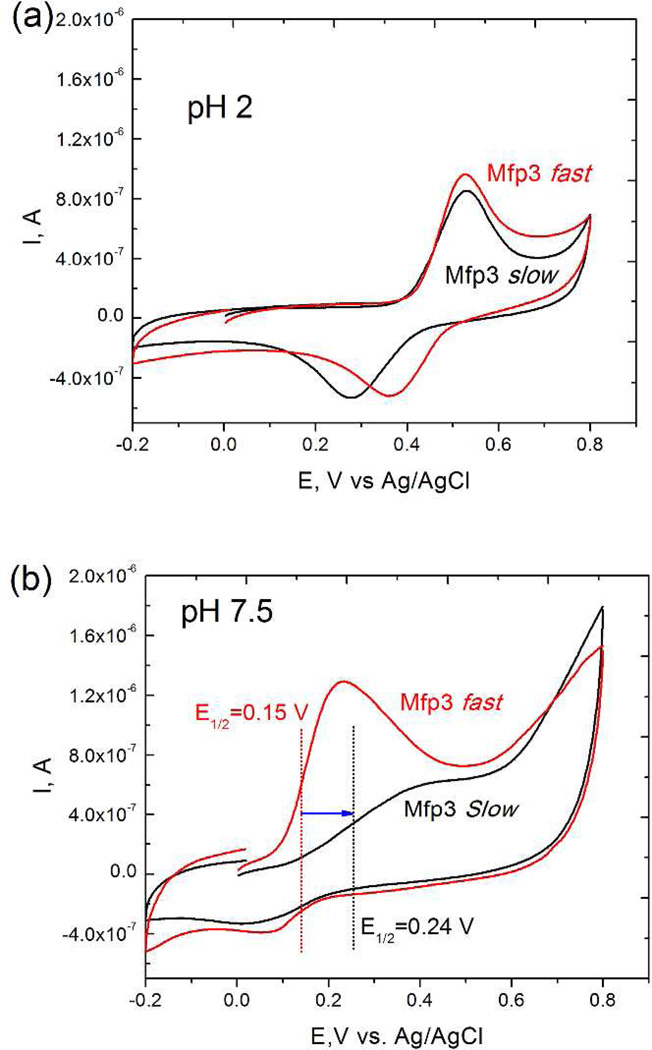
To compare the redox potential of Mfp3 slow and fast, cyclic voltammetry measurements for both proteins were carried out at pH 2 and pH 7.5 (Figure 4). At acidic pH, Mfp3 fast and slow showed similar redox peaks and the same half-wave oxidation potential (E1/2=0.41 V). However at pH 7.5, E1/2 of Mfp3 slow (0.24 V) was significantly higher than that of Mfp3 fast (0.15 V), suggesting that Dopa residues of Mfp3 slow are less prone to auto-oxidation at this pH than those in the fast variants.
Figure 4.
Cyclic voltammograms of (a) Mfp3 fast and slow in pH 2 acetic acid solution; (b) Mfp3 fast and slow in pH 7.5 phosphate buffer. At pH 7.5, before measurement, Mfp3 fast and slow solutions were dropped onto the working electrode surface, and dried at the ambient temperature. The working electrode was then immersed in 0.1 M phosphate buffer for further measurement.
The cohesive (self-interaction) of Mfp3 slow
The selfinteraction or cohesion of Mfp3 slow layers was measured using a symmetric configuration, i.e., depositing monomolecular Mfp3 slow films on both mica surfaces (Figure 5). At pH 3, moderately strong adhesion of Ead ~−1.8 mJ/m2 (Figure 5b) was measured upon separating the surfaces. Surprisingly, the adhesion forces measured at higher pHs did not exhibit a significant pH dependence. The strong adhesion forces persisted, with adhesion energies of −1.8 and −1.4 mJ/m2, respectively, (Figure 5c, d) at the two higher pHs studied (pH 5.5 and 7.5). At all three pH conditions, a longer contact time prior to separation resulted in a stronger interaction. With a 65 min contact, for example, Mfp3 slow interaction energies increased at pH 5.5 (−3.7 mJ/m2) and 7.5 (−3.0 mJ/m2) compared with that at pH 3 (−2.9 mJ/m2). The ability to maintain strong interactions at pH 5.5 and pH 7.5 distinguishes Mfp3 slow cohesive self-interactions from those of Mfp524, Mfp3 fast, and Mfp3 slow on mica surfaces. In the asymmetric configuration experiments, the adhesion strength − attributed to the formation of H-bonds between Dopa residues in Mfp3 slow and mica − decreased as pH-dependent Dopa autoxidation increased.
Figure 5.
Interaction between Mfp3 slow layers absorbed onto mica at (b) pH 3; (c) pH 5.5; (d) pH 7.5. Contact times were as shown. Approach (unfilled symbols); separation (filled symbols).
A tally of possible noncovalent interactions between symmetric films of Mfp3 slow includes chiefly Dopa-mediated, electrostatic, and hydrophobic interactions. The apparent pH-independence of cohesion in Mfp3 slow effectively eliminates contributions based on electrostatic attraction given the very low charge density in Mfp3 slow and the slightly alkaline pI of 8–9. Indeed, as the pH approaches the protein pI, the electrostatic repulsion between two symmetric protein surfaces would be eliminated leading to enhanced hydrophobic interactions. Hydrophobic interactions may thus account for the strong cohesion between symmetric Mfp3 slow films when the pH increases from 3 to 5.5 then 7.5, but before concluding that we addressed the Dopa contribution using periodate oxidation.
Excess periodate was introduced between the mica surfaces with symmetrically deposited Mfp3 slow at pH 3. Almost no attractive interaction (Ead) and a 3-fold greater hardwall (repulsion) were observed (Figure S2). At first blush, this result suggests that cohesion does not depend on hydrophobic interactions, but rather on Dopa mediated protein-protein interactions. In order to conclude this, however, one must know where the failure has occurred. If the hydrophobic contribution to the protein-protein interaction is a strong one, then the weak link may shift to the interface between Mfp3 slow and mica. .However, the measured Ead of the asymmetrically deposited Mfp3 slow at pH 3 was −0.89 mJ/m2 (Figure 3), only half that for the symmetric configuration (Ead = −2.3 mJ/m2 Figure S2), hence it is not clear that the Mfp3-mica interface became the new weak link. Evidently, periodate must form a bidentate chelate with the catechol moiety of Dopa in order to oxidize it.25 Such chelates appear to form with both bound and unbound Dopa, in contrast to pH-induced oxidations where bound Dopa is at least partially shielded from oxidation (Figure 3)16. Results from an experiment in which the symmetric configuration was exposed to periodate after contact suggest that Dopa binding to mica may not be stable to periodate (Figure S3). Considering the widespread use of periodate as a Dopa oxidant, this merits further scrutiny.
ATR-FTIR
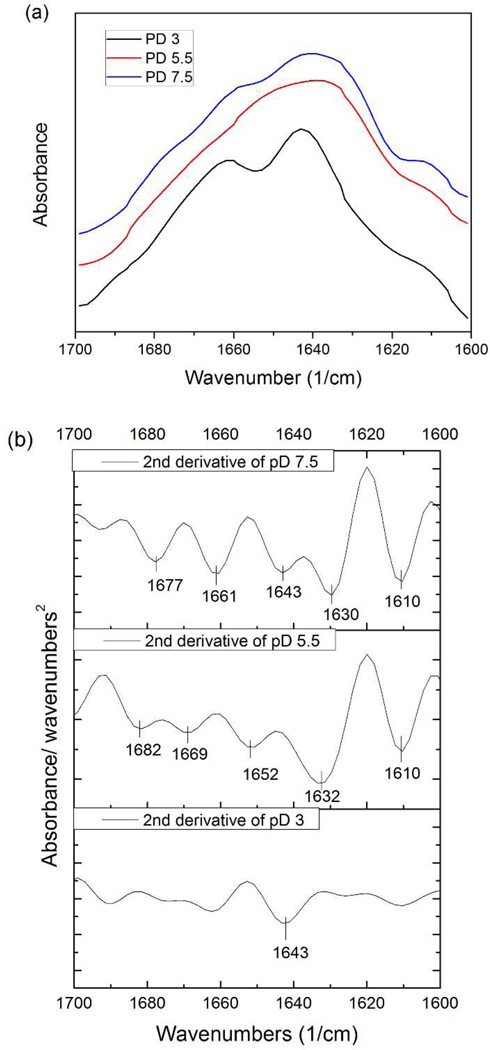
H-bonding is an important interaction that is abundantly common within a protein molecule as well as between different molecules; it also stabilizes protein secondary structure. Correspondingly, secondary structure in proteins could be an indication of H-bonding between chains or side-groups. In order to investigate the impact of pH on secondary structure of Mfp3 slow, FTIR spectra of Mfp3 slow were taken at pD 3, 5.5 and 7.5 in deuterated acetate or phosphate-buffered saline (Figure 6). The results are shown in Figure 6. The primary peak for pD 3 was at 1643 cm−1, which can be assigned to a random coil secondary structure.
Figure 6.
(a) ATR-FTIR spectra of 0.5mg/ml Mfp3 slow in deuterated buffers at 3 different pH conditions used for SFA experiments. The signal of pD 3 and 7.5 were magnified 20 and 5 times respectively for the convenience of comparison. (b) Second derivative of spectra (a): The primary peak at pD 3 at 1643 cm−1 can be assigned to random coils. At pD 5.5, The prominent peak 1632 cm−1 is associated with β sheet, and 1682 cm−1 with high β sheet. Band at 1669 cm−1 is generally assigned to turns. Peak at 1652 cm− 1 indicates the existence of α-helices. At pD 7.5, low β sheet slightly shifted to 1630 cm−1 was observed. Peak at 1661 cm−1 can be associated with both α-helices and extended turns. A shift was also observed for the high frequency β sheet feature at 1677 cm−1.
At both pD 5.5 and 7.5, the proteins phase-separate from the solvent, some of which is deposited on the surface of ATR crystal forming a patchy hydrated layer. ATR-FTIR spectra at both pD reveal an increase in ordered secondary structure compared with the random coil configuration under pD 3 conditions.
Significant differences in the amide I′ peak were observed in the pD 5.5 spectrum compared to that of pD 3: notably, there was a complete loss of the random coil signature (1643 cm−1), and the appearance of new bands at 1632 cm−1, 1652 cm−1, 1669 cm−1 and 1682 cm−1. The prominent peak at 1632 cm−1 and smaller peak at 1682 cm−1 are both associated with β sheets. The peak at 1669 cm−1 is generally assigned to reverse turns23, 26–29 whereas the 1652 cm−1 peak indicates the presence of α-helices. Upon further increasing pD to 7.5, a slight shift of low frequency β sheet from 1632 cm−1 to 1630 cm−1 was observed, together with a smaller peak at 1661 cm−1 which is associated with both α-helices and extended turns.21 A shift was also observed for the high frequency β-sheet feature at 1677 cm−1 (1682 cm−1 for pD 5.5). The frequency of a given vibrational mode corresponds directly to the "strength" of the molecular bond being probed. Thus, any change in bond stability (e.g. changes in inter-residue hydrogen bonding) can result in wavenumber shifts for a given structural element. It follows that the shifts described herein may be due to different amounts of H-bonding in MFp3 slow at pD 5.5 and 7.5.
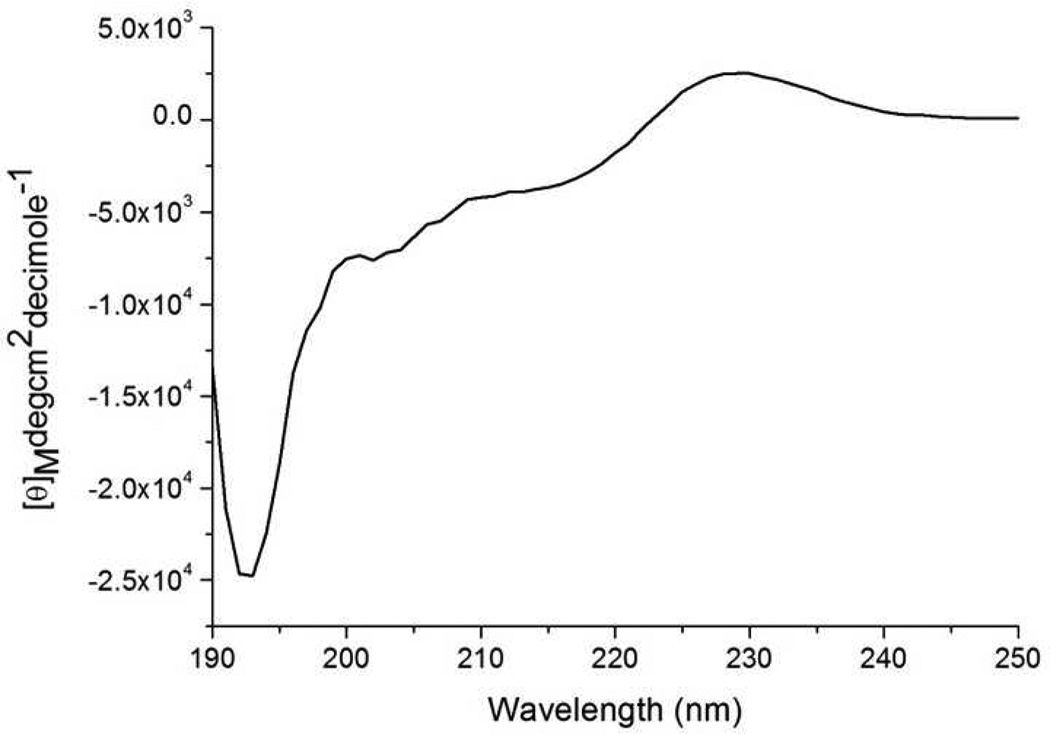
To summarize, as pD was increased from 3 to 5.5, Mfp3 slow transformed from a random coil to highly ordered structure, which consists mostly of β-sheet and turns mixed with some α-helices. At pD 7.5, β-sheet and turns prevail as well, accompanied by α-helices and random coils. The random structure of soluble Mfp3 slow at pH 3 was confirmed by CD spectroscopy (Figure 7). CD measurements at the other two pH conditions were not possible due to light scattering by the phase separated proteins. According to ATR-FTIR results, H-bonds contribute more to protein secondary structure at pH 5.5 and 7.5 than at pH 3.
Figure 7.
CD spectrum of Mfp3 slow in acetic acid solution at pH 3.
Combining the analyses above, the cohesion between Mfp3 slow molecules can be associated with inter-residue H-bonds (including H-bonds of Dopa/Dopa and Dopa/other amino acid residue) and hydrophobic interactions. As the pH increases from 3 to 5.5 then to 7.5, Dopa-mediated interactions decrease due to Dopa oxidation, whereas inter-residue H-bonding and hydrophobic interactions are favored. As Mfp3 slow gets closer to the pI, there are fewer charges on the molecules thus leading to less electrostatic repulsion enabling closer inter-molecular distance. These interactions would complement each other at different pHs to give similar total cohesion energy at the pHs that were measured in SFA.
DISCUSSION
Dopa has emerged as a crucial component for both protein adhesion (to substrates) and cohesion between mussel adhesive proteins. As a result, Dopa has been incorporated into diverse synthetic systems for mimicking biological wet adhesion. However, in a number of synthetic polymers and purified adhesive proteins, the failure to adhere to various substrates on account of Dopa oxidation is well known, especially at neutral to basic pH. The decrease in adhesion to mica with increasing pH from 3 to 5.5 then 7.5 was also observed for Mfp3 slow, but was less pronounced than its electrophoretically fast variant. For example, for Mfp3 fast, adhesion at pH 7.5 was virtually abolished whereas for Mfp3 slow a third of the adhesion energy at pH 3 was retained. CV analyses suggest that Dopa is more difficult to oxidize (Figure 4) in that the oxidation potential of Mfp3 slow is distinctly higher than that of Mfp3 fast at pH 7.5.
Mfp3 slow is the most hydrophobic protein in mussel plaques. At pH 7.5, which is close to its pI, the Mfp3 slow protein molecules will therefore tend to pack more closely due to the absence of electrostatic repulsion and the enhanced hydrophobic interaction. We propose that the Dopa residues in Mfp3 slow are nested in a hydrophobic microenvironment, which prevents them from being naturally exposed to the surrounding aqueous environment. As such, the measured redox potential of Mfp3 slow is higher than that of Mfp3 fast. This could also explain why in the SFA data, the loss of Dopa/mica hydrogen-bonding interaction in Mfp3 slow is much less than in Mfp3 fast when the pH is raised from moderately acidic to slightly basic.
For practical adhesive bonding, cohesion within the adhesive joint is as important as the interfacial adhesion. For marine mussels, while the interfacial adhesion between plaques and various surfaces influences how strongly mussel adhesive proteins bind to surfaces, cohesive interactions are also needed to maintain the integrity of byssal plaques and threads. Up to now, two Mfps have been shown to contribute to the cohesive strength of mussel plaques by SFA measurements in which proteins are deposited on both surfaces: Mfp1 forms Dopa-Fe3+ coordination complexes, which increase both the stiffness and hardness of the plaque cuticle;30, 31 Mfp2 binds to Mfp5 at the interface and to other Mfp2 in the presence of Ca2+ and Fe3+ in the plaque, functioning as a bridge between the plaque interface and the scaffolding collagens and associated matrix proteins that join the thread to the plaque.32 From these two cases, Dopa-metal ion coordination has emerged as a readily translatable cohesive strategy for synthetic systems. Our studies have shown that Mfp3 slow also contributes to the cohesion of mussel plaque that is independent metal ions. Given the strong cohesive interaction of Mfp3 slow over the pH range of 3–7.5, it is not unreasonable to suppose hydrophobic interactions to be dominant.
The hydrophobic interaction is known to be a driving factor in protein folding and in biomacromolecular structure generally, 33–35 but its contribution to wet adhesion has barely been investigated36 especially with regard to the cohesive reinforcement of the bonded adhesive. Considering the high content of amino acids with hydrophobic side chains in Mfp3 slow, we propose that inter- and intra-molecular hydrophobic interactions play a significant role in the cohesion measured by SFA. At this point, it is not clear how pH affects the hydrophobic interactions37. The evolution of pH-dependent secondary structure in Mfp3 slow as detected by by FTIR suggests increasing inter-residue H-bonding as the pH increases from 3 to 5.5 and 7.5. In general, as the pH approaches the protein pI, the electrostatic repulsion between protein molecules is reduced, thereby enhancing hydrophobic interactions, followed by localized H-bonding.
CONCLUSION
We have shown that Dopa nested in hydrophobic aromatic sequences not only enhances adhesion at neutral pH (pI or IEP), but also contributes significantly to the cohesive interactions between adhesive proteins. The high proportion of hydrophobic amino acid residues in Mfp3 slow sequence provides Dopa with a microenvironment that retards oxidation by shielding the amino acids from the solvent and endows the protein with the ability to maintain adhesion at neutral to slightly basic pH.
More importantly, hydrophobic interactions and inter-residue H-bonding combine to result in strong cohesion within Mfp3 slow layers over a relatively wide range of pH. This strategy provides an alternative to Dopa/metal ion chelation, and compensates in part for limitations imposed by facile Dopa-autoxidation. By exploring the adhesive and cohesive mechanisms of bonding by Mfp3 slow, these studies reveal that the wet adhesion of mussels is more complicated than a simple Dopa-mediated recipe, and provide a rationale for engineering Dopa into a new generation of bio-inspired synthetic adhesive polymers.
Supplementary Material
ACKNOWLEDGMENT
Funding Sources
This work was supported by the Materials Research Science and Engineering Centers Program of the National Science Foundation under Award No. DMR 1121053 and the National Institutes of Health under Grant R01-DE018468.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Mfp3 slow content estimation in mussel plaque. SFA results of the influence of periodate on Mfp3 slow adhesion to mica surface, of periodate on Mfp3 slow cohesion, and interactions between Mfp3 slow and other Mfps. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lee H, Dellatore SM, Miller WM, Messersmith PB. Science. 2007;318:426. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee H, Lee BP, Messersmith PB. Nature. 2007;448:338. doi: 10.1038/nature05968. [DOI] [PubMed] [Google Scholar]
- 3.Lee H, Rho J, Messersmith PB. Adv Mater. 2009;21:431. doi: 10.1002/adma.200801222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ham HO, Liu ZQ, Lau KHA, Lee H, Messersmith PB. Angew Chem Int Edit. 2011;50:732. doi: 10.1002/anie.201005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westwood G, Horton TN, Wilker JJ. Macromolecules. 2007;40:3960. [Google Scholar]
- 6.Matos-Perez CR, White JD, Westwood G, McCarron HE, Wilker JJ. Abstr Pap Am Chem S. 2011;241 [Google Scholar]
- 7.Anderson C, Atlar M, Callow M, Candries M, Milne A, Town-sin RL. J Mar Design Operations. 2003;B4:23. [Google Scholar]
- 8.Cho YI, Choi BG. Inter J of Heat and Mass Trans. 1999;8:1491. [Google Scholar]
- 9.Ratner BDJ. Biomed. Mater. Res. 1993;27:283. doi: 10.1002/jbm.820270302. [DOI] [PubMed] [Google Scholar]
- 10.Wisniewski N, Reichert M. Colloids Surf., B. 2000;18:197. doi: 10.1016/s0927-7765(99)00148-4. [DOI] [PubMed] [Google Scholar]
- 11.Bryers JD. Colloids Surf., B. 1994;2:9. [Google Scholar]
- 12.Waite JH, Andersen NH, Jewhurst S, Sun CJ. J Adhesion. 2005;81:297. [Google Scholar]
- 13.Yu J, Wei W, Danner E, Israelachvili JN, Waite JH. Adv Mater. 2011;23:2362. doi: 10.1002/adma.201003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chirdon WM, O'Brien WJ, Robertson RE. J Biomed Mater Res B. 2003;66B:532. doi: 10.1002/jbm.b.10041. [DOI] [PubMed] [Google Scholar]
- 15.Lee H, Scherer NF, Messersmith PB. P Natl Acad Sci USA. 2006;103:12999. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Wei W, Danner E, Ashley RK, Israelachvili JN, Waite JH. Nat Chem Biol. 2011;7:588. doi: 10.1038/nchembio.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao H, Robertson NB, Jewhurst SA, Waite JH. J Biol Chem. 2006;281:11090. doi: 10.1074/jbc.M510792200. [DOI] [PubMed] [Google Scholar]
- 18.Lin Q, Gourdon D, Sun CJ, Holten-Andersen N, Anderson TH, Waite JH, Israelachvili JN. P Natl Acad Sci USA. 2007;104:3782. doi: 10.1073/pnas.0607852104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nozaki Y, Tanford C. J Biol Chem. 1971;246:2211. [PubMed] [Google Scholar]
- 20.Israelachvili JN, Adams GE. J Chem Soc Farad T 1. 1978;74:975. [Google Scholar]
- 21.McGuiggan PM, Israelachvili JN. J Mater Res. 1990;5:2232. [Google Scholar]
- 22.Israelachvili JNEd., editor. Intermolecular And Surface Forces. 3rd ed. Academic Press; 2010. [Google Scholar]
- 23.Byler DM, Susi H. Biopolymers. 1986;25:469. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 24.Danner E, Kan YJ, Hammer MU, Israelachvili JN, Waite JH. Biochemistry. 2012;51:6511. doi: 10.1021/bi3002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weidman SW, Kaiser ET. J Am Chem Soc. 1966;24:5820. [Google Scholar]
- 26.Goormaghtigh E, Cabiaux V, Ruysschaert JM. Eur J Biochem. 1990;193:409. doi: 10.1111/j.1432-1033.1990.tb19354.x. [DOI] [PubMed] [Google Scholar]
- 27.Zurdo J, Guijarro JI, Dobson CM. J Am Chem Soc. 2001;123:8141. doi: 10.1021/ja016229b. [DOI] [PubMed] [Google Scholar]
- 28.Torrent J, Alvarez-Martinez MT, Harricane MC, Heitz F, Liautard JP, Balny C, Lange R. Biochemistry-Us. 2004;43:7162. doi: 10.1021/bi049939d. [DOI] [PubMed] [Google Scholar]
- 29.Serrano V, Liu W, Franzen S. Biophys J. 2007;93:2429. doi: 10.1529/biophysj.106.100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington MJ, Masic A, Holten-Andersen N, Waite JH, Fratzl P. Science. 2010;328:216. doi: 10.1126/science.1181044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng HB, Hwang DS, Israelachvili JN, Waite JH. P Natl Acad Sci USA. 2010;107:12850. doi: 10.1073/pnas.1007416107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang DS, Zeng HB, Masic A, Harrington MJ, Israelachvili JN, Waite JH. J Biol Chem. 2010;285:25850. doi: 10.1074/jbc.M110.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dill KA. Biochemistry-Us. 1990;29:7133. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 34.Dill KA. Protein Sci. 1999;8:1166. doi: 10.1110/ps.8.6.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh K, Dill KA. J Am Chem Soc. 2009;131:2306. doi: 10.1021/ja808136x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamino K, Masahiro N, Kanai S. FEBS J. 2012;10:1750. doi: 10.1111/j.1742-4658.2012.08552.x. [DOI] [PubMed] [Google Scholar]
- 37.Meyer EE, Rosenberg KJ, Israelachvili J. P Natl Acad Sci USA. 2006;103:15739. doi: 10.1073/pnas.0606422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.