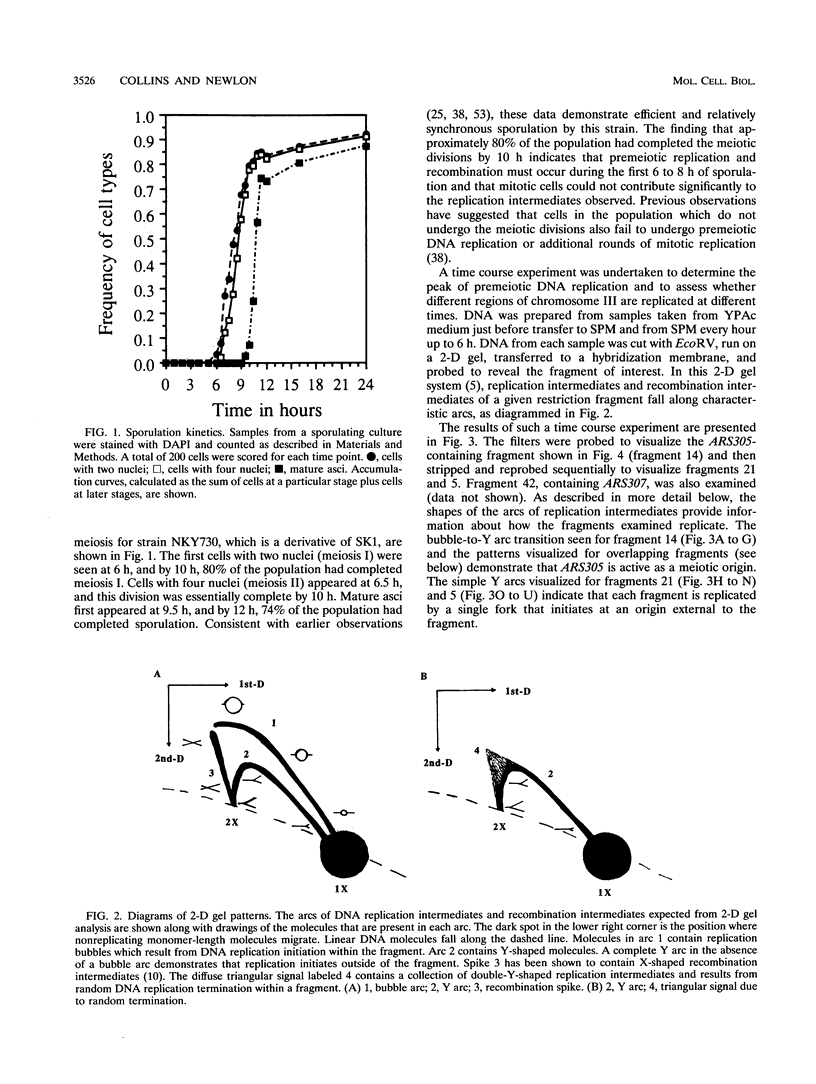
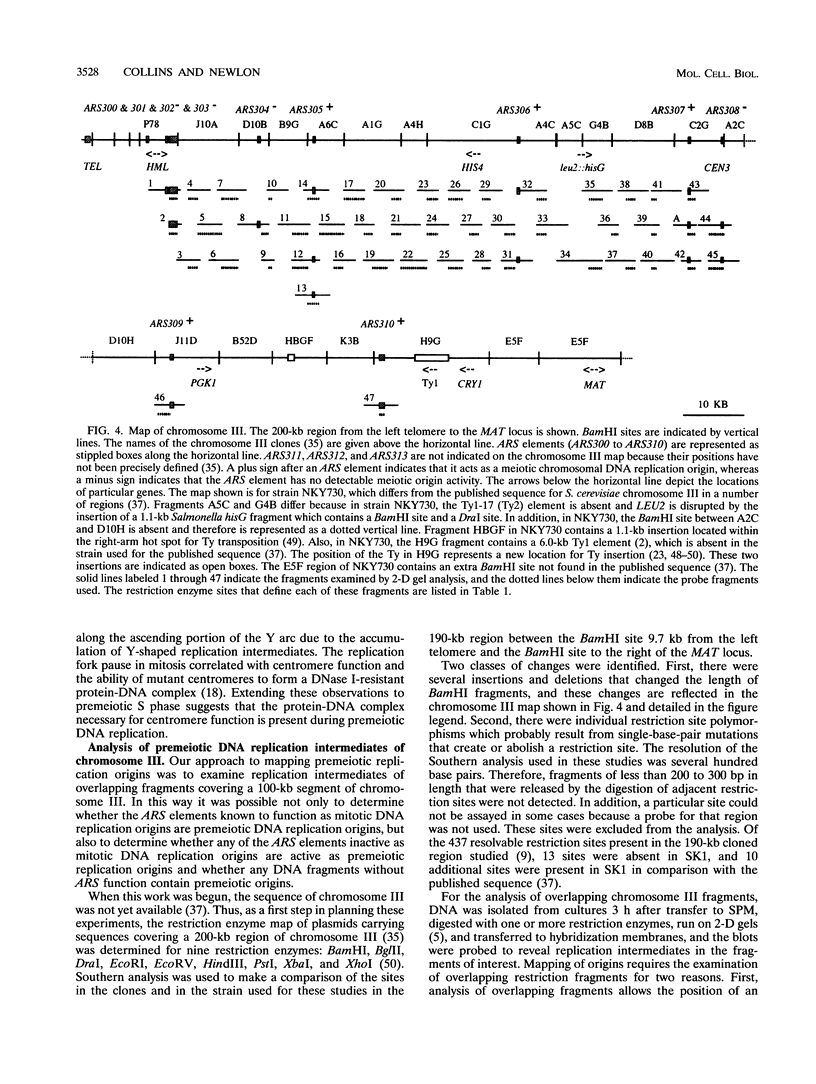
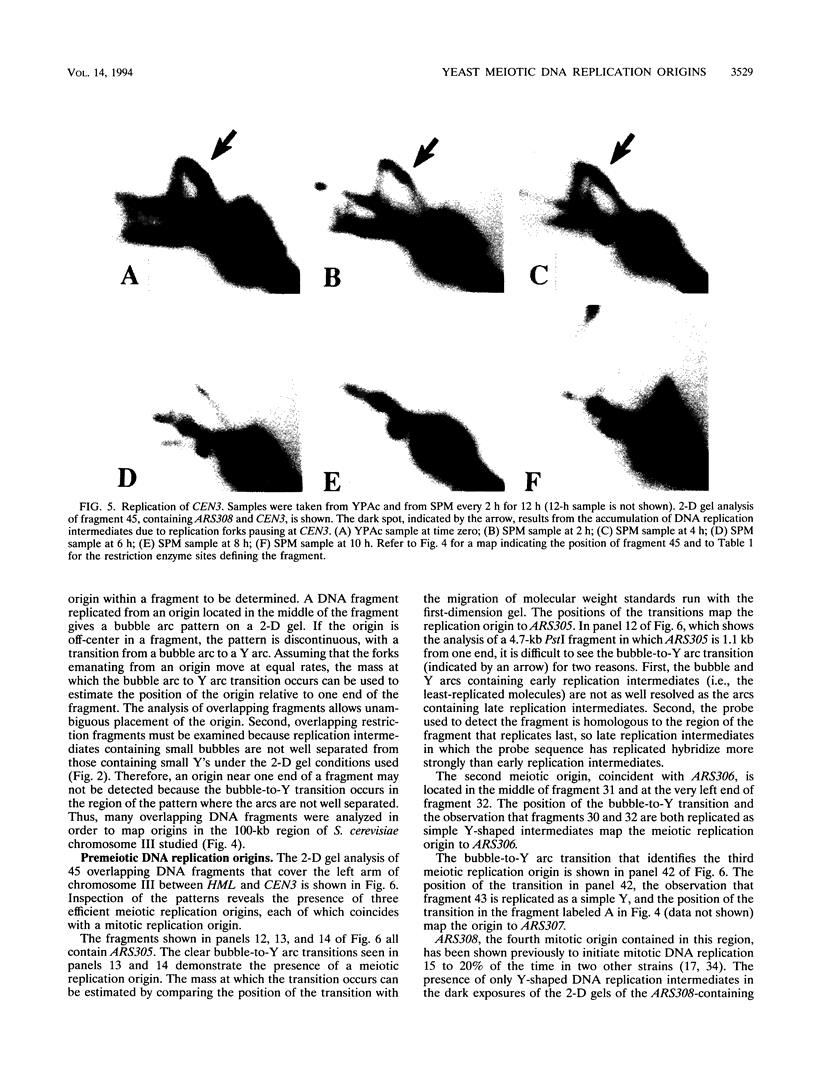
Abstract
Autonomously replicating sequence (ARS) elements are identified by their ability to promote high-frequency transformation and extrachromosomal replication of plasmids in the yeast Saccharomyces cerevisiae. Six of the 14 ARS elements present in a 200-kb region of Saccharomyces cerevisiae chromosome III are mitotic chromosomal replication origins. The unexpected observation that eight ARS elements do not function at detectable levels as chromosomal replication origins during mitotic growth suggested that these ARS elements may function as chromosomal origins during premeiotic S phase. Two-dimensional agarose gel electrophoresis was used to map premeiotic replication origins in a 100-kb segment of chromosome III between HML and CEN3. The pattern of origin usage in premeiotic S phase was identical to that in mitotic S phase, with the possible exception of ARS308, which is an inefficient mitotic origin associated with CEN3. CEN3 was found to replicate during premeiotic S phase, demonstrating that the failure of sister chromatids to disjoin during the meiosis I division is not due to unreplicated centromeres. No origins were found in the DNA fragments without ARS function. Thus, in both mitosis and meiosis, chromosomal replication origins are coincident with ARS elements but not all ARS elements have chromosomal origin function. The efficiency of origin use and the patterns of replication termination are similar in meiosis and in mitosis. DNA replication termination occurs over a broad distance between active origins.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Chlebowicz-Sledziewska E., Fangman W. L. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2529–2531. doi: 10.1128/mcb.4.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988 Nov 18;55(4):637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Callan H. G. DNA replication in the chromosomes of eukaryotes. Cold Spring Harb Symp Quant Biol. 1974;38:195–203. doi: 10.1101/sqb.1974.038.01.023. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Collins I., Newlon C. S. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 1994 Jan 14;76(1):65–75. doi: 10.1016/0092-8674(94)90173-2. [DOI] [PubMed] [Google Scholar]
- Dershowitz A., Newlon C. S. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993 Jan;13(1):391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. M., Newlon C. S. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4305–4313. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D. D., Davis L. R., Greenfeder S. A., Ong L. Y., Zhu J. G., Broach J. R., Newlon C. S., Huberman J. A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991 Oct;11(10):5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Brewer B. J. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- Greenfeder S. A., Newlon C. S. A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol Biol Cell. 1992 Sep;3(9):999–1013. doi: 10.1091/mbc.3.9.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S. A., Newlon C. S. Replication forks pause at yeast centromeres. Mol Cell Biol. 1992 Sep;12(9):4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth R. E., Jr, Sclafani R. A. Yeast pre-meiotic DNA replication utilizes mitotic origin ARS1 independently of CDC7 function. Chromosoma. 1993 Jun;102(6):415–420. doi: 10.1007/BF00360406. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Zhu J. G., Davis L. R., Newlon C. S. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14A):6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Moore D. P., Blomberg M. A., Braiterman L. T., Voytas D. F., Natsoulis G., Boeke J. D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993 Jun 4;73(5):1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., Williamson D. H., Johnson A. L., Fennell D. J. On the mechanism of premeiotic DNA synthesis in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1982 Sep;141(1):53–62. doi: 10.1016/0014-4827(82)90067-2. [DOI] [PubMed] [Google Scholar]
- Kane S. M., Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974 Apr;118(1):8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Real-time interactive frequency filtering of molecular dynamics trajectories. J Mol Biol. 1991 Jul 5;220(1):1–4. doi: 10.1016/0022-2836(91)90373-e. [DOI] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Ambiguities in results obtained with 2D gel replicon mapping techniques. Nucleic Acids Res. 1990 Feb 11;18(3):647–652. doi: 10.1093/nar/18.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Parras L., Hernández P., Martínez-Robles M. L., Schvartzman J. B. Initiation of DNA replication in ColE1 plasmids containing multiple potential origins of replication. J Biol Chem. 1992 Nov 5;267(31):22496–22505. [PubMed] [Google Scholar]
- McCarroll R. M., Fangman W. L. Time of replication of yeast centromeres and telomeres. Cell. 1988 Aug 12;54(4):505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- Newlon C. S., Lipchitz L. R., Collins I., Deshpande A., Devenish R. J., Green R. P., Klein H. L., Palzkill T. G., Ren R. B., Synn S. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991 Oct;129(2):343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Theis J. F. The structure and function of yeast ARS elements. Curr Opin Genet Dev. 1993 Oct;3(5):752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- Newlon C. S. Yeast chromosome replication and segregation. Microbiol Rev. 1988 Dec;52(4):568–601. doi: 10.1128/mr.52.4.568-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991 Sep 20;66(6):1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., McCarroll R. M., Newlon C. S., Fangman W. L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989 Oct;9(10):4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivin C. J., Fangman W. L. Cell cycle phase expansion in nitrogen-limited cultures of Saccharomyces cerevisiae. J Cell Biol. 1980 Apr;85(1):96–107. doi: 10.1083/jcb.85.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Byers B. Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma. 1978 Dec 21;70(1):109–130. doi: 10.1007/BF00292220. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shuster E. O., Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989 Sep;123(1):29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., Hirschberg J. Effects of the mitotic cell-cycle mutation cdc4 on yeast meiosis. Genetics. 1977 May;86(1):57–72. doi: 10.1093/genetics/86.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M. L., Sharrow S. O., Gart J. J. Cell cycle of Saccharomycescerevisiae in populations growing at different rates. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3850–3854. doi: 10.1073/pnas.74.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Copy number control by a yeast centromere. Gene. 1983 Aug;23(2):221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- Warmington J. R., Anwar R., Newlon C. S., Waring R. B., Davies R. W., Indge K. J., Oliver S. G. A 'hot-spot' for Ty transposition on the left arm of yeast chromosome III. Nucleic Acids Res. 1986 Apr 25;14(8):3475–3485. doi: 10.1093/nar/14.8.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicksteed B. L., Collins I., Dershowitz A., Stateva L. I., Green R. P., Oliver S. G., Brown A. J., Newlon C. S. A physical comparison of chromosome III in six strains of Saccharomyces cerevisiae. Yeast. 1994 Jan;10(1):39–57. doi: 10.1002/yea.320100105. [DOI] [PubMed] [Google Scholar]
- Williamson D. H., Fennell D. J. The use of fluorescent DNA-binding agent for detecting and separating yeast mitochondrial DNA. Methods Cell Biol. 1975;12:335–351. doi: 10.1016/s0091-679x(08)60963-2. [DOI] [PubMed] [Google Scholar]
- Williamson D. H., Johnston L. H., Fennell D. J., Simchen G. The timing of the S phase and other nuclear events in yeast meiosis. Exp Cell Res. 1983 Apr 15;145(1):209–217. doi: 10.1016/s0014-4827(83)80022-6. [DOI] [PubMed] [Google Scholar]
- Williamson D. H. The timing of deoxyribonucleic acid synthesis in the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1965 Jun;25(3):517–528. doi: 10.1083/jcb.25.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Newlon C. S., Huberman J. A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]