Abstract
HIV-associated sensory neuropathy remains an important complication of combination antiretroviral therapy (CART) and HIV infection. Mitochondrial DNA haplogroups and single nucleotide polymorphisms (SNPs) have previously been associated with symptomatic neuropathy in clinical trial participants. We examined associations between mitochondrial DNA variation and HIV-associated sensory neuropathy in CHARTER. CHARTER is a U.S. based longitudinal observational study of HIV-infected adults who underwent a structured interview and standardized examination. HIV-associated sensory neuropathy was determined by trained examiners as ≥1 sign (diminished vibratory and sharp-dull discrimination or ankle reflexes) bilaterally. Mitochondrial DNA sequencing was performed and haplogroups were assigned by published algorithms. Multivariable logistic regression of associations between mitochondrial DNA SNPs, haplogroups and HIV-associated sensory neuropathy were performed. In analyses of associations of each mitochondrial DNA SNP with HIV-associated sensory neuropathy, the two most significant SNPs were at positions A12810G (odds ratio [95% confidence interval] = 0.27 [0.11-0.65]; p = 0.004) and T489C (odds ratio [95% confidence interval] = 0.41 [0.21-0.80]; p = 0.009). These synonymous changes are known to define African haplogroup L1c and European haplogroup J, respectively. Both haplogroups are associated with decreased prevalence of HIV-associated sensory neuropathy compared with all other haplogroups (odds ratio [95% confidence interval] = 0.29 [0.12-0.71]; p = 0.007 and odds ratio [95% confidence interval] = 0.42 [0.18-1.0]; p = 0.05, respectively). In conclusion, in this cohort of mostly combination antiretroviral therapy-treated subjects, two common mitochondrial DNA SNPs and their corresponding haplogroups were associated with a markedly decreased prevalence of HIV-associated sensory neuropathy.
Keywords: genetics, mitochondria, HIV-related neurological diseases, peripheral neuropathy
Introduction
Morbidity and mortality from HIV infection have been reduced by potent combination antiretroviral therapy (CART). Though the incidence of many neurologic complications of HIV has declined (Sacktor, 2002), peripheral nerve disorders remain common (R. Ellis, 2010). HIV is associated with a predominantly sensory polyneuropathy that is attributed to HIV infection itself, or a toxic neuropathy associated with specific dideoxynucleoside analogue reverse transcriptase inhibitors (“D-drugs,” principally stavudine or didanosine). Together these conditions are designated HIV-associated sensory neuropathy (HIV-SN). HIV-SN adversely affects quality of life, including sleep and diverse aspects of physical and emotional functioning (R. J. Ellis et al., 2010). Spontaneous pain is common, and this pain often does not respond fully to analgesic medications (Verma et al., 2004). Additional symptoms include paresthesias and sensory loss. Although the recognition that D-drug therapy can be neurotoxic has led providers in developed countries to substitute alternatives, stavudine remains a commonly used component of fixed-dose generic drugs being used in the worldwide initiative to treat HIV. Thus, the study of HIV-SN susceptibility remains relevant.
HIV-SN caused by HIV infection or D-drugs are clinically similar, making them difficult to differentiate in individual patients. Although the pathophysiology of HIV-SN is not yet clearly defined, abnormal mitochondrial structure and mitochondrial DNA (mtDNA) depletion have been seen in nerve specimens of the HIV-SN model in Simian immunodeficiency virus (SIV)-infected macaques who have not received D-drugs (Lehmann et al., 2011), and in specimens from patients with D-drug therapy-associated peripheral neuropathy (Dalakas et al., 2001). D-drug-containing CART has known adverse effects on mitochondria (Brinkman et al., 1999). Peripheral neuropathies are also common findings in inherited mtDNA diseases (DiDonato S., 2009). Mitochondrial DNA is distinct from the nuclear genome, encodes 13 subunits of the electron transport chain, and exhibits abundant genetic variation across its >16,000 base pairs. Human mtDNA sequences have diverged over approximately the last 150,000 years, resulting in distinct patterns of single nucleotide polymorphisms (SNPs), called haplogroups. In addition to their role in cellular energy production, mitochondria also are involved in free radical generation and apoptosis. MtDNA variation may lead to distinctive mitochondrial electron transport chains, each with perhaps different capacities for energy production, free radical generation and apoptosis (D. C. Wallace et al., 1999). There is epidemiological evidence for differences among mtDNA haplogroups in studies of longevity (Niemi et al., 2003) and neurodegenerative disorders (Van Der Walt et al., 2003), and recent in vitro studies have demonstrated different oxidative phosphorylation capacities by haplogroup (Gomez-Duran et al., 2010).
Susceptibility to HIV-SN varies, with prevalence rates among HIV-affected persons reported to range from 10–35% (Lehmann et al., 2011). This variability suggests a role for human genomic variation. Similarities between the clinical manifestations of inherited mtDNA diseases and NRTI toxicities have prompted us to look for mtDNA variations that may explain susceptibility to peripheral neuropathy among HIV-infected persons. Previously, we examined treatment-naïve participants in a clinical trial (AIDS Clinical Trials Group [ACTG] study 384) of CART (including D-drugs), and identified associations between peripheral neuropathy and specific European (Hulgan et al., 2005) and African (Canter et al., 2010) mitochondrial haplogroups. We report here the results of an analysis of full mtDNA sequence from a subgroup of participants enrolled in CNS HIV Antiretroviral Therapy Effects Research (CHARTER), a multicenter observational study of the neurologic effects of HIV in the CART era. CHARTER included persons both on and off CART and utilized more sensitive and specific methods for characterizing HIV-SN than the ACTG study. Because HIV-SN even in the absence of D-drug therapy may be the result of mitochondrial dysfunction (Lehmann et al., 2011), our hypothesis was that variation in mtDNA from these subjects would also be associated with susceptibility to HIV-SN.
Materials and Methods
Study population
The CHARTER Study is a prospective, observational study conducted at six U.S. locations: The Johns Hopkins University, Baltimore, Maryland; Mount Sinai School of Medicine, New York, New York; University of California, San Diego; The University of Texas Medical Branch, Galveston; University of Washington, Seattle; and Washington University, St Louis, Missouri. Institutional review boards at each site approved this research, and each participant gave informed consent. Data were collected according to a protocol of comprehensive neuromedical, neurobehavioral, and laboratory assessments that were standardized across sites. These data reported herein are from a genetics sub-study within CHARTER.
Phenotype definitions
Physicians and nurses trained in neurological AIDS disorders performed a standardized, targeted examination to evaluate HIV-SN signs, including diminished ability to recognize vibration and reduced sharp-dull discrimination in the feet and toes or reduced ankle reflexes. For these analyses, the presence of ≥one bilateral and symmetric sign was considered to be evidence of HIV-SN. Neuropathy symptoms also were assessed in the legs, feet, and toes and included bilateral neuropathic pain and dysesthesias (burning, aching, or shooting), paresthesias, and loss of sensation. Using a standardized form and a structured interview, clinicians also classified neuropathic pain into the following five severity levels that were used for secondary analyses: none, slight (occasional, fleeting), mild (frequent), moderate (frequent, disabling), and severe (constant, daily, disabling, requiring analgesic medication or other treatment).
Mitochondrial DNA Isolation and Sequencing
Isolation of DNA from whole blood samples was performed using PUREGENE (Gentra Systems Inc., Minneapolis, MN, USA). Full mtDNA sequencing was performed using the GeneChipR Human Mitochondrial Resequencing Array v2.0 (Affymetrix, Inc., Santa Clara, CA, USA). From full-sequence data, mtDNA variants were identified using the revised Cambridge Reference Sequence (rCRS) (S. Anderson et al., 1981). Haplogroups were assigned using a published method (Herrnstadt et al., 2002).
Many of the same subjects included in these analyses also underwent nuclear genotyping for ongoing genome-wide association studies using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc., Santa Clara, CA, USA). To adjust for population substructure, we excluded 17 participants without nuclear genotype data. Quality control filters were applied sequentially to the remaining 549 samples using the PLatform for the Analysis, Translation, and Organization of large scale data (PLATO) (Grady et al., 2010). Samples or SNPs that did not reach the following thresholds were excluded from analyses: 95% SNP genotyping efficiency, 95% sample genotyping efficiency, and minor allele frequency of at least 1%.
Statistical Analysis
Univariable logistic regression was used to calculate unadjusted odds ratios (OR) and 95% confidence intervals (CI) to assess the association of clinical and demographic variables with HIV-SN status. Adjusted ORs were calculated by performing multivariable logistic regression with a model that included all variables selected via backward-elimination step-wise regression (variables with p-value >0.1 were removed from the model in a step-wise fashion). The first four components of a multidimensional scaling (MDS) analysis were generated using genome-wide nuclear genotype data, which were added to the model as continuous covariates in our regression model to control for potential population stratification. Briefly, MDS is a method similar to principal components analysis that takes high-dimensional data and reduces its dimensionality while preserving the overall genetic similarity. Using a defined number of indices generated from an MDS analysis in regression analysis is a common way of controlling for confounding due to population substructure and has previous been described in detail (Purcell et al., 2007).
We performed multivariable logistic regression to test the association of each mtDNA SNP with HIV-SN status while controlling for the following variables: age, D-drug therapy, nadir CD4 count, and population substructure via the MDS components. We also performed multivariable logistic regression to assess the association of each mtDNA haplogroup with HIV-SN and neuropathic pain adjusting for the same variables as the SNP association analysis. PLINK was used to perform MDS analysis and multivariable logistic regression for the mtDNA SNP association analysis (Purcell et al., 2007). STATA (Stata, Inc.; College Station, TX) was used to perform the univariable logistic regression, stepwise logistic regression, and multivariable logistic regression for the mitochondrial haplogroup association analyses.
Results
The mean age of the population was 44 years, 21% were female, and 44% self-reported non-Hispanic white ethnicity (Table 1). At cohort entry, median CD4 cell count was 435 cells/mm3, nadir CD4 cell count was 175 cells/mm3, and plasma HIV-1 RNA was 1.9 log10 copies/mL, 403 (73%) were receiving CART at the time of cohort entry, and 299 (55%) had previous or current exposure to D-drug therapy. A total of 332 (59%) subjects met the definition of HIV-SN; these were older, had a lower CD4 nadir, and were more likely to have been previously or currently exposed to D-drug therapy (p<0.001 for all; Table 1). Individuals with HIV-SN had lower HIV-1 RNA levels (p=0.001) and were more likely to have hepatitis C infection (p=0.01).
Table 1.
Demographic and clinical characteristics of CHARTER subjects included in this analysis, total and by HIV-SN status. HIV-SN is defined as the presence of at least one sign of HIV-SN. No HIV-SN is defined as no signs. Adjusted odds ratio is calculated from a multivariable logistic regression model that includes age, nadir CD4 count, and D-drug therapy.
| Total (N=549) |
HIV-SN (N=323) |
No HIV-SN (N=226) |
Unadjusted | Adjusted | |||
|---|---|---|---|---|---|---|---|
| OR (95%CI) | P | OR (95%CI) | p | ||||
| Age, mean (sd), years | 44 (8.5) | 45 (8.1) | 40 (7.8) | 1.10 (1.07-1.12) | < 0.001* | 1.09 (1.06-1.12) | < 0.001 |
| Female sex, n(%) | 116 (21.1) | 63 (19.5) | 53 (23.4) | 0.79 (0.52-1.20) | 0.27 | ||
| Ethnicity, n(%)a White Black Hispanic Other |
242 (44.1) 238 (43.4) 57 (10.4) 12 (2.2) |
142 (44.0) 156 (45.2) 29 (9.0) 4 (1.2) |
100 (44.2) 90 (39.8) 28 (12.4) 8 (3.5) |
0.99 (0.70-1.39) 1.28 (0.90-1.80) 0.70 (0.40-1.21) 0.34 (0.10-1.15) |
0.95 0.16 0.20 0.08 |
||
| Nadir CD4, median(IQR), cells/µL |
175 (52-300) |
109 (30-240) |
239 (120-388) |
0.996 (0.995-0.998) |
<0.001* | 0.997 (0.996-0.999) |
< 0.001 |
| Current CART use, n(%) Yes No |
403 (73.4) 146 (26.6) |
270 (83.6) 53 (16.4) |
133 (58.8) 93 (41.2) |
3.6 (2.4-5.3) |
<0.001 |
||
| D-drug therapy, n(%) Ever Never |
299 (54.5) 250 (45.5) |
216 (66.9) 107 (33.1) |
83 (36.7) 143 (63.3) |
3.5 (2.4-5.0) |
<0.001* |
2.2 (1.4-3.2) |
< 0.001 |
| Plasma HIV- RNA, median(IQR), log10 copies/mL |
1.9 (1.7-3.8) |
1.7 (1.7-3.3) |
2.6 (1.7-4.2) |
0.79 (0.69-0.90) |
0.001 |
||
| HCV status, n(%) Positive Negative Missing |
132 (24.0) 408 (74.3) 9 (1.6) |
90 (28.3) 228 (71.7) 5 (1.5) |
42 (18.9) 180 (81.1) 4 (1.8) |
1.7 (1.1-2.6) |
0.01 |
||
Indicates that variables remained significant after backward-elimination step-wise regression at p<0.1
Self-identified ethnicity
A total of 429 out of 679 mitochondrial SNPs from 549 subjects remained after quality control filtering. The top 20 results for the mitochondrial SNP association analyses are shown in Fig. 1. Note the effect sizes in the Fig. 1 are presented as beta coefficients, not odds ratios. Twenty-six of the 429 SNPs (6%) were significantly associated with HIV-SN status at p ≤ 0.05. The two most strongly associated SNPs were at positions A12810G (adjusted odds ratio [aOR]=0.27; 95% CI=0.11-0.65; p=0.004) and T489C (aOR=0.41; 0.21-0.80; p=0.009), representing synonymous changes in the ND5 gene and D-loop of the mitochondrial genome, respectively. These SNPs are also included in African haplogroup L1c and European haplogroup J definitions, respectively. Many of the other significant SNPs also defined major haplogroups, thus were highly correlated with one another.
Fig. 1.
Results for association of mitochondrial SNPs with HIV-SN using multivariable logistic regression analyses adjusting for age, D-drug therapy, nadir CD4 count, and population substructure. Each variant is denoted by the base pair position and the base pair change. The haplogroup(s) with which these variants are associated are shown in parentheses. The top row displays the –log10(p-value) with a triangle that denotes the direction of effect. The second row displays the log odds and corresponding 95% confidence interval. The third row displays the minor allele frequency (MAF) of each variant.
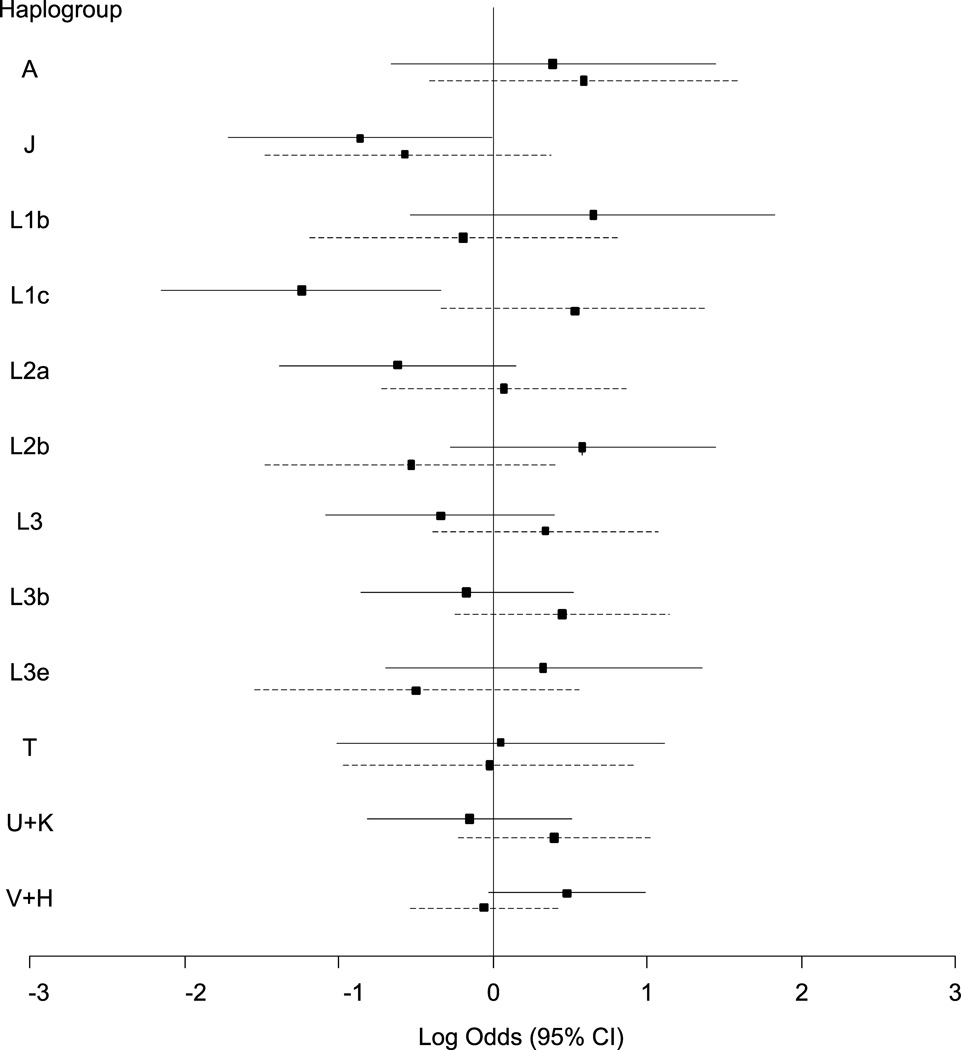
Haplogroup distribution of the population is shown in Table 2. The results from haplogroup association analyses are shown in Fig. 2. First, we used multivariable logistic regression to test for an association between mitochondrial haplogroup and HIV-SN while controlling for age, D-drug therapy, nadir CD4 count, and population stratification. Two haplogroups were significantly associated with HIV-SN status at p ≤ 0.05: haplogroups L1c (aOR=0.29; 0.12-0.71; p=0.007) and J (aOR=0.42; 95% CI=0.18-1.00; p=0.05). We also used multivariable logistic regression to test for an association with the presence of neuropathic pain controlling for the same variables as the HIV-SN analysis. None of the haplogroups were significantly associated with neuropathic pain.
Table 2.
Distribution of haplogroups among study subjects.
| Haplogroup | Total (N=549), n(%) |
Cases (N=323), n(%) |
Controls (N=226), n(%) |
|---|---|---|---|
| A | 24 (4.4) | 15 (4.6) | 9 (4.0) |
| J | 30 (5.5) | 12 (3.7) | 18 (8.0) |
| L1b | 23 (4.2) | 18 (5.6) | 5 (2.2) |
| L1c | 27 (4.9) | 11 (3.4) | 16 (7.1) |
| L2a | 42 (7.6) | 24 (7.4) | 18 (8.0) |
| L2b | 35 (6.4) | 24 (7.4) | 11 (4.9) |
| L3 | 39 (7.1) | 20 (6.2) | 19 (8.4) |
| L3b | 49 (8.9) | 29 (9.0) | 20 (8.8) |
| L3e | 24 (4.4) | 18 (5.6) | 6 (2.6) |
| T | 21 (3.8) | 14 (4.3) | 7 (3.1) |
| U+K | 58 (10.6) | 32 (9.9) | 26 (11.5) |
| V+H | 142 (25.9) | 85 (26.3) | 57 (25.2) |
| Other | 34 (6.2) | 21 (6.5) | 13 (5.8) |
Fig. 2.
Results for association of mitochondrial haplogroups with HIV-SN (solid line) and neuropathic pain (dashed line) using multivariable logistic regression analyses adjusting for age, D-drug therapy, nadir CD4 count, and population substructure.
To ensure that the exclusion of the participants who did not have nuclear genotype data did not significantly impact our results, we performed additional analyses that included these 17 individuals and adjusted for ethnicity as a categorical variable. The results were similar to those for the MDS component adjusted analysis (L1c: aOR=0.37; 95% CI=0.16-0.88; p=0.02 and J: aOR=0.39; 95% CI=0.17-0.91; p=0.03). For the mitochondrial SNP analyses, the most significant SNPs at base positions A12180G and T489C also remained statistically significant with effects in the same direction and of similar magnitudes
Discussion
We report associations between mtDNA SNPs and HIV-SN in a large subgroup of CHARTER study participants. Many of these SNPs define mtDNA haplogroups in persons of either European or African descent. Not unexpectedly, we also identified associations between these relatively common haplogroups and HIV-SN. The associations were independent of other predictors of HIV-SN, including age, CD4 cell nadir, and D-drug therapy. Although mechanisms underlying these associations are unknown, recent data suggests that rates of oxidative phosphorylation differentiate haplogroups (Gomez-Duran et al., 2010). In addition, a primate model of SIV-associated peripheral neuropathy has affirmed a primary role of mitochondrial dysfunction in this process (Lehmann et al., 2011). Together with the adverse effects on peripheral nerves of D-drugs that are toxic to mitochondria, these data provide indirect biologic evidence for differences in HIV-SN susceptibility conferred by mtDNA variants within populations.
The critical and complex roles of mitochondria in energy production, reactive oxygen species homeostasis, and apoptotic regulation make them key mediators of cellular damage in response to environmental stresses. Mitochondrial DNA variants, including those represented as haplogroups, are associated with metabolic and neurodegenerative diseases, and likely influence uccessful aging (D. C. Wallace, 2005). The prominence of neurodegeneration, including peripheral neuropathies, in mtDNA diseases also provides support for the importance of mtDNA variation in HIV-SN. A growing number of studies have reported associations between mtDNA haplogroups and HIV- or ART-associated outcomes, including peripheral neuropathy (Canter et al., 2010; Hulgan et al., 2005), neuroretinal disease (Hendrickson et al., 2010), metabolic derangements (Hendrickson et al., 2009; Hulgan et al., 2011, 2008), and AIDS outcomes (Hendrickson et al., 2008).
A previous study found an association between haplogroup L1c and increased risk for incident symptoms and/or signs of neuropathy (Canter et al., 2010), an association opposite our finding in the CHARTER study. There are several possible explanations for this difference. The previous analysis included treatment-naïve subjects initiating CART as part of a clinical trial, ACTG 384. Peripheral neuropathy was identified using screening questions and a brief neuropathy screening examination, with self-reported symptoms or a new clinical diagnosis of neuropathy without documented signs included as a neuropathy case. CHARTER diagnosed HIV-SN in persons with ≥1 symmetric sign, irrespective of neuropathic pain or other symptoms. Thus the discrepancy in results may result from inclusion of different neuropathic phenotypes as “cases”. Indeed, substantial discordance between neuropathic symptoms and signs has been recognized in the CHARTER cohort (Robinson-Papp et al., 2010). In an attempt to address this possibility, we also examined the association of African mtDNA haplogroup L1c with neuropathic pain in CHARTER. While L1c was not significantly associated with neuropathic pain, the point estimate odds ratio was in the same direction as that from the ACTG analysis (OR=1.71, 95% CI=0.73-4.00, p=0.22; Fig. 2). We also calculated the concordance of HIV-SN signs and neuropathic symptoms in our subgroup: 61% of CHARTER subjects in this analysis with at least one sign of HIV-SN had no neuropathic pain and 24% of individuals with neuropathic pain had no HIV-SN signs.
Another possible explanation for the results from these two studies is differences in the study populations. Whereas the ACTG analysis included CART-naïve subjects with relatively short-term (median follow-up ~3 years) CART exposure, CHARTER includes a more heterogeneous population of persons with longer durations of HIV-infection, more extensive and varied CART regimens (including untreated subjects), and longer cumulative CART exposure among treated subjects (median [IQR]=10 years [8-25]).
Two independent studies have now identified the L1c African haplogroup as influencing HIV-associated and/or toxic peripheral neuropathy. Taken together, these findings suggest that a) underlying genomic risk factors for signs of HIV-SN and neuropathic symptoms may differ, b) the population studied may substantially influence observed associations, and c) additional study in independent cohorts with uniform populations and phenotyping, and/or replication using a validation sample from within CHARTER are needed to clarify these findings. To date, no studies have linked haplogroup L1c to other disease phenotypes in HIV-infected or uninfected populations.
In our analysis, haplogroup J was modestly associated with a decreased prevalence of HIV-SN. Previous studies have found haplogroup J and multi-haplogroup categories that include J to be associated with decreased prevalence of Parkinson’s disease (Van Der Walt et al., 2003), neuroretinal disorder in HIV patients (Hendrickson et al., 2010), and acquired demyelinating syndrome (Venkateswaran et al., 2011). Conversely, findings from other studies suggest that haplogroup J is a risk factor for or not associated with these and other neurologic disorders (Jones et al., 2007; Kalman et al., 1999; Mehta et al., 2009; Reynier et al., 1999; Ross et al., 2003). In particular, several studies have shown that an increased risk of Leber’s hereditary optic neuropathy (LHON) is associated with mtDNA mutations or chemical toxins in persons with haplogroup J (Brown et al., 2002, 1997; Shafa Shariat et al., 2006; Torroni et al., 1997). Haplogroup J has been associated with with increased longevity in some populations, but not in others (DeBenedictis et al., 1999; Niemi et al., 2003; Shlush et al., 2008). Together, these findings suggest that the role of haplogroup J in disease risk is complex and dependent on interactions with other factors, such as mtDNA mutations, nuclear genome variation, and the environment.
Several statistically significant associations were with non-synonymous (NS) mitochondrial SNPs. These are of particular interest because the resulting amino acid change could potentially alter protein function. The NS SNP G10398A has previously been associated with other human traits (Ruiz-Pesini et al., 2007). This SNP results in an alanine to threonine amino acid change in the NADH dehydrogenase subunit 3 of Complex I. In our study, the A allele was considered the minor allele with a relatively high frequency of 44%. The A allele is actually more common in whites (79%), Hispanics (62%), and other (75%) racial/ethnic groups and less common only among blacks (4%). Interestingly, associations with different alleles of this SNP have been inconsistent. The A allele has been associated with increased risk for breast cancer and more invasive cancer-cell phenotype (Canter et al., 2005), lower pH levels in the mitochondrial matrix (Kazuno et al., 2006), and increased risk of Type II diabetes (Rai et al., 2007). The G allele has been associated with increased risk for oral (Datta et al., 2007) and breast cancer (Pezzotti et al., 2009), metabolic syndrome (Juo et al., 2010), longevity (Niemi et al., 2005), and decreased risk of Parkinson’s disease (Van Der Walt et al., 2003). Our results show an association with increased risk for HIV-SN for 10398A (aOR=2.01; 95% CI=1.16-3.49; p=0.01). Because the frequency of this allele is highly variable across racial/ethnic groups, different results could be due to population differences, variable methods used to control for population substructure (e.g. stratification or adjustment), and/or interactions with other genetic factors. For example, nuclear genetic variants in cytokine (Cherry et al., 2008) and iron-metabolism genes (Kallianpur et al., 2006) have been shown to be associated with HIV-SN prevalence in previous analysis. Future studies should explore the possibility of nuclear-mitochondrial interactions playing a role in HIV-SN susceptibility.
Many of the significant SNPs in our analysis were highly correlated with one another and defined the same haplogroups, as shown by the same or similar allele frequencies and effect sizes in Fig. 1. These correlated SNPs may all be defining a “functional” variant that is driving the observed association. Pinpointing the causative variant will require molecular experiments to delineate the functional effects of individual SNPs.
A potential limitation of these analyses is the relatively small sample size. Although this study represents the largest analysis of full mtDNA sequence in an HIV-infected population to date, several subhaplogroups were of inadequate size to reliably identify associations. Because of the sample size and exploratory nature of these analyses, we did not correct for multiple comparisons. A conservative Bonferroni correction for the haplogroup analysis would have resulted in p-values greater than the 0.05 cut-off with the L1c association trending toward significance (p= 0.08). Nonetheless, our findings are notable and potentially important due to the repeat finding of a haplogroup L1c association. Due to incomplete overlap with genome-wide nuclear DNA data, we were unable to identify ancestry-derived ethnicity variables for all persons with mtDNA sequence. Sensitivity analyses including all persons and using other methods for adjustment for ethnicity did not substantially affect results. We also performed haplogroup analyses classifying HIV-SN by the presence of ≥two signs. Although no longer statistically significant (perhaps due to smaller case group sample sizes), the point estimate ORs for haplogroups L1c and J were in the same direction (aOR=0.66; 95% CI=0.24-1.83; p=0.43 and aOR=0.47; 95% CI=0.16-1.34; p=0.16, respectively).).
In summary, full mtDNA sequence data identified several mtDNA SNPs and haplogroups associated with HIV-SN in this well-characterized neuroAIDS cohort. HIV-SN was more frequent in persons from both non-Hispanic black and white populations with relatively common mtDNA SNPs and African and European haplogroups. The African L1c haplogroup was again identified as a susceptibility factor in a HIV-SN, although the direction of the association differed from that seen in a clinical trial of CART initiation. Importantly, these associations must be validated statistically in independent data sets and biologically using in vitro and in vivo study designs. Specifically, these results also highlight the need for replication of studies in comparable populations, ideally using similar study designs, and for mechanistic studies, particularly those focused on functional effects of mtDNA SNPs within the L1c haplogroup.
Acknowledgements
The CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) group is affiliated with the Johns Hopkins University, Mount Sinai School of Medicine, University of California, San Diego, University of Texas, Galveston, University of Washington, Seattle, Washington University, St. Louis and is headquartered at the University of California, San Diego and includes: Director: Igor Grant, M.D.; Co-Directors: Scott L. Letendre, M.D., Ronald J. Ellis, M.D., Ph.D., Thomas D. Marcotte, Ph.D.; Center Manager: Donald Franklin, Jr.; Neuromedical Component: Ronald J. Ellis, M.D., Ph.D. (P.I.), J. Allen McCutchan, M.D., Terry Alexander, R.N.; Laboratory, Pharmacology and Immunology Component: Scott Letendre, M.D. (P.I.), Edmund Capparelli, Pharm.D.; Neurobehavioral Component: Robert K. Heaton, Ph.D. (P.I.), J. Hampton Atkinson, M.D., Steven Paul Woods, Psy.D., Matthew Dawson; Virology Component: Davey M. Smith, M.D. (P.I.), Joseph K. Wong, M.D.; Imaging Component: Christine Fennema-Notestine, Ph.D. (Co-P.I.), Michael J. Taylor, Ph.D. (Co-P.I.), Rebecca Theilmann, Ph.D.; Data Management Component: Anthony C. Gamst, Ph.D. (P.I.), Clint Cushman,; Statistics Component: Ian Abramson, Ph.D. (P.I.), Florin Vaida, Ph.D.; Johns Hopkins University Site: Justin McArthur (P.I.), Mary Smith; Mount Sinai School of Medicine Site: Susan Morgello, M.D. (Co-P.I.) and David Simpson, M.D. (Co-P.I.), Letty Mintz, N.P.; University of California, San Diego Site: J. Allen McCutchan, M.D. (P.I.), Will Toperoff, N.P..; University of Washington, Seattle Site: Ann Collier, M.D. (Co-P.I.) and Christina Marra, M.D. (Co-P.I.), Trudy Jones, M.N., A.R.N.P.; University of Texas, Galveston Site: Benjamin Gelman, M.D., Ph.D. (P.I.), Eleanor Head, R.N., B.S.N.; and Washington University, St. Louis Site: David Clifford, M.D. (P.I.), Muhammad Al-Lozi, M.D., Mengesha Teshome, M.D.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the United States Government.
Funding:
The CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) study is supported by awards [N01 MH22005, HHSN271201000027C, and HHSN271201000030C] from the National Institutes of Health.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Brinkman K, Smeitink JA, Romijn JA, Reiss P. Mitochondrial toxicity induced by nucleoside-analogue reversetranscriptase inhibitors is a key factor in the pathogenesis of antiretroviral-therapy-related lipodystrophy. Lancet. 1999;354:1112–1115. doi: 10.1016/S0140-6736(99)06102-4. [DOI] [PubMed] [Google Scholar]
- Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, Mikhailovskaya IE, Sukernik RI, Wallace DC. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup. J. Hum Genet. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- Brown MD, Sun F, Wallace DC. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am J Hum Genet. 1997;60:381–387. [PMC free article] [PubMed] [Google Scholar]
- Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- Canter JA, Robbins GK, Selph D, Clifford DB, Kallianpur AR, Shafer R, Levy S, Murdock DG, Ritchie MD, Haas DW, Hulgan T. African mitochondrial DNA subhaplogroups and peripheral neuropathy during antiretroviral therapy. J Infect.Dis. 2010;201:1703–1707. doi: 10.1086/652419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry CL, Rosenow A, Affandi JS, McArthur Justin C, Wesselingh SL, Price P. Cytokine genotype suggests a role for inflammation in nucleoside analog-associated sensory neuropathy (NRTI-SN) and predicts an individual’s NRTI-SN risk. AIDS Res. Hum. Retroviruses. 2008;24:117–123. doi: 10.1089/aid.2007.0168. [DOI] [PubMed] [Google Scholar]
- Dalakas MC, Semino-Mora C, Leon-Monzon M. Mitochondrial alterations with mitochondrial DNA depletion in the nerves of AIDS patients with peripheral neuropathy induced by 2’3’-dideoxycytidine (ddC) Lab Invest. 2001;81:1537–1544. doi: 10.1038/labinvest.3780367. [DOI] [PubMed] [Google Scholar]
- Datta S, Majumder M, Biswas NK, Sikdar N, Roy B. Increased risk of oral cancer in relation to common Indian mitochondrial polymorphisms and Autosomal GSTP1 locus. Cancer. 2007;110:1991–1999. doi: 10.1002/cncr.23016. [DOI] [PubMed] [Google Scholar]
- DeBenedictis G, Rose G, Carrieri G, De LM, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- Van Der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Goetz CG, Small GW, Mastaglia F, Stajich JM, McLaurin AC, Middleton LT, Scott BL, Schmechel DE, Pericak-Vance MA, Vance JM. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato S. Multisystem manifestations of mitochondrial disorders. J Neurol. 2009;256:693–710. doi: 10.1007/s00415-009-5028-3. [DOI] [PubMed] [Google Scholar]
- Ellis R. HIV and antiretroviral therapy: impact on the central nervous system. Prog Neurobiol. 2010:185–187. doi: 10.1016/j.pneurobio.2009.10.016. 2009/10/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Rosario D, Clifford DB, McArthur JC, Simpson D, Alexander T, Gelman BB, Vaida F, Collier A, Marra CM, Ances B, Atkinson JH, Dworkin RH, Morgello S, Grant I. Continued high prevalence and adverse clinical impact of human immunodeficiency virus-associated sensory neuropathy in the era of combination antiretroviral therapy: the CHARTER Study. Arch Neurol. 2010;67:552–558. doi: 10.1001/archneurol.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, Diez-Sanchez C, Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010:3343–3353. doi: 10.1093/hmg/ddq246. 2010/06/23. [DOI] [PubMed] [Google Scholar]
- Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S, Wallace DC, O’Brien SJ. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson SL, Jabs DA, Van NM, Lewis RA, Wallace DC, O’Brien SJ. Mitochondrial haplogroups are associated with risk of neuroretinal disorder in HIV-positive patients. J Acquir.Immune.Defic.Syndr. 2010;53:451–455. doi: 10.1097/QAI.0b013e3181cb8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, Palella FJ, Bream JH, Wallace DC, O’Brien SJ. Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. J Acquir.Immune.Defic.Syndr. 2009;51:111–116. doi: 10.1097/QAI.0b013e3181a324d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulgan T, Haas DW, Haines JL, Ritchie MD, Robbins GK, Shafer RW, Clifford DB, Kallianpur AR, Summar M, Canter JA. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS. 2005;19:1341–1349. doi: 10.1097/01.aids.0000180786.02930.a1. [DOI] [PubMed] [Google Scholar]
- Hulgan T, Haubrich R, Riddler SA, Tebas P, Ritchie MD, McComsey GA, Haas DW, Canter JA. European mitochondrial DNA haplogroups and metabolic changes during antiretroviral therapy in AIDS Clinical Trials Group Study A5142. AIDS. 2011;25:37–47. doi: 10.1097/QAD.0b013e32833f9d02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulgan T, Tebas P, Canter JA, Mulligan K, Haas DW, Dube M, Grinspoon S, Robbins GK, Motsinger AA, Kallianpur AR. Hemochromatosis gene polymorphisms, mitochondrial haplogroups, and peripheral lipoatrophy during antiretroviral therapy. J Infect.Dis. 2008;197:858–866. doi: 10.1086/528697. [DOI] [PubMed] [Google Scholar]
- Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM. Mitochondrial DNA haplogroups and age-related maculopathy. Arch Ophthalmol. 2007;125:1235–1240. doi: 10.1001/archopht.125.9.1235. [DOI] [PubMed] [Google Scholar]
- Juo SH, Lu MY, Bai RK, Liao YC, Trieu RB, Yu ML, Wong LJ. A common mitochondrial polymorphism 10398A>G is associated metabolic syndrome in a Chinese population. Mitochondrion. 2010;10:294–299. doi: 10.1016/j.mito.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Kallianpur AR, Hulgan T, Canter JA, Ritchie MD, Haines JL, Robbins GK, Shafer RW, Clifford DB, Haas DW. Hemochromatosis (HFE) gene mutations and peripheral neuropathy during antiretroviral therapy. AIDS. 2006;20:1503–1513. doi: 10.1097/01.aids.0000237366.56864.3c. [DOI] [PubMed] [Google Scholar]
- Kalman B, Li S, Chatterjee D, O’Connor J, Voehl MR, Brown MD, Alder H. Large scale screening of the mitochondrial DNA reveals no pathogenic mutations but a haplotype associated with multiple sclerosis in Caucasians. Acta Neurol.Scand. 1999;99:16–25. doi: 10.1111/j.1600-0404.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- Kazuno AA, Munakata K, Nagai T, Shimozono S, Tanaka M, Yoneda M, Kato N, Miyawaki A, Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS.Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann HC, Chen W, Borzan J, Mankowski JL, Hoke A. Mitochondrial dysfunction in distal axons contributes to human immunodeficiency virus sensory neuropathy. Ann Neurol. 2011:100–110. doi: 10.1002/ana.22150. 2011/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Mellick GD, Rowe DB, Halliday GM, Jones MM, Manwaring N, Vandebona H, Silburn PA, Wang JJ, Mitchell P, Sue CM. Mitochondrial DNA haplogroups J and K are not protective for Parkinson’s disease in the Australian community. Mov Disord. 2009;24:290–292. doi: 10.1002/mds.22389. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Moilanen JS, Tanaka M, Hervonen A, Hurme M, Lehtimaki T, Arai Y, Hirose N, Majamaa K. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet. 2005;13:166–170. doi: 10.1038/sj.ejhg.5201308. [DOI] [PubMed] [Google Scholar]
- Pezzotti A, Kraft P, Hankinson SE, Hunter DJ, Buring J, Cox DG. The mitochondrial A10398G polymorphism, interaction with alcohol consumption, and breast cancer risk. PLoS.One. 2009;4:e5356. doi: 10.1371/journal.pone.0005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai E, Sharma S, Koul A, Bhat AK, Bhanwer AJ, Bamezai RN. Interaction between the UCP2-866G/A, mtDNA 10398G/A and PGC1alpha p.Thr394Thr and p.Gly482Ser polymorphisms in type 2 diabetes susceptibility in North Indian population. Hum Genet. 2007;122:535–540. doi: 10.1007/s00439-007-0421-4. [DOI] [PubMed] [Google Scholar]
- Reynier P, Penisson-Besnier I, Moreau C, Savagner F, Vielle B, Emile J, Dubas F, Malthiery Y. mtDNA haplogroup J: a contributing factor of optic neuritis. Eur J Hum Genet. 1999;7:404–406. doi: 10.1038/sj.ejhg.5200293. [DOI] [PubMed] [Google Scholar]
- Robinson-Papp J, Morgello S, Vaida F, Fitzsimons C, Simpson DM, Elliott KJ, Al-Lozi M, Gelman BB, Clifford D, Marra CM, McCutchan JA, Atkinson JH, Dworkin RH, Grant I, Ellis R. Association of self-reported painful symptoms with clinical and neurophysiologic signs in HIV-associated sensory neuropathy. Pain. 2010;151:732–736. doi: 10.1016/j.pain.2010.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross OA, McCormack R, Maxwell LD, Duguid RA, Quinn DJ, Barnett YA, Rea IM, El-Agnaf OM, Gibson JM, Wallace A, Middleton D, Curran MD. mt4216C variant in linkage with the mtDNA TJ cluster may confer a susceptibility to mitochondrial dysfunction resulting in an increased risk of Parkinson’s disease in the Irish. Exp Gerontol. 2003;38:397–405. doi: 10.1016/s0531-5565(02)00266-8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor N. The epidemiology of human immunodeficiency virus-associated neurological disease in the era of highly active antiretroviral therapy. J Neurovirol. 2002;8(Suppl 2):115–121. doi: 10.1080/13550280290101094. [DOI] [PubMed] [Google Scholar]
- Shafa Shariat PM, Houshmand M, Tabassi AR. Mitochondrial D-loop variation in leber hereditary neuropathy patients harboring primary G11778A, G3460A, T14484C mutations: J and W haplogroups as high-risk factors. Arch Med Res. 2006;37:1028–1033. doi: 10.1016/j.arcmed.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Atzmon G, Weisshof R, Behar D, Yudkovsky G, Barzilai N, Skorecki K. Ashkenazi Jewish centenarians do not demonstrate enrichment in mitochondrial haplogroup. J. PLoS.One. 2008;3:e3425. doi: 10.1371/journal.pone.0003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D’Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De NA, Scozzari R. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran S, Zheng K, Sacchetti M, Gagne D, Arnold DL, Sadovnick AD, Scherer SW, Banwell B, Bar-Or A, Simon DK. Mitochondrial DNA haplogroups and mutations in children with acquired central demyelination. Neurology. 2011;76:774–780. doi: 10.1212/WNL.0b013e31820ee1bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Estanislao L, Mintz L, Simpson D. Controlling neuropathic pain in HIV. Curr HIV/AIDS Rep. 2004:136–141. doi: 10.1007/s11904-004-0020-0. 2005/08/11. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005:359–407. doi: 10.1146/annurev.genet.39.110304.095751. 2005/11/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]