Abstract
Facile, two-step synthesis and kinetic characterization of new chemical probes for selective labeling of sulfenic acid (–SOH) in proteins are presented. The synthesis route relies on the simple and highly efficient Michael addition of thiol containing tags or linkers to 4-cyclopentene-1,3-dione, the unsaturated derivative of 1,3-cyclopentanedione.
Cysteine oxidative modification to sulfenic acid (–SOH), by various types of reactive oxygen species (ROS) (e.g. H2O2), has emerged as an important post-translational modification in proteins and was shown to play a pivotal role in regulating protein functions under both physiological and oxidative stress conditions.1–2 During the modification process redox sensitive cysteines are first oxidized to –SOH as an initially formed product. Depending on the microenvironment where the cysteine is located, this metastable intermediate is further transformed into more stable products like disulfides and sulfenamides—the sometimes cyclic condensation product of sulfenic acid. These are readily reversed to yield the reduced thiol form via the action of cellular reductants like thioredoxin, glutaredoxin and glutathione.3 Hyperoxidation to sulfinic or sulfonic acid may occur if sulfenic acids encounter high oxidant concentrations, a process which is typically irreversible in the cell. However, an exception to this is now recognized. A specialized repair protein, sulfiredoxin, can rescue at least a subset of cysteine-based peroxidases (peroxiredoxins) in which hyperoxidation to sulfinic acid occurs.4 Given the significance of cysteine oxidation in proteins, various analytical methods and probes have been developed to identify the –SOH proteins.5–10
The main strategy of detecting –SOH modified proteins relies on the reactivity of dimedone (5,5-dimethyl-1,3-cyclohexanedione) with –SOH (Scheme 1), a reaction first recognized by Allison et al. in 1974.11 Several dimedone-based derivatives have recently been synthesized to label –SOH proteins, featuring biotinylated dimedone analogues5,10,12 for affinity purification, azide “tailed” dimedone for “click” chemistry or Staudinger ligation,13 and fluorophore-linked dimedone analogues for visualization.5,14 While proven to be useful in labeling –SOH proteins, these reagents are all centered on the classic reactive group of dimedone, which imparts similar reactivity to its derivatives. One particular disadvantage of the dimedone moiety is the derivatization process which requires long chemical synthetic steps with an overall low yield.5,10,13 Alternative reagents that may allow for a more facile derivatization have not been reported. In this study we investigated whether other 1,3-diketones, like 4-cyclopentene-1,3-dione, could be used for synthesis of protein –SOH chemical probes. The envisioned advantage of the 4-cyclopentene-1,3-dione is that it could be directly conjugated to thiol-containing tags or linkers through the highly efficient Michael addition.15 Protection of the enol isomer, which is routinely employed in dimedone derivatization strategies, would not be required. Furthermore, fluorophores or enrichment tags (e.g. biotin) could be attached in two easy steps and with high yield as it will be shown below.
Scheme 1.
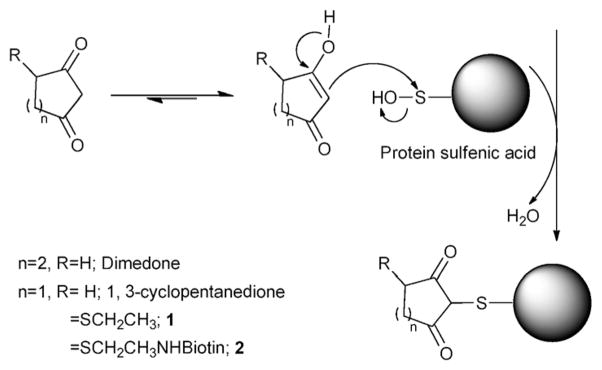
Reaction of sulfenic acid modified protein with dimedone, 1,3-cyclopentanedione and its derivatives (1 and 2).
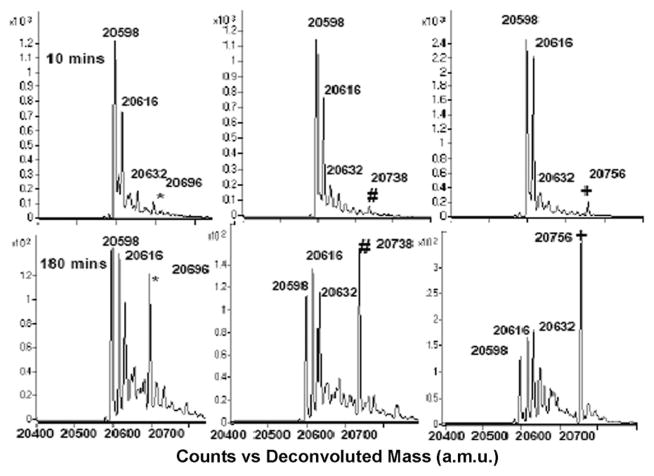
To test whether 1,3-cyclopentanedione derivatives exhibit similar reactivity and selectivity towards –SOH as dimedone, we first synthesized 4-(ethylthio)cyclopentane-1,3-dione compound (1, Scheme 1, R=SCH2CH3) following the procedure described in Supplementary Material.† The C165S mutant of AhpC was used as the prototype protein in these studies.5,9 AhpC is a cysteine-based peroxidase from bacteria known to form –SOH at the reactive C46, by H2O2 oxidation. Mutation of C165 to serine stabilizes the –SOH species at C46 and allows for quantitative analysis of covalent adduct formation with chemical probes. Labeling of C165S AhpC–SOH by 1 was monitored as a function of reaction time, and compared with dimedone and 1,3-cyclopentanedione using electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). The three major peaks observed at 20 598, 20 616, and 20 632 a.m.u. correspond to sulfenamide (likely formed in gas phase from sulfenic acid after loss of one molecule of H2O), –SOH, and –SO2H C165S AhpC species, respectively. The individual AhpC product adduct peaks at a.m.u. 20 696 (*), 20 738 (#) and 20 756 (+) increased over time and correspond to 1,3-cyclopentanedione, dimedone and 1 adducts, respectively (Fig. 1). The adduct formation was then plotted against the reaction time, as shown in Fig. 2A and Fig. S1, which in addition includes the data for AhpC sulfenamide, –SOH and –SO2H species. Data were modeled using KinTek Explorer with the following pseudo-first-order rates of adduct formation: 0.0018 min−1 (1,3-cyclopentanedione), 0.0024 min−1 (dimedone), and 0.0036 min−1 (1), indicating higher reactivity of 1 towards –SOH compared with dimedone or 1,3-cyclopentanedione (shown also in Fig. S1†).
Fig. 1.
Representative ESI-TOF MS spectra of C165S AhpC–SOH reaction with 1,3-cyclopentanedione (left), dimedone (middle), and 1 (right) (AhpC: 50 μM, labeling reagent: 5 mM; buffer: 50 mMbis-tris-citric acid pH 5.5).
Fig. 2.
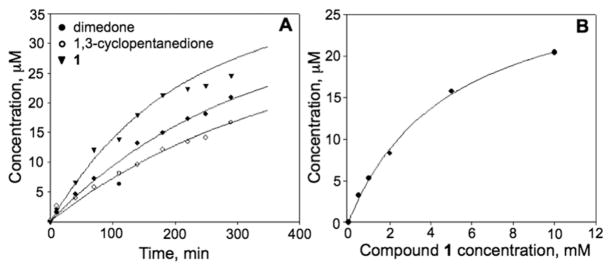
(A) Plot of adduct formation of 1,3-cyclopentanedione (open circles), dimedone (closed circles), and 1 (inverted triangle) with C165S AhpC-SOH (50 μM). The adduct concentration was calculated based on adduct abundance among the total ion abundances of the four prominent species in MS spectrum as indicated in Fig. 1. (B). Concentration dependence of AhpC–SOH labeling by 1. AhpC–SOH (50 μM) was incubated with different concentration of 1 (0, 0.5, 1, 2, 5, 10 mM) at pH 5.5 for 85 min and at rt.
Further studies were aimed at determining the dependence of the reaction rate on the concentration of 1 (Fig. 2B). Data were fit to a hyperbolic equation to obtain a K0.5 of 5 ± 0.4 mM. Formation of a covalent adduct was confirmed by nanoLC-MS/MS. A peptide containing the C46 modified by 1 (Fig. S2A†) was identified, thus confirming the covalent bond formation between the sulfur at C46 in C165S AhpC and 1.
To address the selectivity of the probe with regard to other cysteine oxidation states and amino acid side chains of proteins, the compound 1 was incubated with the reduced form (–SH) of C165S AhpC protein, the C165S AhpC–SO2H and the disulfide bond (–S–S–) linked dimer of wt AhpC, respectively, at pH 5.5 and pH 7.5 conditions. Results showed that 1 did not react with –SH, –SO2H and –S–S– forms of AhpC (Fig. S3†), and is therefore selective for the –SOH oxidation state. Since 1 was synthesized by introducing a thioether group through Michael addition which is potentially reversible, we tested the stability of the AhpC-1 adduct to other thiols. Data in Fig. S4A† show that treatment of AhpC-1 adduct with 50 mM DTT for 1 h did not alter the probe as evidenced by lack of a mass shift, consistent with previous reports.16,17 The thioether group in AhpC-1 was also resistant to oxidation with 5 and 10 mM H2O2 for 1 h (Fig. S5B†). Oxidation of thioether to sulfoxide and sulfone was observed, however, at higher concentrations of H2O2 (50 and 100 mM, 1 h) (Fig. S5B) and 17 h incubation (Fig. S5D).† These oxidized species were resistant to treatment with other thiols as well (Fig. S4B).†
Previous labeling studies using dimedone and dimedone derivatives did not show a pH dependence for Escherichia coli f RMsr.9 To determine whether this occurs with 1 as well, the reaction of 1 with C165S AhpC–SOH was monitored using buffers at pH 6.5, 5.5, 4.5 and 3.5. A strong increase of reaction rate was observed at lower pH (Fig. 3), indicating that the enol form instead of the enolate may act as the primary species reacting with the –SOH as the reaction proceeds through acid catalysis. Therefore, labeling and kinetics experiments were performed at slightly acidic pH (pH 5.5) where protein loss was limited while the reactivity of 1,3-cyclopentanedione derivatives 1 and 2 with –SOH proteins was increased relative to higher pH. It should be noted that the stock solution of 1 was made in 0.5 M Bis-Tris and DMSO (v : v = 1 : 1) to pre-buffer the acidity of 1 to around 6.5 before adding it to the reaction buffer, and final pH values of 5.5 were confirmed. Final DMSO concentration during labeling was 0.5%.
Fig. 3.
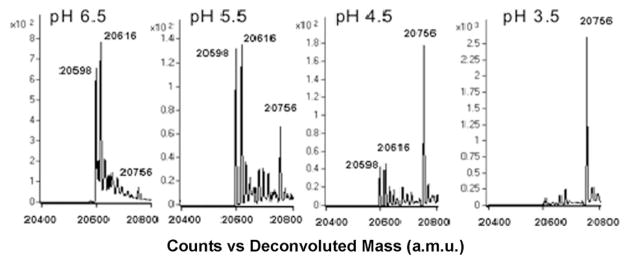
MS spectra of C165S AhpC–SOH labeling with 1 at 40 min reaction time and pH 3.5–6.5. Increased adduct formation (a.m.u. 20 756) was observed as pH was decreased from 6.5 to 3.5.
We then synthesized a biotin analogue of 1 (2, Scheme 2) as described in the Supplementary Material.† First, cysteinamine was reacted with 4-cyclopentene-1,3-dione to yield an intermediate (Int, Scheme 2) that was then conjugated with biotin– NHS to produce the biotin-tagged product, 2. The biotin analogue 2 can therefore be constructed in two simple reactions with an overall yield of 59–68%.
Scheme 2.

Synthesis of a biotin-tagged 1,3-cyclopentanedione, 2.
Its labeling of sulfenic acid proteins was also confirmed by reaction with C165S AhpC–SOH, which was nearly all converted to the adduct species within 4 h and at pH 5.5 (Fig. 4A).MS/MS analysis of the labeled protein digests confirmed that C46 was modified by 2 (Fig. S2B†). To further demonstrate the application of 2 in labeling –SOH containing proteins and profiling the –SOH proteome, we incubated lysates obtained from NIH 3T3 cells with increased concentrations of H2O2 in the presence of 2. The selectivity of 2 for –SOH was examined again by prereducing the lysates with TCEP, a disulfide reductant, which was also shown to reduce SOH to SH.18 Pre-reduced C165S AhpC was added to lysates as a positive control. Lysates were resolved by SDS-PAGE, transferred to nitrocellulose membrane and probed for labeling with 2 using streptavidin– HRP (Fig. 4B). The results showed that treatment with TCEP almost completely prevented labeling of cellular proteins with 2 (Fig. 4B, lane 2). The slight labeling in the TCEP control may have been due to the incomplete reduction of some highly abundant –SOH proteins, which were heavily labeled in the non-TCEP samples. Biotin signal intensity increased with H2O2 concentration up to 100 μM and leveled off at higher H2O2 concentrations (500 μM). Interestingly, equal labeling of C165S AhpC was observed at all H2O2 concentrations (Fig. 4B, lanes 3, 4, 5, and 6). This is likely because of the rapid disulfide bond formation between C165S AhpC and other free thiols (proteins or low molecular weight thiols like GSH). To identify some of the proteins that showed increased labeling with 2 in the presence of H2O2, the labeled lysate was incubated with streptavidin–agarose and probed for PLCγ1, a signaling protein recently identified as redox sensitive using a biotin derivative of dimedone (DCP-Bio1).2 These results show that diverse oxidation patterns exist for individual proteins depending on the sensitivity to oxidation of specific cysteine sites and reactivity of cysteine sulfenic acids with 2 or other thiols.
Fig. 4.
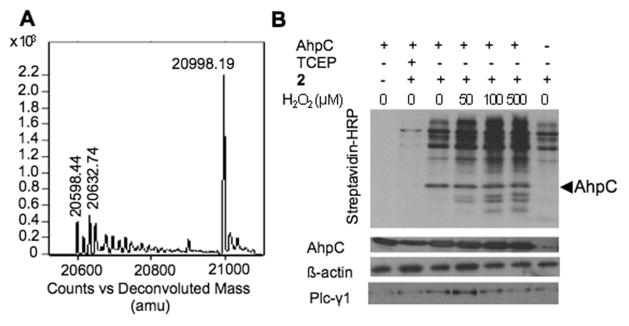
Reaction products of pure protein and cell lysates with 2. (A) ESI-TOF MS spectrum showing C165S AhpC-2 covalent adduct formation. (B) Western-blot analysis of proteins from NIH 3T3 cell lysates labeled with 2. The blot was probed with streptavidin-HRP, and antibodies for AhpC and β-actin. Samples after enrichment in proteins labeled with 2 were probed for PLCγ1 (see Supporting Material†).
In summary, we present a new and facile route for the synthesis of chemical probes to label SOH modified proteins. The comparative kinetic studies show that 1 has improved reactivity compared to dimedone. The reactivity of 1 with –SOH was enhanced under acidic conditions. Compound 2, the biotinylated derivative of 1, was easily synthesized in two steps and shown to label C165S AhpC–SOH and –SOH containing proteins in cell lysates. In this regard, we envision that labeling using this biotinylated derivative is well suited to monitor the –SOH formation in cells under oxidative conditions.
Supplementary Material
Acknowledgments
This research was supported by the NIH R01 CA136810 (CMF), and R33 CA126659/CA126659Z (LBP).
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c1cc12127h
Notes and references
- 1.Poole LB, Nelson KJ. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 4.Jonsson TJ, Tsang AW, Lowther WT, Furdui CM. J Biol Chem. 2008;283:22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Bioconjugate Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo YH, Carroll KS. Bioorg Med Chem Lett. 2009;19:356–359. doi: 10.1016/j.bmcl.2008.11.073. [DOI] [PubMed] [Google Scholar]
- 7.Saurin AT, Neubert H, Brennan JP, Eaton P. Proc Natl Acad Sci U S A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu SE, King SB, Furdui CM, Poole LB. Methods Enzymol. 2010;473:77–94. doi: 10.1016/S0076-6879(10)73003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles RL, Schroder E, May G, Free P, Gaffney PRJ, Wait R, Begum S, Heads RJ, Eaton P. Mol Cell Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Benitez LV, Allison WS. J Biol Chem. 1974;249:6234–6243. [PubMed] [Google Scholar]
- 12.Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Angew Chem, Int Ed. 2011;50:4423–4427. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- 13.Leonard SE, Reddie KG, Carroll KS. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 14.Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Bioconjugate Chem. 2005;16:1624–1628. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- 15.Qian J, Cole RB, Cai Y. J Mass Spectrom. 2011;46:1–11. doi: 10.1002/jms.1854. [DOI] [PubMed] [Google Scholar]
- 16.Lin D, Saleh S, Liebler DC. Chem Res Toxicol. 2008;21:2361–2369. doi: 10.1021/tx800248x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith MEB, Schumacher FF, Ryan CP, Tedaldi LM, Papaioannou D, Waksman G, Caddickand S, Baker JR. J Am Chem Soc. 2010;132:1960–1965. doi: 10.1021/ja908610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansuy D, Dansette PM. Arch Biochem Biophys. 2011;507:174–185. doi: 10.1016/j.abb.2010.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.