Abstract
Circadian rhythms govern a wide variety of physiological and metabolic functions in many organisms, from prokaryotes to humans. We previously reported that silent information regulator 1 (SIRT1), a NAD+-dependent deacetylase, contributes to circadian control. In addition, SIRT1 activity is regulated in a cyclic manner in virtue of the circadian oscillation of the coenzyme NAD+. Here we used specific SIRT1 activator compounds both in vitro and in vivo. We tested a variety of compounds to show that the activation of SIRT1 alters CLOCK:BMAL1-driven transcription in different systems. Activation of SIRT1 induces repression of circadian gene expression and decreases H3 K9/K14 acetylation at corresponding promoters in a time-specific manner. Specific activation of SIRT1 was demonstrated in vivo using liver-specific SIRT1-deficient mice, where the effect of SIRT1 activator compounds was shown to be dependent on SIRT1. Our findings demonstrate that SIRT1 can fine-tune circadian rhythm and pave the way to the development of pharmacological strategies to address a broad range of therapeutic indications.
Keywords: chromatin, circadian clock, metabolism, sirtuins, epigenetic
The circadian clock is a highly conserved system constituted by a network of transcriptional/translational feedback loops that modulate the expression of up to 15% of mammalian genes in each given tissue (1–3). The core transcription factors CLOCK and BMAL1 act as positive regulators of the circadian machinery, by forming heterodimers and driving transcription of several clock-controlled genes (CCGs), through direct binding to E-box DNA elements located in their promoters (4). Period (Per1, Per2, Per3) and cryptochrome genes (Cry1, Cry2) are circadian clock genes that encode proteins that translocate to the nucleus to inhibit CLOCK:BMAL1-mediated transcription. This cyclic feedback loop occurs during a period of about 24 h and controls the expression of many different genes involved in a wide array of cellular processes.
The complex program of gene expression that characterizes circadian physiology is possible through dynamic changes in chromatin transition (5). Activation of CCGs by CLOCK:BMAL1 is associated with circadian changes in histone modifications at their promoters (5). CLOCK itself is an enzyme with histone acetyltransferase (HAT) activity, specifically targeting H3 K9/K14 in the chromatin and also nonhistone targets, such as its own partner BMAL1 (6). The HAT activity of CLOCK is counterbalanced by silent information regulator 1 (SIRT1), a member of the sirtuin family of NAD+-dependent histone deacetylases (HDACs) (7, 8). Besides its deacetylase activity on histone tails, especially H3K9, SIRT1 also targets various nonhistone proteins that are important regulators of various cellular processes, from inflammation, to cancer development, to metabolism (9). Targeting SIRT1 with small-molecule modulators has been a significant area of interest for several years, primarily based on the promise this approach holds for the discovery of new therapeutic agents for multiple diseases of aging (10, 11).
SIRT1 appears to operate as a rheostat of the circadian system by physically associating with CLOCK and being recruited to the CLOCK:BMAL1 chromatin complex to circadian promoters (7, 8). Importantly, intracellular levels of NAD+ oscillate with a 24-h rhythm driven by the circadian clock (12, 13). CLOCK:BMAL1 regulate the circadian expression of nicotinamide phosphoribosyltransferase (Nampt), the gene encoding the rate-limiting step enzyme in the NAD+ salvage biosynthetic pathway, the expression of which modulates the levels of NAD+ and its by-product nicotinamide (NAM). SIRT1 is recruited to the Nampt promoter with CLOCK:BMAL1, thus contributing to the synthesis of its own coenzyme (12).
Here we used several SIRT1 activator compounds (STACs) to further elucidate its role in circadian gene expression. By using a pharmacological approach, we demonstrate how modulation of SIRT1 determines changes in circadian gene expression in vitro and in vivo. These data demonstrate how SIRT1 finely regulates the circadian system and reveal how SIRT1 could be efficiently targeted to alter circadian physiology.
Results
Absence of SIRT1 Affects NAD+ Oscillation.
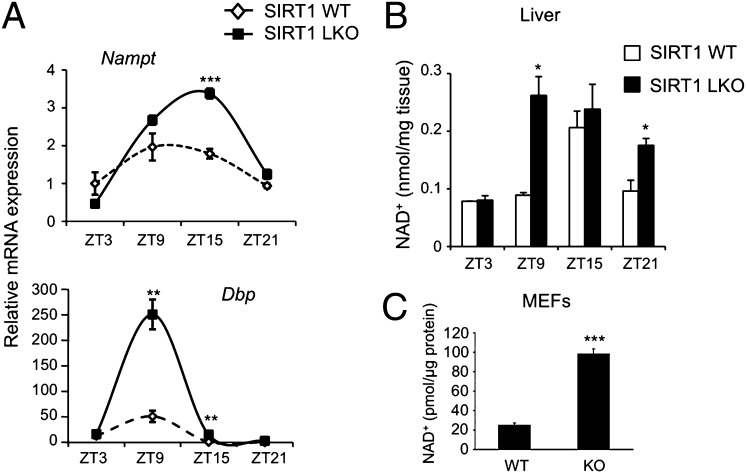
We initially analyzed gene expression in livers from WT and liver-specific Sirt1Δex4 mice (LKO). Ablation of SIRT1 results in altered circadian gene expression in the liver of these mice (7). In particular, we observed increased levels of expression of the CCGs Dbp and Nampt, the amplitude in oscillation of which is significantly larger in the absence of SIRT1 (Fig. 1A).
Fig. 1.
Absence of SIRT1 alters NAD+ cellular levels. (A) Livers from SIRT1Δex4 mice (LKO) and isogenic WT were collected at four different times of the circadian cycle. Nampt and Dbp mRNA expression profiles were analyzed by quantitative PCR. The values are relative to those of 18S mRNA levels at each ZT. All values are the mean ± SEM (n = 3); **P < 0.01, ***P < 0.001. (B) NAD+ was extracted from WT and SIRT1Δex4 livers (LKO) at the indicated time points and analyzed by LC/MSn. Results were normalized for mg of liver tissue. All values are the mean ± SD (n = 3); *P < 0.05. (C) Cellular NAD+ was extracted from WT and Sirt1−/− MEFs and analyzed by LC/MSn. Results were normalized for micrograms of protein. All values are the mean ± SD (n = 3); ***P < 0.001.
We then assessed whether lack of SIRT1 may control the amount of NAD+ available in the cell. As lack of SIRT1 results in increased Nampt expression (Fig. 1A), and because of the critical function of NAMPT in the circadian control of the NAD+ biosynthesis pathway (12, 13), we expected an increase in NAD+ levels in the absence of SIRT1. Indeed, we measured cellular NAD+ levels by liquid chromatography coupled to tandem mass spectrometry (LC/MSn). This process revealed a clear oscillation of NAD+ levels in the liver from WT mice, with maximum peak at zeitgeber time (ZT) 15. The oscillation was enhanced in livers from Sirt1 LKO mice, where the levels of NAD+ showed a significant increase, without losing its oscillatory course (Fig. 1B). Other metabolic mechanisms in addition to the NAD+ salvage pathway could contribute to NAD+ accumulation at specific circadian times, as suggested by the peaks of Nampt mRNA expression (ZT15) and that of NAD+ production (ZT9–ZT15).
This result was confirmed in mouse embryo fibroblasts (MEFs) derived from WT or Sirt1−/− mice, as NAD+ levels were significantly higher in Sirt1−/− MEFs compared with WT MEFs, with an average of fourfold difference (Fig. 1C). This result further consolidates the concept that SIRT1 directly controls the levels of its own coenzyme NAD+ (9, 12).
Activation of SIRT1 Represses Clock Genes Both in Vitro and in Vivo.
The effect of SIRT1 on the transcriptional efficacy of the CLOCK:BMAL1 complex is not fully understood. Although ablation of Sirt1 results in increased amplitude in circadian gene expression (Fig. 1A) (7, 12, 13), ectopic overexpression of SIRT1 may lead to variable outcomes depending on the efficiency and quality of the expression system used (8). To overcome this problem, we sought to further clarify the role of SIRT1 in circadian gene transcription by using a pharmacological approach.
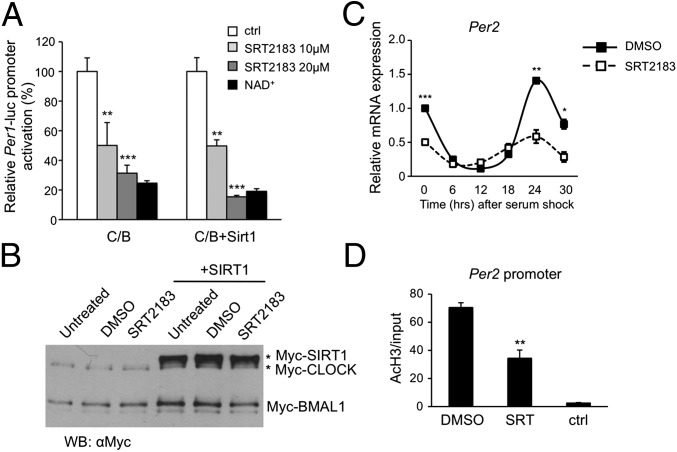
To do this approach, we activated both endogenous and overexpressed SIRT1 with different concentrations of the previously described STAC SRT2183 (11) or with NAD+. Both methods of activating SIRT1 reproducibly repressed CLOCK:BMAL1-driven gene expression in a luciferase assay (Fig. 2A). Moreover, SRT2183 showed a dose–response effect in suppressing CLOCK:BMAL1, an effect amplified by the presence of exogenous SIRT1 (Fig. 2A). Importantly, the expression levels of the core proteins CLOCK and BMAL1 were not altered by the treatment (Fig. 2B), indicating that SIRT1 activation did not affect the ability of the tagged proteins to be expressed or their stability.
Fig. 2.
Modulation of SIRT1 alters CLOCK:BMAL1-mediated transcription. (A) JEG3 cells were transfected with 50 ng of mPer1-luciferase promoter together with CLOCK (C; 100 ng) and BMAL1 (B; 50 ng), with or without SIRT1 (100 ng). Six hours after transfection, cells were treated with SRT2183 10 and 20 μM or with NAD+ 1 mM. DMSO treatment was added in control cells. The effect of the treatment on CLOCK:BMAL1-induced transcription was evaluated 24 h after transfection. After normalization for transfection efficiency using β-galactosidase activity, reporter gene activities were expressed as percentage of relative activation. Value of transcriptional activation in controls (ctrl) was set to 100%. All of the values are the mean ± SD (n = 3); **P < 0.01, ***P < 0.001. (B) Immunoblot analysis of protein extracts obtained from HEK 293 cells transfected with CLOCK:BMAL1 expression vectors alone or with SIRT1. Six hours after transfection, cells were left untreated or treated with DMSO or SRT2183 for 8 h. Nuclear extracts were prepared and loaded on a 6% acrylamide gel. CLOCK and BMAL1 amount was evaluated using Myc antibody. (C) WT MEFs were pretreated with SRT2183 10 μM or DMSO for 16 h before serum shock. Per2 mRNA expression profile was analyzed by quantitative PCR. The values are relative to those of 18S mRNA levels at each circadian time. All of the values are the mean ± SEM (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001. (D) Histone H3 (K9/K14) acetylation at Per2 promoter. WT MEFs were treated with SRT2183 10 μM at time 15 (CT15) after serum shock for 1 h. Cross-linked cell extracts were isolated and subjected to ChIP analysis with antiacetyl histone H3 (K9/K14) and control IgG (ctrl), and analyzed by quantitative PCR. All of the values are the mean ± SD (n = 3); **P < 0.01.
To explore the effect of SIRT1 activation on circadian gene expression, MEFs from WT mice were pretreated with 10 μM SRT2183 (11) for 16 h, and then synchronized with high-serum treatment. The mRNA expression profile showed a robust repression of Per2 transcription and a significant decrease in amplitude (Fig. 2C) in cells treated with SRT2183, compared with DMSO-treated control cells. Similar results were obtained with 1-h treatment with 10 μM SRT2183 (Fig. S1).
Next, we sought to determine whether the overall reduction in Per2 transcription was associated with a decrease in histone acetylation at its promoter. WT MEFs were serum-shocked and then treated with SRT2183 for 1 h at circadian time (CT) 15 during the circadian cycle, or with DMSO. ChIP was performed to monitor H3 K9/K14 acetylation. In samples treated with SRT2183, acetylation at the Per2 promoter was significantly reduced in comparison with DMSO-treated samples (Fig. 2D), suggesting that enhanced histone deacetylation is likely to be responsible for the effects on gene transcription, consistent with a HDAC function of SIRT1. Similar results were obtained for the Dbp promoter (Fig. S2). We conclude that SIRT1 activation with SRT2183 leads to a decrease of circadian gene expression by reducing H3 acetylation at circadian gene promoters.
Chromatin Modifications Induced by SRT1720 in Mouse Liver.
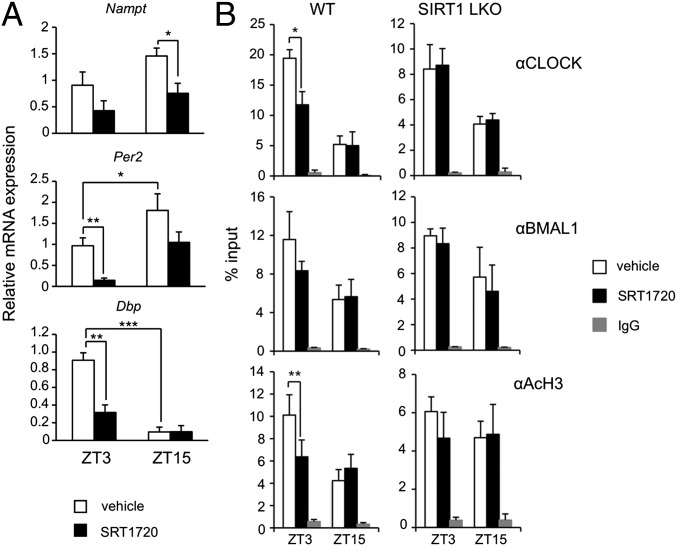
SIRT1 activity has been shown to have a high tissue-specificity (14). To explore how SIRT1 activation could specifically affect liver circadian transcription, we fed WT mice with a standard chow diet supplemented with the STAC SRT1720 for 3 wk, compared with vehicle-supplemented chow (15). The average dosage for each animal was 100 mg SRT1720/kg body weight per day. SRT1720 was used in these experiments instead of SRT2183 based on its more appropriate in vivo properties (16). No difference in body weight and food intake was observed between the two groups. After 3 wk, livers were collected at two different times of the circadian cycle (ZT). The mRNA expression levels of various CCGs were evaluated (Fig. 3A). In mice treated with SRT1720, we observed a robust suppression of the expression peak for Per2, Dbp, and Nampt, compared with the control chow-fed group, confirming that pharmacological activation of SIRT1 activity alters circadian transcription in vivo.
Fig. 3.
Modification of circadian expression in WT mice treated with SRT1720. (A) Change in circadian expression of Nampt, Per2, and Dbp clock genes in livers from mice treated with SRT1720 for 3 wk, compared with vehicle-treated mice. The values are relative to those of 18S mRNA levels at each ZT. Three animals for each condition were used. All of the values are the mean ± SD (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001. (B) ChIP from livers of WT and SIRT1Δex4 mice (LKO) treated with SRT1720 or with vehicle for 3 wk. Livers were collected at two ZT (ZT3 and ZT15) and subjected to dual cross-link and DNA was immunoprecipitated with anti-CLOCK, anti-BMAL1, anti-H3(Lys9/Lys14), and rabbit IgG. Primers for Dbp promoter UP region were used for quantitative PCR. All of the values are the mean ± SD (n = 3); *P < 0.05, **P < 0.01.
We sought to determine if this effect was mediated by changes at the chromatin level. To test this theory, we performed a dual cross-linking ChIP assay in livers from WT and Sirt1 LKO mice treated with SRT1720 or with the vehicle. We observed a significant time-specific reduction of CLOCK recruitment on E-box elements of the Dbp gene promoter (Fig. 3B) in the mice treated with SRT1720, and a reduction of H3 K9/K14 acetylation at the same promoter region (Fig. 3B) compared with vehicle-treated mice. No specific signal was observed in the 3′ UTR region of the same promoter (Fig. S3). Importantly, no differences between SRT1720-treated and vehicle mice were observed in Sirt1 LKO mice (Fig. 3B), thus demonstrating a SIRT1-dependent effect of SRT1720 in the modulation of circadian transcription in vivo.
NAD+ Levels Are Not Affected by SRT1720 Treatment.
To understand the mechanism of action of the SIRT1 activators in circadian physiology, we measured NAD+ levels in livers from mice treated with SRT1720 (Fig. S4). We did not observe any significant change in treated samples compared with the controls, consistent with the mechanism of SIRT1 activation by this compound that acts by changing the affinity of SIRT1 for the substrate, without affecting the Km for the coenzyme NAD+ (11). We also analyzed whether SRT1720 treatment could affect the locomotor activity of the mice. WT mice entrained on a 12-h light:12-h dark cycle were treated for 1 wk and then transferred to constant darkness (free running). No detectable changes in the rest/activity pattern were observed during and after the treatment.
Unique Class of SIRT1 Activators.
A unique chemical class of STACs, designated SRTCD1023, SRTCL1015, and SRTCE1022, were identified and evaluated in a biochemical assay measuring the SIRT1 deacetylase activity against two peptide substrates, as described previously (17) (Table S1). The dose-dependent direct activation of SIRT1 by these compounds was evaluated where the EC1.5 (concentration at which the compound increases SIRT1 activity 50% above DMSO control) was determined and used as a measure of activation efficiency (17). In addition to the biochemical assays, a cell-based assay measuring the SIRT1-dependent deacetylation of p53 (18) was used to characterize the compounds. SRTCE1022 had no activity in either the biochemical or cell-based assays and served as a chemically equivalent inactive control. SRTCL1015 had the highest activity in the cell-based assay and directly activated SIRT1 using this substrate, whereas SRTCD1023, although active in the cell-based assay, only activated SIRT1 using the TAMRA-peptide substrate (Table S1). The biochemical profiles for SRTCD1023 and SRTCL1015 using the two distinct peptide substrates was expected based on a proposed allosteric mechanism of activation, which predicts the extent and efficiency of activation that is critically dependent on the structure of the substrate and subsequent binding energetics (17).
Unique STACs Repress Circadian Transcription at the Chromatin Level.
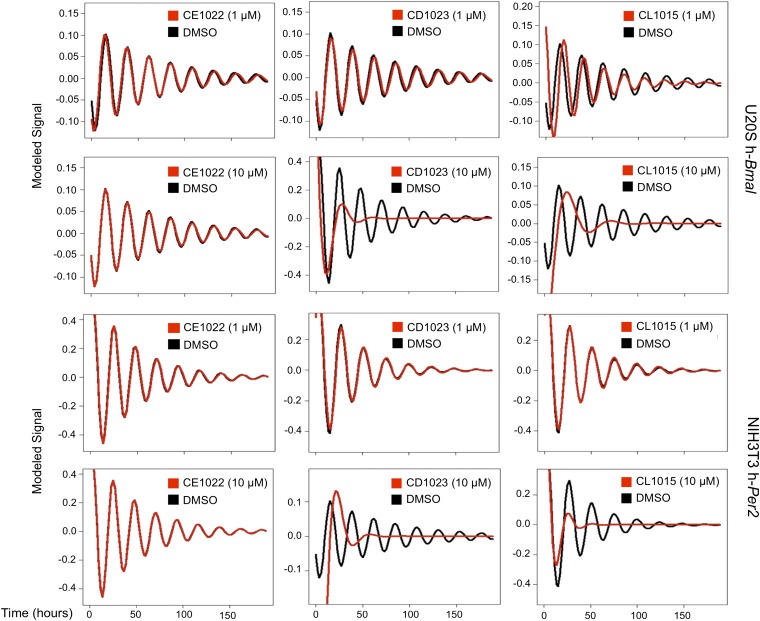
The unique SIRT1 activators were then used to reproduce and confirm the effects of SRT1720 and SRT2183 on circadian expression. We first recorded in real time the bioluminescence from both U2OS cells stably expressing the human-Bmal1 promoter (U2OS-h-Bmal) and NIH 3T3 stably expressing the human-Per2 promoter (NIH-h-Per2) linked to luciferase. Cells were synchronized with 2 h of dexamethasone treatment and then treated with the compounds. Treatment with SRTCD1023 and SRTCL1015, compared with the DMSO control or to treatment with the inactive equivalent SRTCE1022, resulted in a dramatic reduction in a dose-dependent manner in the amplitude of the oscillation of both promoters (Fig. 4 and Fig. S5). SRTCL1015 affected the oscillation of hBmal1 even at 1 μM, a concentration at which no cell toxicity was observed.
Fig. 4.
SIRT1 activators decrease the amplitude of the bioluminescence oscillations. U2OS cells stably transfected with Bmal1-luciferase promoter (U2OS-h-Bmal) and NIH 3T3 stably transfected with Per2-luciferase promoter (NIH-h-Per2) were synchronized by Dex treatment and the bioluminescence was red every 90 min for 5 consecutive days in DMSO-treated cells (black line) compared with cells treated with SRTCE1022, CD1023, CL1015 (red line) at the concentration of 1 μM and 10 μM.
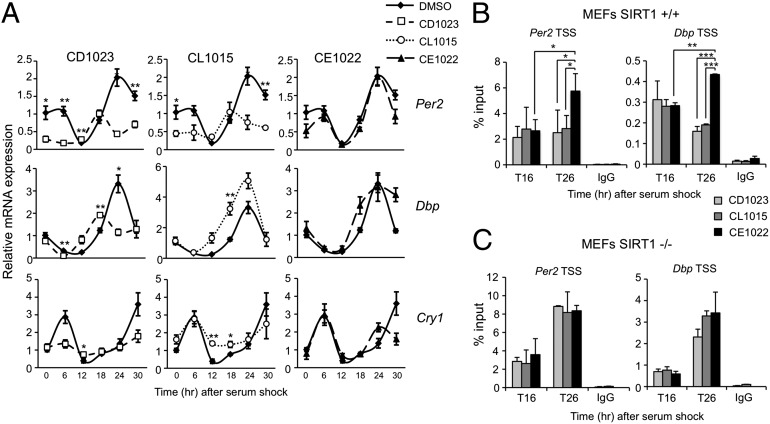
The finding that SIRT1 activation inhibits the transcriptional potential of CLOCK:BMAL1 prompted us to investigate if the effect of the unique STACs might influence circadian endogenous gene expression as well. MEFs generated from WT mice were thereby pretreated with SRTCD1023, CL1015, CE1022, or DMSO, before serum-induced synchronization. RNA was extracted from cells at various times after serum-shock (T0) and analyzed using quantitative PCR. The analysis revealed that SIRT1 activation alters the expression of Per2, Cry1, and Dbp, compared with the cells treated with the inactive SRTCE1022 or with DMSO (Fig. 5A). In particular, repression of Per2, Cry1, and Dbp mRNA expression was observed in cells treated with SRTCD1023, as well as Per2 and Cry1 expression in cells treated with SRTCL1015. Conversely, the amplitude of Dbp mRNA oscillation was increased by treatment with SRTCL1015, suggesting that the activity of transcription factors acting on the Dbp promoter, other than CLOCK and BMAL1, could be affected by the increased SIRT1 activity, thus causing enhancement of Dbp gene expression, instead of repression. Overall, the results confirm that modulation of SIRT1 activity has a profound effect on circadian transcription.
Fig. 5.
SIRT1 activators repress circadian gene expression in a SIRT1-specific manner. (A) Per2, Dbp, and Cry1 mRNA expression profiles in WT MEFs pretreated with DMSO, SRT CD1023, CL1015, and CE1022 (10 μM), before serum shock synchronization, were analyzed by quantitative PCR. The values are relative to those of Gapdh mRNA levels at each circadian time. Time 0 value in DMSO-treated cells was set to 1. All of the values are the mean ± SEM (n = 3); *P < 0.05, **P < 0.01. (B) Cross-linked cell extract were isolated at the indicated time points after serum shock from WT MEFs pretreated with SRTCD1023, CL1015, and CE1022 (10 μM). The samples were subjected to ChIP assay with anti-CLOCK and anti-IgG (ctr), and analyzed by quantitative PCR with primers for Per2 and Dbp promoter. All of the values are the mean ± SD (n = 3); *P < 0.05, **P < 0.01, ***P < 0.001. (C) Same experimental conditions as in B, but using SIRT1−/− MEFs.
We sought to decipher the mechanism by which the unique SIRT1 activators exert their action. To do so, we tested CLOCK recruitment by dual cross-linking ChIP at the promoter of Dbp and Per2 genes at two different times of the circadian cycle (Fig. 5B). A time-specific reduction of CLOCK recruitment was observed in cells pretreated with SRTCD1023 and SRTCL1015 compared with cells pretreated with the inactive compound SRTCE1022 (Fig. 5B). Importantly, this effect was totally abolished in Sirt1−/− MEFs (Fig. 5C), demonstrating that this effect is SIRT1-dependent. These results further demonstrate that modulation of SIRT1 activity largely affects circadian regulation at the chromatin level.
Discussion
Disruption of circadian rhythms can have a profound influence on human health and has been linked to depression, insomnia, jet lag, coronary heart disease, neurodegenerative and metabolic disorders, as well as cancer (19). At the heart of the circadian regulatory pathway is the clock machinery, a remarkably coordinated system that operates in all cells via transcriptional feedback autoregulatory loops. A large fraction of the genome is under circadian control, a process that implicates a series of chromatin remodeling events, some of which have been elucidated (5). Interestingly, accumulating evidence reveals intimate connections between the epigenetic control of the clock system and a variety of metabolic processes (9). Indeed, a relevant fraction of CCGs encode proteins with known enzymatic activity involved in a number of metabolic pathways (2, 20, 21). One of these proteins, the enzyme NAMPT, is responsible for the oscillatory pathway of NAD+. SIRT1 is a NAD+-dependent deacetylase and its activity oscillates in response to the cyclic levels of NAD+ in circadian cells. By taking part in Nampt circadian expression, we already demonstrated how SIRT1 is directly involved in controlling the intracellular levels of its own coenzyme NAD+ (9, 12). These studies identified SIRT1 as an important modulator of circadian gene expression, functioning as a molecular link between metabolic control and circadian rhythms.
Studies in genetically modified mice where SIRT1 gene function has been knocked-out or overexpressed have been critical in linking this enzyme to beneficial and pathological responses to metabolic stress (22–26). This process has led to the proposal that pharmacologically increasing levels of SIRT1 activity would be an ingenious and effective approach for the treatment of age-related metabolic diseases (27–30). In keeping with these notions, a mouse model with noncircadian NAD+ levels displays altered behavioral and metabolic rhythms (31).
Based on these notions, a variety of synthetic compounds has been developed that directly activate SIRT1 (32). These compounds have been shown to have beneficial effects when tested in vivo in models of metabolic dysfunction (10, 11, 28, 33, 34).
Here we investigated whether the absence of SIRT1 or, on the other hand, SIRT1 activation, could interfere with circadian transcription in vivo. The hypothesis that SIRT1 activation could repress circadian transcription prompted us to test a series of compounds with known effects as SIRT1 activators (10, 11, 17, 33, 34). We have demonstrated that modulating SIRT1 activity significantly affects circadian transcription both in cultured cells as well as in vivo, in the mouse liver. Using mice with targeted mutation of Sirt1 in the liver, we have also demonstrated specificity of the pharmacological effect of STACs. Importantly, both in vitro and in vivo approaches confirm that activation of SIRT1 results in a robust decrease in the amplitude of circadian expression of all of the genes tested.
As epigenetic control has been shown to play an important role in circadian gene expression (5), the role of SIRT1 as a HDAC was explored in response to STACs. Importantly, the repressive effect of SIRT1 activation by STACs on gene expression was always paralleled by a time-specific decrease in H3 acetylation at K9/K14 (Fig. 3). The acetylation of these lysine residues is commonly associated to gene activation (5) and SIRT1 has already been shown to function as an efficient HDAC for these residues (14). Our results provide further support to the notion that activated SIRT1 deacetylates histone H3 at the K9/K14 residues and indicate that pharmacological activation of this HDAC has direct physiological consequence on the epigenome. An interesting observation made here is that activation of SIRT1 by SRT1720 also leads to a decreased recruitment of CLOCK:BMAL1 to a circadian promoter. Because CLOCK:BMAL1 and SIRT1 have been found to be associated in the nucleus as a complex (7), this finding suggests that the formation of the complex may be regulated by the enzymatic activity of SIRT1. Further experiments are needed to establish the dynamics of complex formation along the circadian cycle.
In conclusion, we believe that the findings described herein further define the role of SIRT1 in modulating the amplitude of circadian gene expression (9). This notion strengthens the concept of SIRT1 functioning as an enzymatic rheostat, leading to strategies of pharmacological intervention in the modulation of circadian physiology.
Methods
Reagents and Antibodies.
SRT2183, SRTCD1023, SRTCL1015, and SRTCE1022 (Sirtris, a GlaxoSmithKline Company) were dissolved in DMSO and used at the indicated dosage. SRT1720 was suspended in 40% PEG400/0.5% Tween80 (wt/vol) in deionized water and incorporated into the diet (Research Diet), as previously described (10, 15). NAD+ was purchased from Sigma-Aldrich. Antibodies against Myc and acetyl-histone H3 were purchased from Millipore; antibodies against CLOCK and rabbit IgG were from Santa Cruz Biotechnology. Antibody against BMAL1 was as previously described (35).
Animals.
WT and liver-specific Sirt1−/− mice were housed under 12-h light/dark cycles over 2 wk. Mice were killed at specified circadian times and livers were isolated and processed for RNA extraction or ChIP as described below. For experiment using SRT1720, animals were fed a standard diet supplemented or not with SRT1720 (Research diet) for 3 wk (15). The average dosage for each animal was 100 mg SRT1720/kg body weight per day. The same vehicle (dosing solution without SRT compound) was used as a negative control group. Experiments were performed in conformity with the Institutional Animal Care and Use Committee (IACUC) guidelines at the University of California at Irvine.
Cell Culture and Transfection.
JEG3 cells (ATCC) were cultured in Basal Medium Eagle supplemented with 10% FBS and antibiotics. HEK 293 cells (ATCC) were cultured in DMEM supplemented with 10% NCS and antibiotics. Cells were transfected with indicated plasmids using BioT transfection reagent (Bioland Scientific) according to the manufacturer’s recommendations. MEFs were generated from WT or homozygous Sirt1−/− sibling mice and cultured in DMEM supplemented with 7.5% NCS, 2.5% FBS, and antibiotics.
Plasmids.
Plasmids expressing mouse Flag-Myc-SIRT1/pcDNA, Myc-tagged CLOCK and Flag-Myc–tagged BMAL1 have been described previously (12, 36). Plasmids expressing both β-galactosidase (pGL3-lacZ) for transfection control, and luciferase (luc) for luminometry based expression, pGL3-mPer1-Luc promoter, were as described previously (37).
RNA Analysis by Real-Time Quantitative PCR.
Each quantitative real-time RT-PCR was performed using the Chromo4 real time detection system (Bio-Rad). The PCR primers for murine Per2, Cry1, Dbp, Bmal1 mRNA, 18S rRNA, Per2 transcription start site (TSS) promoter region, Dbp UP promoter region, Dbp TSS promoter region, Dbp 3′ UTR promoter region were as described previously (7). Primers for murine Gapdh were the following: Forward: 5′-TGTAGACCATGTAGTTGAGGTCA-3′; Reverse: 5′- AGGTCGGTGTGAACGGATTTG-3′. For a 20 µL PCR, 25–50 ng of cDNA template were mixed with 300 nM primers and 10 µl of iQ SYBR Green Supermix (Bio-Rad), respectively. The reaction was first incubated at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15s and 60 °C for 45s.
ChIP Assay.
The dual cross-linking ChIP assay (38) was used as previously described (7). For the livers, 100 mg of fresh liver tissue were chopped into small pieces in 5 mL of PBS containing 1 mM MgCl2. Disuccinimidyl glutarate (DSG, Pierce) was added to a final concentration of 2 mM for cross-linking and the mix was incubated 45 min at room temperature. Formaldehyde was added to a final concentration of 1% (vol/vol) and samples were incubated for 15 min for dual cross-linking. The cross-linking was quenched by adding glycine to a final concentration of 0.1 M and the reaction was incubated for 10 min. Liver pieces were then centrifuged and pellets were homogenized in 2 mL of ice-cold PBS. After centrifugation, pellets were then resuspended in 1 mL of ice-cold cell lysis buffer [5 mM Hepes pH 8.0, 85 mM KCl, 0.5% Nonidet P-40, 1 mM PMSF, 1× protease inhibitor mixture (Roche Diagnostics)] and incubated for 15 min on ice. Nuclei were precipitated by centrifugation (2,300 × g for 5 min), resuspended in 500 µL ice-cold RIPA buffer (50 mM Tris•HCl pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycolate, 1 mM PMSF, 1× protease inhibitor mixture) and incubated on ice for 30 min. Sonication was performed to obtain DNA fragments 100–600 bp in length.
Generation and Transduction of Human Per2 and Bmal1-Luciferase Promoters.
The human-Bmal1 promoter (U2OS-h-Bmal) and human-Per2 promoter (NIH-h-Per2) linked to luciferase were transfected in U2OS cells and NIH 3T3 cells and lines stably integrating the two promoters in these two cell lines were selected. To measure their oscillations, the cells were grown in T175 flasks in DMEM-F12/M1 media (Gibco) containing either 100 μg/mL (for U2OS-h-Bmal) or 200 μg/mL (for NIH-hper2) hygromycin B (Invitrogen). One day prior the assay, the cells were seeded in 384-well opaque white plates at a density of 50 K cells per well in 50 μL media. The following day the media was aspirated and 50 μL assay media (Phenol Free media-DMEM-M1) (Gibco) containing 10% FBS and 100 nM Dexamethasone (Dex) was added to each well to synchronize the cells. After 2 h, the synchronization media was aspirated and cells were washed once with 80 μL assay media without Dex. Then 40 μL assay media containing 100 μg/mL (for U2OS-h-Bmal) or 200 μg/mL (for NIH-h-Per2) hygromycin and 2 mM Hepes (Sigma) were added to each well. Then 10 μL of compounds diluted in assay media were added. The compound stock concentration was 5× fold higher than that of final concentration. The plates were then sealed using TopSeal (PerkinElmer) and were placed in the Envision. The plates were read every 90 min continuously for 5 d. The data were then analyzed using software developed at GlaxoSmithKline, which models the detrended signal using a sine function. This process provides estimates of the clock parameters: amplitude, period, phase, and damping. The four parameters were estimated as established (39) by fitting the cosine wave function log(y) = a × exp(b × t) × cos[(2 × π × t × 24)/c + d], which includes an exponential term for damping (a = amplitude, b = damping, c = period, d = phase).
NAD+ Measurement.
NAD+ and NAM analysis was performed by LC/MSn and NAD+ and NAM were extracted as previously described (40). For the livers, 50 mg of liver tissue were homogenized in 500 μL of extraction buffer [99% 5 mM ammonium formate/1% methanol containing 1 μM 2-chloroadenosine (41)] and sonicated. After centrifugation (18,000 × g, 10 min, 4 °C), 20 μL of resultant supernatant was kept for protein measurement, the rest of supernatant was filtered with a 0.22-μm filter followed by a regenerated cellulose 3,000 molecular weight cutoff Microcon YM-3 filter (Millipore) to remove the cellular debris and large molecules. NAD+ was identified and quantified by LC/MSn, using a 1100-LC system equipped with a iontrap XCT and electrospray as ionization source (Agilent Technologies). Analytes were separated using a ZORBAX SB-CN column (2.1 × 150 mm i.d., 5 μm; Agilent Technologies) maintained at 30 °C. The Mobile phase was water containing 5 mM ammonium acetate and 0.25% acetic acid (A) and methanol containing 5 mM ammonium acetate and 0.25% acetic acid (B). A gradient from 0 to 50% B in 10 min and then to 70% B from 10 to 15 min was applied at a flow rate of 0.15 mL/min. Total run time was 19 min and posttime was 15 min (100% A). Injection volume was 10 μL. Detection was set in the positive mode, capillary voltage was 4.0 kV, skim1 40 V, and capillary exit 140 V. N2 was used as drying gas at a flow rate of 10 L/min, temperature of 350 °C, and nebulizer pressure of 60 psi. Helium was used as collision gas. Cell-derived NAD+ was identified by comparison of its LC retention time and MS2 fragmentation pattern with those of authentic standards. Full-scan MS2 spectra of NAD+ and 2-chloroadenosine was acquired using multiple reaction monitoring with isolation width of 2 and fragmentation voltage of 1.1 V. Ion charge control was on, smart target set at 100,000 and maximum accumulation time at 200 ms at 26,000 m/z per second. Extracted ion chromatograms were used to quantify NAD+ (m/z 664.3 > 523.8) using 2-chloroadenosine (m/z 302.3 > 170.5) as an internal standard. Detection and analysis were controlled by Agilent/Bruker Daltonics software v5.2.
Supplementary Material
Acknowledgments
We thank Akiko Kawai for assisting with animal care, Sherry Dilag for laboratory assistance, and all members of the P.S.-C. laboratory for help and discussions. This study was supported in part by postdoctoral fellowships from Associazione Italiana per la Ricerca sul Cancro (M.M.B.) and the Della Martin Foundation (M.M.B.); the National Institutes of Health (P.S.-C.); and the Institut National de la Sante et de la Recherche Medicale, France (P.S.-C.).
Footnotes
Conflict of interest statement: M.B., E.W., D.E.M., K.A.E., C.L., J.L.E., and G.P.V. are employees of GlaxoSmithKline. P.S.-C. is a member of the Sirtris Scientific Advisory Board.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214266110/-/DCSupplemental.
References
- 1.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12(7):540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 2.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 3.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12(7):551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 4.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 5.Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010;13(11):1324–1329. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 9.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism—The epigenetic link. J Cell Sci. 2010;123(Pt 22):3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8(5):347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450(7170):712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: Implications for metabolic diseases. Trends Pharmacol Sci. 2010;31(5):212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feige JN, Lagouge M, Auwerx J. Dietary manipulation of mouse metabolism. Curr Protoc Mol Biol. 2008 doi: 10.1002/0471142727.mb29b05s84. Chapter 29: Unit 29B.5. [DOI] [PubMed] [Google Scholar]
- 16.Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11(6):443–461. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai H, et al. SIRT1 activation by small molecules: Kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285(43):32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robers MB, et al. Measurement of the cellular deacetylase activity of SIRT1 on p53 via LanthaScreen® technology. Mol Biosyst. 2011;7(1):59–66. doi: 10.1039/c0mb00026d. [DOI] [PubMed] [Google Scholar]
- 19.Sahar S, Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer. 2009;9(12):886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 20.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 22.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6(6):759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 23.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105(28):9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang RH, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9(4):327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 28.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 29.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477(7365):482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 31.Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 2011;3(8):794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blum CA, et al. SIRT1 modulation as a novel approach to the treatment of diseases of aging. J Med Chem. 2011;54(2):417–432. doi: 10.1021/jm100861p. [DOI] [PubMed] [Google Scholar]
- 33.He W, et al. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. 2010;120(4):1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 36.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 37.Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci USA. 2006;103(16):6386–6391. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39(5):715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 39.Maier B, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23(6):708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans J, Wang TC, Heyes MP, Markey SP. LC/MS analysis of NAD biosynthesis using stable isotope pyridine precursors. Anal Biochem. 2002;306(2):197–203. doi: 10.1006/abio.2002.5715. [DOI] [PubMed] [Google Scholar]
- 41.Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem. 2006;352(2):282–285. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.