Abstract
Proteasomes are essential and ubiquitous ATP-dependent proteases that function in eukarya, archaea, and some bacteria. These destructive but critically important proteolytic machines use a 20S core peptidase and a hexameric ATPase associated with a variety of cellular activities (AAA+) unfolding ring that unfolds and spools substrates into the peptidase chamber. In archaea, 20S can function with the AAA+ Cdc48 or proteasome-activating nucleotidase (PAN) unfoldases. Both interactions are stabilized by C-terminal tripeptides in AAA+ subunits that dock into pockets on the 20S periphery. Here, we provide evidence that archaeal Cdc48 also uses a distinct set of near-axial interactions to bind 20S and propose that similar dual determinants mediate PAN–20S interactions and Rpt1–6–20S interactions in the 26S proteasome. Current dogma holds that the Rpt1–6 unfolding ring of the 19S regulatory particle is the only AAA+ partner of eukaryotic 20S. By contrast, we show that mammalian Cdc48, a key player in cell-cycle regulation, membrane fusion, and endoplasmic-reticulum–associated degradation, activates mammalian 20S and find that a mouse Cdc48 variant supports protein degradation in combination with 20S. Our results suggest that eukaryotic Cdc48 orthologs function directly with 20S to maintain intracellular protein quality control.
Keywords: AAA+ protease, p97
AAA+ (ATPase associated with a variety of cellular activities) enzymes use cycles of ATP binding and hydrolysis to exert mechanical forces on substrates and power a diverse array of biological functions (1). In all cells, protein-unfolding machines of the AAA+ family form hexameric rings that cooperate with associated peptidases to execute intracellular protein degradation (2). The proteolytic sites of AAA+ proteases are sequestered within the interior of a barrel-shaped, self-compartmentalized peptidase, accessible only through narrow axial entrance tunnels that exclude folded proteins. Peptidase access is controlled by the AAA+ unfoldase, which docks with the peptidase, pulls the native structure of target proteins apart, and then translocates the polypeptide through its axial channel and into the degradation chamber.
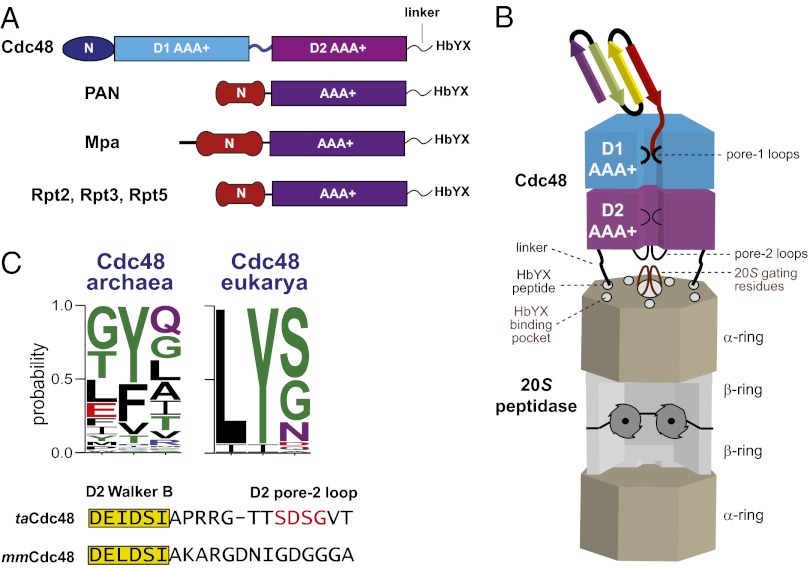
The 20S peptidase is the degradation module for proteasomes in all domains of life, but functions with different AAA+ unfoldases, including the Rpt1–6 unfolding ring of the eukaryotic 26S proteasome, the double-ring Cdc48 or single-ring proteasome-activating nucleotidase (PAN) enzymes in archaeal proteasomes, and the Mpa unfoldase in Actinobacteria (Fig. 1A) (3–6). In all organisms, the 20S peptidase has a four-ring α7β7β7α7 structure and uses N-terminal residues of the α subunits to gate entrance of substrates into its degradation chamber (Fig. 1B) (5, 7–9).
Fig. 1.
Model for bipartite interactions between Cdc48 and 20S. (A) Domain architectures of the Cdc48, PAN, Mpa, and 19S Rpt AAA+ ATPases. Cdc48 consists of two highly homologous AAA+ domains (D1 and D2), a family-specific N-terminal domain, and a flexible and unstructured C-terminal tail consisting of a linker and the HbYX motif. Different family-specific N-domains and flexible unstructured C-termini terminating with an HbYX tripeptide are also found in the AAA+ single-ring unfoldases Mpa, PAN, and some 19S Rpt subunits. (B) Cartoon model of the archaeal Cdc48-20S proteasome. The N domains of Cdc48 are omitted for clarity. The HbYX tripeptides of Cdc48 dock into conserved binding pockets on the periphery of the 20S α ring. Near-axial interactions mediated by the pore-2 loops of the D2 ring of Cdc48 and the N-terminal gating residues of 20S also appear possible. (C) Upper, a WebLogo (28) representation of the C-terminal HbYX motif from archaeal and eukaryotic Cdc48. Lower, the sequence of pore-2 residues following the D2-ring Walker-B motif in archaeal and eukaryotic Cdc48. For functional studies, we deleted the central part of the loop (red) in archaeal Cdc48.
The subunits of the AAA+ ring have C-terminal tripeptides that dock into conserved pockets on the peptidase α ring and help stabilize an open-gate conformation to allow 20S degradation of proteins or large peptides (Fig. 1B) (10). These tripeptides generally match a hydrophobic, tyrosine, any residue motif (HbYX). Indeed, HbYX-peptidase interactions have been proposed to be both necessary and sufficient for docking and regulation (10). However, we recently discovered that the HbYX motif of archaeal Cdc48 was not essential for functional collaboration with archaeal 20S, suggesting that additional interactions between the AAA+ ring and 20S must exist (4). Eukaryotic Cdc48, which is also called p97 or valosin-containing protein (VCP), is an essential enzyme that plays roles in cell-cycle regulation, postmitotic membrane fusion, endoplasmic-reticulum-associated degradation, and the ubiquitin-proteasome system (11–13). Cdc48 is generally thought to function upstream of the 26S proteasome, for example by powering retro translocation of substrates across the endoplasmic reticulum membrane or disassembling macromolecular complexes (14, 15). However, Cdc48 has also been proposed to function more directly with the 26S proteasome (16). Despite having HbYX tails (Fig. 1C), eukaryotic Cdc48 has never been reported to interact directly with 20S.
Here, we identify residues in loops near the bottom of the axial channel of archaeal Cdc48 that appear to function in combination with HbYX interactions to mediate 20S binding. We also show that mouse Cdc48 interacts functionally with the cognate mouse 20S peptidase as well as with archaeal 20S and demonstrate that a variant of mouse Cdc48 collaborates with mouse or archaeal 20S in protein degradation. Our results deepen understanding of the functional determinants of 20S interactions with AAA+ unfolding rings, suggest that the use of peripheral and near-axial contacts may be a conserved feature of many AAA+ proteases, and support a potential role for Cdc48 in higher organisms in direct 20S-dependent protein quality control.
Results
Probing Non-HbYX Interactions Between Archaeal Cdc48 and 20S.
Cdc48 from the archaeon Thermoplasma acidophilum (taCdc48) and orthologs in archaea and eukarya contain a family-specific N domain, D1 and D2 AAA+ rings, and a flexible linker that connects the D2 ring to the C-terminal HbYX motif (Fig. 1A). Recently, we found that a taCdc48 variant lacking the N domain and C-terminal HbYX tripeptide (taCdc48∆N/∆C3) bound ta20S only ∼25-fold less tightly than the parental taCdc48∆N enzyme, indicating that non-HbYX residues must also make significant contributions to 20S binding (4). To identify potential non-HbYX contacts, we built a simple model of a Cdc48–20S complex by initially aligning the axis of the hexameric ring of mouse Cdc48/p97 (Protein Data Bank ID code 3CF1; ∼50% homology with taCdc48; ref. 13) with the axis of a heptameric α ring from the crystal structure of T. acidophilum 20S (Protein Data Bank ID code 1YAU; ref. 17). The Cdc48 ring was then moved along and rotated around the common axis until the two rings were close and the C-terminal residues of each Cdc48 subunit (the linker and HbYX motif were disordered) were close to one of the α-binding pockets. In this model, loops at the bottom of the axial pore of the D2 ring (pore-2 loops) were in proximity to the expected positions of N-terminal gating residues of the α ring. These potential interactions are shown in cartoon form in Fig. 1B. The linkers that connect the D2 ring to the HbYX docking motifs would also be in the vicinity of the α ring.
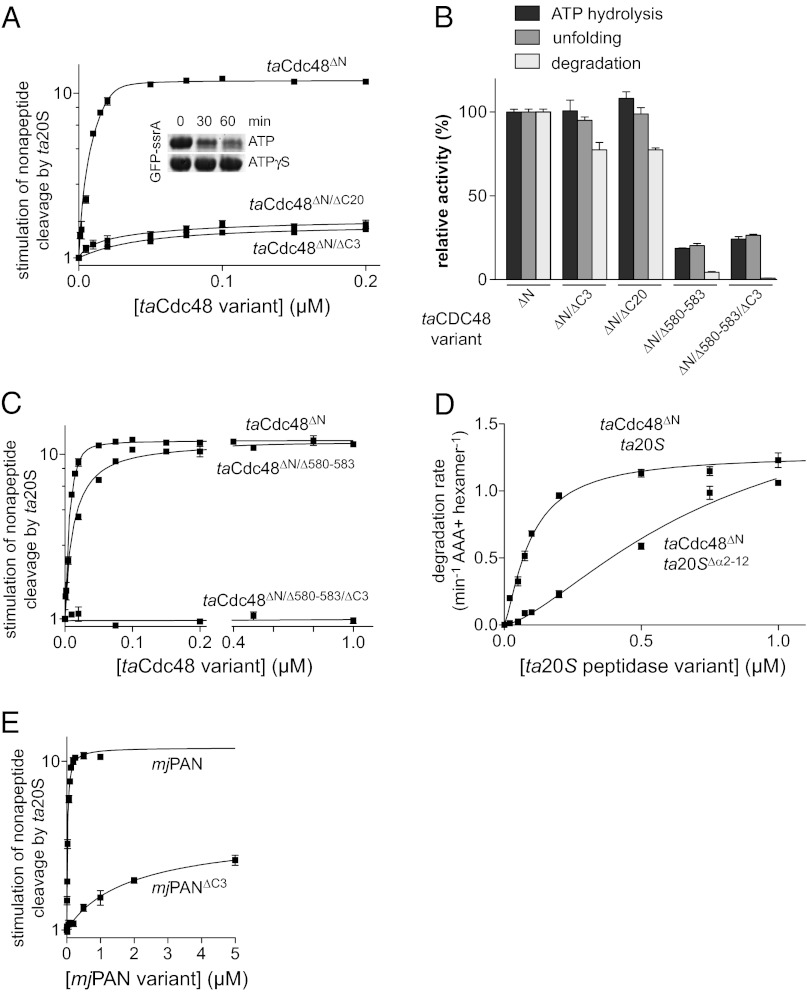
To test the importance of the linkers, we deleted the C-terminal 20 residues of taCdc48∆N (taCdc48∆N/∆C20), which removes the HbYX motif and the complete linker region. As assayed by stimulation of peptide degradation (Fig. 2A), the ∆C20 deletion did not weaken ta20S binding substantially (Kapp = 41 ± 8 nM) compared with the ∆C3 deletion (Kapp = 24 ± 4 nM). However, both variants bound more weakly than the parental taCdc48∆N enzyme (Kapp = 2 ± 0.5 nM) and supported lower maximal levels of peptide degradation. By contrast, taCdc48ΔN/ΔC20 and taCdc48∆N displayed similar activities in ATP hydrolysis and unfolding of GFP-ssrA in the absence of ta20S (Fig. 2B). taCdc48ΔN/ΔC20 also supported ta20S degradation of GFP-ssrA (Fig. 2A, Inset; Fig. 2B). We conclude that linker residues do not make major non-HbYX interactions with archaeal 20S or play roles in ATP-dependent protein unfolding or translocation.
Fig. 2.
AAA+ D2 pore loops contribute to 20S binding. (A) Nonapeptide cleavage by ta20S peptidase (10 nM) as a function of increasing concentrations of taCdc48 variants. Solid lines are fits to a quadratic equation for near-stoichiometric binding. Values are averages (n = 3) ± SEM. Fitted Kapp values for taCdc48ΔN, taCdc48ΔN/ΔC3, and taCdc48ΔN/ΔC20 were 2 ± 0.5, 24 ± 4, and 42 ± 8 nM, respectively. The inset shows an SDS/PAGE assay of GFP-ssrA (5 µM) degradation by 1.2 µM taCdc48ΔN/ΔC20 and 0.4 µM ta20S. Reactions were performed in the presence of 2 mM ATP and an ATP regeneration system or in the presence of 2 mM ATPγS. (B) ATP-hydrolysis rates, GFP-ssrA unfolding rates, and GFP-ssrA degradation rates are plotted for taCdc48 variants relative to these activities for taCdc48ΔN. ATP hydrolysis was measured at 45 °C in the presence of an ATP regeneration system and 2 mM ATP. Unfolding of GFP-ssrA (5 µM) by taCdc48ΔN variants (0.3 µM) was assayed at 60 °C in the presence of 10 mM ATP. Degradation of GFP-ssrA (5 µM) by taCdc48ΔN variants (0.3 µM) and ta20S (0.9 µM) was measured at 45 °C in the presence of 5 mM ATP and an ATP regeneration system. Values are plotted as averages (n = 3) ± SEM. (C) Nonapeptide cleavage by ta20S as a function of increasing concentrations of taCdc48∆N variants. Experiments were performed and analyzed as described in (A). For taCdc48ΔN/∆580–583, the fitted Kapp was 24 ± 4 nM. (D) Degradation of GFP-ssrA (5 µM) at 45 °C was assayed by changes in fluorescence in the presence of taCdc48ΔN (50 nM) and increasing concentrations of ta20S or ta20S∆α2–12. Values are averages (n = 3) ± SEM. (E) Nonapeptide cleavage by ta20S (10 nM) was assayed at 45 °C as a function of increasing concentrations of mjPAN or mjPAN∆C3 in the presence of 100 µM ATPγS. Solid lines are fits to a quadratic equation for near-stoichiometric binding for mjPAN (Kapp 54 ± 7 nM) and to a hyperbolic equation for mjPAN∆C3 (Kapp 3.8 ± 1 µM). Values are averages (n = 3) ± SEM.
To test the importance of the D2 pore-2 loop of taCDC48, we deleted the central amino acids (residues 580–583) of this loop. The taCdc48∆N/∆580–583 mutant displayed ∼10-fold lower ta20S affinity (∼20 nM) than the taCdc48∆N parent, as assayed by stimulation of peptide cleavage (Fig. 2C). Strikingly, although taCdc48∆N/∆580–583 and taCdc48∆N/∆C3 both interacted with 20S with interaction constants of ∼20 nM, combining the ∆580–583 and ∆C3 mutations resulted in a variant (taCdc48∆N/∆580–583/∆C3) that displayed no activation of ta20S peptide cleavage at concentrations up to 1 µM (Fig. 2C). These results suggest that the pore-2 loops of the D2 ring and the HbYX motifs make independent contributions to ta20S recognition. One caveat is that taCdc48∆N/∆580–583 and taCdc48∆N/∆580–583/∆C3 had reduced ATPase activity compared with the parental enzyme (Fig. 2B). Although this phenotype could reflect changes in protein conformation and thus be an indirect effect on 20S binding, several observations favor direct effects. First, ATP hydrolysis is not required for the interaction of taCdc48∆N with ta20S (4). Second, taCdc48∆N/∆580–583 fully activated peptide cleavage by ta20S (Fig. 2C). Third, taCdc48∆N/∆580–583 and taCdc48∆N/∆580–583/∆C3 had protein-unfolding activity commensurate with their reduced ATP hydrolysis activities (Fig. 2B) and thus are functional enzymes. Fourth, although taCdc48∆N/∆580–583 unfolded GFP-ssrA at ∼20% of the parental rate, it supported 20S degradation of GFP-ssrA at only ∼5% of the parental rate (Fig. 2B), indicating a substantially larger defect in coordination of 20S protein degradation than in protein unfolding.
In our model of the complex, the pore-2 loops of the D2 ring of Cdc48 were close to the expected positions of the N-terminal gating residues of the 20S α subunits (Fig. 1B). If these regions of Cdc48 and 20S do contact each other, then gating-residue mutations should also diminish binding. Indeed, when we deleted the gating residues in the ta20S∆α2–12 mutant, higher concentrations of this enzyme were required to support GFP-ssrA degradation by a fixed concentration of taCdc48∆N in comparison with degradation by ta20S (Fig. 2D). This result supports a role for the near-axial gating residues of the α ring in binding Cdc48.
Evidence for Non-HbYX Recognition of 20S by PAN.
Given our Cdc48 results, we wondered if non-HbYX interactions might also contribute to PAN–20S recognition. Although we previously failed to detect an interaction between a Methanocaldococcus jannaschii PAN variant missing the HbYX motif (mjPAN∆C3) and ta20S (4), replacing ATP with ATPγS in the peptide-cleavage assay allowed mjPAN∆C3 to stimulate ta20S activity with an apparent affinity of 3.8 ± 1 µM (Fig. 2E). Wild-type mjPAN bound ta20S more tightly (54 ± 7 nM) under the same conditions (Fig. 2E). Thus, the PAN HbYX motif makes significant contributions to 20S binding but is not necessary for recognition. Pore-2 loops similar to those in the D2 ring of Cdc48 are also present in the single AAA+ rings of PAN, Mpa, and the Rpt1–6 subunits of the 26S proteasome (18), raising the possibility that all of these enzymes use a combination of peripheral HbYX contacts and near-axial interactions to collaborate with the 20S peptidase.
Eukaryotic 20S Interacts Functionally with Cdc48.
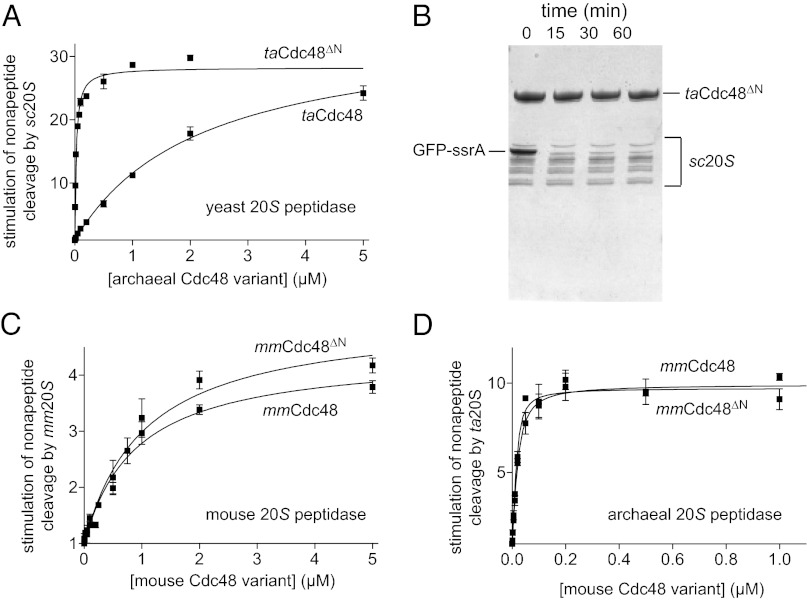
In eukaryotes, both the α7 and β7 rings of 20S consist of seven genetically distinct subunits, whereas the corresponding rings in archaeal 20S are each built from seven identical subunits (5). Moreover, in the 26S proteasome, the C-terminal tails of the Rpt1–6 subunits in the AAA+ unfolding ring differ in sequence and dock with distinct α subunits in the 20S core peptidase (10, 19–21). Did evolution of eukaryotic 20S result in loss of functional interactions with Cdc48? To address this question, we initially used Saccharomyces cerevisiae 20S (sc20S) and tested if archaeal taCdc48 or taCdc48∆N could stimulate peptide cleavage. Indeed, both taCdc48 enzymes enhanced peptide cleavage by yeast sc20S (Fig. 3A). Moreover, robust degradation of GFP-ssrA was observed when we combined taCdc48∆N and sc20S (Fig. 3B). Thus, a homomeric Cdc48 AAA+ ring can functionally collaborate with the heteromeric rings of yeast 20S both in gate opening and protein degradation.
Fig. 3.
Evolutionary conservation of a direct Cdc48–20S interaction. (A) Nonapeptide cleavage by the S. cerevisiae 20S peptidase (10 nM) as a function of increasing concentrations of taCdc48 or taCdc48∆N. Experiments were performed at 37 °C in the presence of 2 mM ATP. Data were fit to a hyperbolic equation for taCdc48 (Kapp 2.2 ± 0.2 µM) or a quadratic equation for near stoichiometric binding for taCdc48∆N (Kapp 24 ± 2 nM). Values are averages (n = 3) ± SEM. (B) SDS/PAGE assay of GFP-ssrA (5 µM) degradation by taCdc48ΔN (1.2 µM) and sc20S (0.4 µM). Reactions were performed at 37 °C in the presence of 2 mM ATP and an ATP regeneration system. (C) Stimulation of nonapeptide cleavage by mouse 20S (5 nM) was assayed at 37 °C in the presence of 100 µM ATPγS and increasing concentrations of mouse Cdc48 or mouse Cdc48∆N. The lines are fits to a hyperbolic equation with Kapp values of 0.9 ± 0.2 µM (mmCdc48) and 1.0 ± 0.2 µM (mmCdc48∆N). Values are averages (n = 3) ± SEM. (D) Stimulation of nonapeptide cleavage by archaeal ta20S peptidase (10 nM) by increasing concentrations of mouse Cdc48 or mouse Cdc48∆N. The lines are fits to a quadratic equation for near stoichiometric binding with Kapp values of 10 ± 2 nM (mmCdc48) and 5 ± 2 nM (mmCdc48ΔN). Experiments were performed at 37 °C in the presence of ATP (2 mM).
Next, we tested if eukaryotic Cdc48, which harbors conserved 20S binding motifs (Fig. 1C), could functionally interact with 20S, either from the same eukaryotic species or from archaea. For these experiments, we used mouse Cdc48 and an ∆N variant of this enzyme, because these Mus musculus enzymes (mmCdc48 and mmCdc48∆N) were better behaved than the comparable yeast enzymes. Importantly, mmCdc48 and mmCdc48∆N enhanced peptide cleavage by mouse 20S (Fig. 3C). The mmCdc48 and mmCdc48∆N enzymes also stimulated peptide cleavage by archaeal ta20S (Fig. 3D). Thus, mouse Cdc48 interacts both with mouse 20S and with the archaeal homolog of this enzyme. Binding of mmCdc48 to mouse 20S (Kapp ∼1 µM) was weaker than binding to ta20S (Kapp ∼10 nM), and the ∆N deletion in mouse Cdc48 did not strengthen 20S affinity as it does in archaeal Cdc48. Nevertheless, mammalian Cdc48 retains the ability to interact functionally with mammalian 20S.
20S-Dependent Degradation by an Altered-Specificity Mouse Cdc48 Variant.
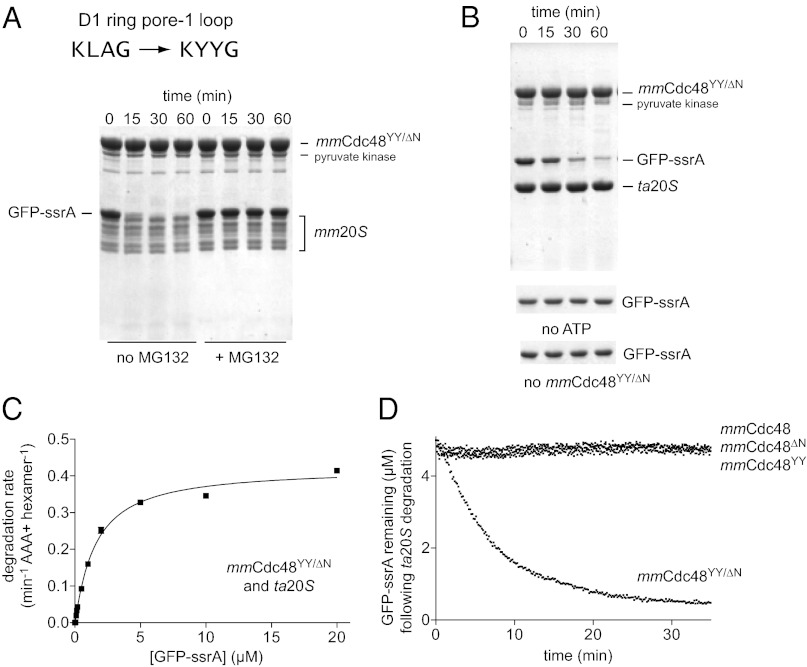
GFP-ssrA is recognized and unfolded by archaeal Cdc48 but not by mouse Cdc48 (22). However, Zwickl and colleagues showed that substituting a Tyr-Tyr (YY) sequence from the D1 pore-1 loop of taCdc48 for the corresponding Leu-Ala (LA) sequence in mouse Cdc48∆N transplanted the ability to recognize and unfold GFP-ssrA (22). We found that mmCdc48YY/∆N was able to collaborate with both mm20S and ta20S to degrade GFP-ssrA (Fig. 4 A and B). No degradation was observed without mmCdc48YY/∆N, without ATP, or when MG132, a 20S peptidase inhibitor, was added. For mmCdc48YY/∆N–ta20S, Vmax and KM for GFP-ssrA degradation (Fig. 4C) were similar to values for taCdc48∆N–ta20S degradation of this substrate (4). Thus, the enzymatic machinery required for protein degradation is still present, at least vestigially, in mammalian Cdc48.
Fig. 4.
Protein degradation by mouse 20S and a mouse Cdc48 variant. (A) In the mmCdc48YY/ΔN variant of mouse Cdc48, the pore-1 loop of the D1 ring is mutated from KLAG to KYYG to match the sequence in taCdc48. The gel shows an SDS/PAGE assay of degradation of GFP-ssrA (5 µM) by mmCdc48YY/ΔN (1.2 µM) and mm20S (0.4 µM) at 37 °C with 2 mM ATP and a pyruvate-kinase based ATP regeneration system. Degradation was inhibited by MG132 (100 µM). (B) SDS/PAGE assay of GFP-ssrA degradation by mmCdc48YY/ΔN and ta20S using the same conditions as in (A). The gel strips on the bottom show that GFP-ssrA is not degraded without ATP or without mmCdc48YY/ΔN. (C) Michaelis-Menten plots of the steady-state degradation of different concentrations of GFP-ssrA by ta20S (0.9 µM) and mmCdc48YY/ΔN (0.3 µM). The fitted KM was 1.7 ± 0.1 µM and Vmax was 0.43 ± 0.01 min−1 Cdc486−1. (D) As assayed by loss of native fluorescence, GFP-ssrA (5 µM) was not degraded by ta20S (0.4 µM) in the presence of mmCdc48, mmCdc48∆N, or mmCdc48YY (1.2 µM) but was degraded when mmCdc48YY/∆N (1.2 µM) was present.
We did not detect GFP-ssrA degradation by mmCdc48 or mmCdc48YY in the presence of ta20S (Fig. 4D). Thus, the N domain of mmCdc48 represses unfolding/degradation, as we previously observed for the N domain of archaeal taCdc48 (4). The structure of the N domain is similar but not identical in archaeal and mammalian Cdc48, but the N-domain sequence is relatively poorly conserved between archaea and eukarya. These variations probably account for the differences observed in N-domain modulation of the activity of the archaeal and mammalian enzymes.
Discussion
Attempts to detect functional interactions between eukaryotic Cdc48 and 20S have not previously been successful (23). For example, it was recently noted that no eukaryotic Cdc48–20S complex has been isolated or successfully reconstituted (24). We revisited this issue because archaeal Cdc48 and 20S comprise a functional proteasome, and the archaeal and eukaryotic orthologs of these enzymes share substantial sequence and structural homology (4). Our current biochemical results demonstrate functional collaboration between the Cdc48 and 20S enzymes from mouse and also show that eukaryotic 20S can function with archaeal Cdc48 and vice versa. Based on these results, we propose that eukaryotic 20S could be a component of a Cdc48–20S complex as an alternative to binding the 19S regulatory particle to form the 26S proteasome.
Whether a eukaryotic Cdc48–20S complex functions as an alternative proteasome remains to be determined but mouse Cdc48 can clearly function as a 20S activator. In addition, our studies show that mouse Cdc48 variants contain the machinery needed to unfold and translocate protein substrates into the 20S degradation chamber. Moreover, degradation by mouse 20S and a Cdc48 variant was inhibited by MG132. Thus, if Cdc48–20S did function as an alternative eukaryotic proteasome, then inhibition of substrate proteolysis by MG132 or related 20S inhibitors would not be sufficient to distinguish between degradation by the 26S proteasome or Cdc48–20S. The role of Cdc48 in eukaryotic protein degradation has generally been thought to involve extracting proteins from membranes or macromolecular complexes for delivery to the 26S proteasome, although some studies suggest that Cdc48 works more directly with 26S (14–16). Indirect roles of Cdc48, direct roles in collaboration with the 26S proteasome, and direct roles of a Cdc48–20S proteasome all remain possible. The challenge will be to identify substrates degraded by one or more of these Cdc48-dependent mechanisms.
Docking of C-terminal HbYX tripeptides into pockets on the periphery of the 20S α ring are important for 20S binding by archaeal PAN and the Rpt1–6 unfolding ring of the 19S eukaryotic regulatory particle (10, 19, 21). HbYX-α interactions also help stabilize binding of Cdc48 to 20S, but archaeal Cdc48 still binds and functionally collaborates with 20S after deletion of this C-terminal tripeptide (4). Our current results show that PAN lacking the HbYX motif also binds archaeal 20S and stimulates peptide cleavage. We also found that near-axial interactions mediated by residues in the pore-2 loop in the Cdc48 D2 ring, which are positioned to interact with N-terminal gating residues of the α subunits, appear to stabilize functional complexes between Cdc48 and 20S. Based on sequence and architectural homology, similar interactions may help stabilize Mpa–20S, PAN–20S, and 19S–20S binding. Thus, 20S binding by the AAA+ rings of Mpa, Cdc48, PAN, and Rpt1–6 may all involve peripheral HbYX-α contacts as well as near-axial contacts between N-terminal α gating residues and pore-2 residues in the AAA+ rings. Interestingly, the length of the pre-HbYX linker varies significantly between archaeal Cdc48 (∼1–19 residues) and eukaryotic Cdc48 (∼29–47 residues). Nevertheless, we find that archaeal 20S functionally interacts with eukaryotic Cdc48 and vice versa. Near-axial interactions may help to make these interactions between enzymes from different domains of life possible despite the observed linker-length differences. Whether the longer linkers of eukaryotic Cdc48 are important for other functions, such as interactions with specific adaptors, remains to be determined.
The hexameric AAA+ ring of the ClpX unfoldase also interacts with the heptameric rings of the ClpP peptidase, which is structurally unrelated to 20S, using peripheral contacts as well as near-axial interactions between pore-2 loops in the ClpX ring and N-terminal residues that surround the entry pore to the ClpP chamber (25). For ClpXP, these near-axial interactions vary dynamically with nucleotide state, help control the rate of ATP hydrolysis, and facilitate efficient protein unfolding (25). Thus, bipartite interactions between AAA+ unfoldases and their cognate peptidases may be common features of many AAA+ proteases. Near-axial contacts may coordinate substrate translocation through the central pore of the unfoldase ring and into the degradation chamber of the peptidase. Crystal structures of 20S with the heptameric ATP-independent activator PA26 also show peripheral interactions between C-terminal tails and pockets in the α ring as well as interactions between an internal activation loop in PA26 and the near-axial gating residues of the α ring (17), further supporting the possibility that bipartite recognition is a universal feature of functional interactions with the 20S peptidase.
Materials and Methods
Plasmids and Strains.
The mouse Cdc48 gene was amplified from Mus musculus cDNA by PCR and cloned into the pET28A (NdeI/SalI) expression vector, resulting in fusion of an N-terminal His6-thrombin tag to Cdc48. Mutations were generated by site-directed mutagenesis and verified by sequencing. The S. cerevisiae strain RJD1144 [MATa, his3Δ200 leu2-3,112 lys2-801 trpΔ63 ura3-52 PRE1-FLAG-6xHIS::Ylplac211 (URA3)] was generously provided by R. Deshaies (California Institute of Technology, Pasadena, CA)].
Expression and Protein Purification.
The taCdc48, ta20S, mjPAN, and GFP-ssrA proteins were expressed and purified as described (4). mmCdc48 proteins were expressed in strain Escherichia coli BL21 (DE3) RIL (Stratagene). Protein expression was induced at OD600 ∼0.8 by adding 1 mM isopropyl β-D-1-thiogalactopyranoside, and growing cells for an additional 4 h at 37 °C. Cells were harvested by centrifugation, resuspended in 50 mM Hepes⋅KOH (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, and 1 mM DTT in the presence of a protease inhibitor mix (Roche), 1,000 units Benzonase (Sigma-Aldrich), and 1 mg/mL lysozyme, and lysed by sonication. Cellular debris were removed by centrifugation at 30,000 × g for 30 min. Polyethylenimine (0.1% vol/vol) was added to the supernatant to precipitate nucleic acids, which were removed by a centrifugation at 30,000 × g for 30 min. The cleared supernatant was incubated with washed Ni++-NTA beads (Thermo Scientific) at 4 °C for 2 h. After washing with 50 mM NaH2PO4 (pH 8.0), 500 mM KCl, 50 mM imidazole, and 0.5 mM EDTA, proteins were eluted with 500 mM imidazole. Samples were dialyzed against 20 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 1 mM DTT, and 1 mM EDTA and loaded onto a MonoQ column (GE Healthcare) followed by elution with a linear salt gradient from 50 mM to 1 M NaCl. After thrombin cleavage overnight, fractions containing Cdc48 were loaded onto a Superdex 200 (16/60) column (GE Healthcare) equilibrated in 50 mM Hepes⋅KOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 1 mM DTT. Cdc48 fractions that eluted at a position expected for hexamers were concentrated and frozen in liquid nitrogen for storage at –80 °C.
The yeast 20S core peptidase was isolated from S. cerevisiae strain RJD1144, which contains a Flag-His6 tag on Pre1, using anti-FLAG M2 resin (Sigma-Aldrich) and size-exclusion chromatography as described (26, 27). Mouse 20S peptidase was purchased from R&D Systems. Cdc48 and PAN concentrations are reported as hexamer equivalents. 20S concentrations are reported as α7β7β7α7 equivalents.
Peptidase Assays.
Cleavage of Mca-AKVYPYPME-Dpa(Dnp)-amide by ta20S in the presence or the absence of taCdc48 was assayed at 45 °C as described (4). For reactions containing mmCdc48 variants, the assay temperature was 37 °C. Cleavage of Mca-RPPGFSAFK-(Dnp)-OH (R&D Systems) by mm20S (5 nM) was measured in the presence or absence of mmCdc48 or taCdc48 at 37 °C. Reactions were performed in 50 µL R buffer (25 mM Hepes⋅KOH, pH 7.5, 50 mM potassium acetate, and 5 mM magnesium acetate) and assayed by changes in fluorescence (excitation 340 nm; emission 405 nm) using a Spectramax 5 spectrofluorometer (Molecular Devices). Cleavage of Mca-RPPGFSAFK-(Dnp)-OH by sc20S (10 nM) was assayed at 37 °C in R buffer plus 0.01% (vol/vol) IGEPAL CA-630.
Protein Unfolding and Degradation Assays.
Protein unfolding was assayed by loss of GFP-ssrA fluorescence (excitation 467 nm; emission 511 nm) in a Spectramax 5 plate reader (Molecular Devices). Reactions were performed in 30 µL 50 mM Hepes⋅KOH (pH 7.5), 100 mM KCl, and 20 mM MgCl2 at 60 °C for taCdc48 and 37 °C for mmCdc48 variants. Reaction components were preincubated for 20 min before adding 10 mM ATP. Protein degradation was assayed by loss of GFP-ssrA fluorescence and SDS/PAGE at 37 °C for mmCdc48 variants, 20S or at 45 °C for taCdc48 and 20S peptidase in 30 µL 50 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 20 mM MgCl2, 3% (vol/vol) glycerol, 5 mM ATP, and an ATP-regenerating system (20 U⋅mL−1 pyruvate kinase; 15 mM phosphoenolpyruvate).
Acknowledgments
We thank R. Deshaies for generously providing the S. cerevisiae strains. Supported by National Institutes of Health Grant AI-16892.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 2.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 3.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20(5):687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Barthelme D, Sauer RT. Identification of the Cdc48⋅20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337(6096):843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voges D, Zwickl P, Baumeister W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 6.Wang T, et al. Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure. 2009;17(10):1377–1385. doi: 10.1016/j.str.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7(11):1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 8.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 9.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41(1):8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27(5):731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458(7237):453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 13.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: Implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16(5):715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21(4):615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rape M, et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107(5):667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 16.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41(1):82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Förster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18(5):589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34(4):473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar B, Kim YC, DeMartino GN. The C terminus of Rpt3, an ATPase subunit of PA700 (19 S) regulatory complex, is essential for 26 S proteasome assembly but not for activation. J Biol Chem. 2010;285(50):39523–39535. doi: 10.1074/jbc.M110.153627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30(3):360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian G, et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011;18(11):1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothballer A, Tzvetkov N, Zwickl P. Mutations in p97/VCP induce unfolding activity. FEBS Lett. 2007;581(6):1197–1201. doi: 10.1016/j.febslet.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 23.Isakov E, Stanhill A. Stalled proteasomes are directly relieved by P97 recruitment. J Biol Chem. 2011;286(35):30274–30283. doi: 10.1074/jbc.M111.240309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matouschek A, Finley D. Cell biology. An ancient portal to proteolysis. Science. 2012;337(6096):813–814. doi: 10.1126/science.1227301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27(1):41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma R, et al. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11(10):3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]