Abstract
Development of therapy against infections caused by antibiotic-resistant pathogens is a major unmet need in contemporary medicine. In previous work, our group chemically modified an antimicrobial peptidomimetic motif for targeted applications against cancer and obesity. Here, we show that the modified motif per se is resistant to proteolytic degradation and is a candidate antiinfective agent. We also show that the susceptibility of microorganisms to the drug is independent of bacterial growth phase. Moreover, this peptidomimetic selectively interferes with the integrity and function of the microbial surface lipid bilayer, data indicative that bacterial death results from membrane disruption followed by dissipation of membrane potential. Finally, we demonstrate two potential translational applications: use against biofilms and synergy with antibiotics in use. In summary, we introduce the mechanism of action and the initial evaluation of a prototype drug and a platform for the development of D-enantiomer antimicrobial peptidomimetics that target bacterial membranes of certain Gram-negative problem pathogens with promising translational applications.
Keywords: antibiotic resistance, Gram-negative bacteria, bacterial infection, antimicrobial synergism
Routine use of antibiotics has reduced the threat of disease and deaths from bacterial infections that were widespread. Although the introduction of these agents was a landmark of modern medicine, their excessive use has resulted in prevalence of resistant microorganisms that have spread through hospitals and, more recently, outside of medical facilities (1, 2). It is now estimated that more than 70% of hospital-acquired infections are resistant to at least one of the antibiotics conventionally used to treat them (3). It is increasingly clear that antimicrobials with new mechanisms of action (4) or alternative strategies, such as phage therapy (5), are needed to combat drug-resistant organisms.
The Infectious Diseases Society of America has identified six “problem pathogens” that urgently require novel therapies (6). These agents are (i) the Gram-positives Enterococcus faecium and Staphylococcus aureus, and (ii) the Gram-negatives Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (6). Notably, the latter group is of extreme concern because of the severely limited number of antimicrobial agents in pharmaceutical industry pipelines to treat infections caused by such selected Gram-negative organisms (6). These pathogens are often treated with “last resort,” toxic drugs that have never been properly assessed by the Food and Drug Administration, such as colistin and polymixin B (7).
Several lines of evidence have suggested that either naturally occurring or synthetic antimicrobial peptides (AMP) may represent a model for the design and generation of new functional classes of antibiotics (8, 9). A major advantage of AMP is their selective activity against microbial agents based largely on the biochemical differences between prokaryotic and eukaryotic cell membrane composition, polarization, and structural features (9, 10). AMP induce bacterial membrane modifications from minor lipid bending to complete membrane dissolution, the latter event resembling a detergent-induced micelle formation process that results in total loss of bacterial membrane integrity (11–14). However, their potential as drugs has been limited by: (i) high susceptibility to proteolytic degradation by endogenous or microbial enzymes, (ii) possible toxicity due to large drug amounts required for treatment, and (iii) manufacturing costs (15).
Attempts to circumvent these hurdles have been centered chiefly on the synthesis of proteolytically resistant versions of natural peptides by either complete or partial substitution of L-residues with nonnatural D- or β-residues (16–18). In particular, lysine-leucine-rich α-helical amphipathic peptides (referred to as Lys-Leu or KL peptides) have been originally used for studying the interactions between peptides and lipid interfaces (19, 20). Further studies have established the peptide (KLAKLAK)2 as an antimicrobial with low toxicity toward mammalian cells (21). Over the past decade, our group has introduced synthetic drug candidates containing a modified version of D(KLAKLAK)2 as part of ligand-targeted agents against cancer (22–26) and obesity (27, 28). However, the value of an untargeted D(KLAKLAK)2 as an antibiotic prototype has not as yet been thoroughly evaluated.
Here we show that: (i) the all-D-enantiomer of (KLAKLAK)2 maintains its antimicrobial activity against bacteria, and (ii) the susceptibility of clinical isolates to the peptidomimetic does not correlate with preexisting resistance to antibiotics. Moreover, in model membranes and bacterial cells, we show that D(KLAKLAK)2-mediated lipid bilayer disruption results in membrane potential dissipation and bacterial death. Finally, we show activity against biofilms, and synergism with conventional antibiotics when used against exponentially growing bacteria. Taken together, our data indicate that D(KLAKLAK)2 is a valuable prototype drug lead for the synthesis and optimization of a class of antimicrobial molecules with a Gram-negative membrane-disruptive mechanism and activity against problem pathogens.
Results
D(KLAKLAK)2 Is Active Against Problem Gram-Negative Pathogens.
We demonstrated that the proapoptotic peptide, D(KLAKLAK)2 preserved its ability to disrupt mitochondrial membranes, which resulted in marked reduction of experimental tumors (22–26) and white adipose tissue (27, 28). We hypothesized that, secondary to enhanced stability of the all-D-enantiomer (Figs. S1 and S2 and SI Materials and Methods), D(KLAKLAK)2 may have improved activity against bacteria and that its persistence may allow treatment with relatively low amounts of the peptidomimetic. We first determined the susceptibility of several species of bacteria to both L(KLAKLAK)2 and D(KLAKLAK)2 by using a standardized broth microdilution assay (29). Complete growth inhibition was observed for the Gram-negative rod bacteria P. aeruginosa, A. baumannii, K. pneumoniae, and Escherichia coli (Table S1). The median minimum inhibitory concentration (MIC) was 150 µg/mL (range, 75–300 µg/mL). In contrast, none of the Gram-positive organisms tested were affected by the compound, potentially indicating a specific mechanism of action. Because cationic peptides interfere with the homeostasis of lipid membranes, we hypothesized that either the absence of an outer membrane in Gram-positive bacteria or the presence of lipopolysaccharides (LPS) in Gram-negative bacteria affects peptidomimetic activity. Peptidoglycan enzymatic degradation rendered both S. aureus and E. faecalis susceptible to D(KLAKLAK)2, suggesting that the thick cell wall protects the cytoplasmic membrane from injury (Fig. S3 A and B). In contrast, binding assays indicated no interaction between the peptidomimetic and LPS (Fig. S3C). Together, these data suggest that, unlike cathelicidins, D(KLAKLAK)2 does not interfere with LPS activity; however, it seems that the peptidomimetic is unable to efficiently diffuse through the Gram-positive cell wall. As such, we focused our investigation on the mechanism of action and effect of D(KLAKLAK)2 against Gram-negative rods.
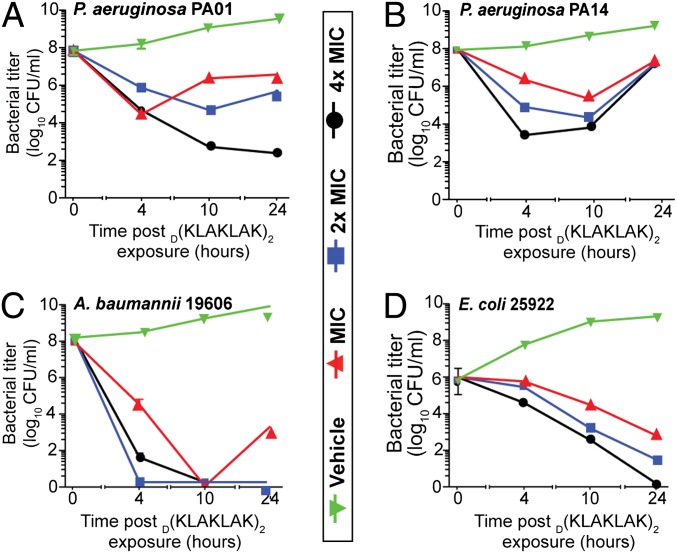
Although widely used to determine antibiotic efficacy and susceptibility, MIC measurements do not provide information about the dynamic interaction between a therapeutic moiety and bacteria (30). To begin to understand the kinetics of D(KLAKLAK)2 interaction with Gram-negative organisms, we first compared the time-kill kinetics of four representative laboratory strains at multiples of MIC for a period of 24 h. D(KLAKLAK)2 induced marked bacterial growth inhibition ranging between two to four orders of magnitude, beginning at 4 h after exposure. The effect of D(KLAKLAK)2 was specific and dose dependent. For instance, P. aeruginosa strains, PAO1 (Fig. 1A) and PA14 (Fig. 1B), revealed different growth inhibition profiles. At MIC, PAO1 growth abruptly decreased within 4 h; however, by 10 h, the rate of multiplication exceeded the drug-induced killing rate, suggesting that not all microorganisms are eliminated by the peptidomimetic at MIC. At concentrations higher than the MIC, we again observed dose- and time-dependent growth inhibition. In contrast, PA14 was less sensitive to D(KLAKLAK)2. Although the growth inhibition of PA14 was initially dose dependent, the overall activity was less substantial after 24 h. Further analysis showed that the decrease in PA14 susceptibility is reversible after two passages in media without antibiotics, suggesting a phenotypic rather than a genotypic change. In contrast, the strains A. baumannii 19606 (Fig. 1C) and E. coli 25922 (Fig. 1D), D(KLAKLAK)2 induced dose- and time-dependent growth inhibition with a bactericidal effect at all concentrations after 10 h and 24 h. Together, these data indicate substantial killing rates against several medically important pathogens.
Fig. 1.
Time-kill kinetics postexposure to D(KLAKLAK)2. P. aeruginosa PAO1 (A), P. aeruginosa PA14 (B), A. baumannii 19606 (C), and E. coli 25922 (D) cultures were supplemented with D(KLAKLAK)2 at the MIC (red line), twice the MIC (2× MIC, blue line), fourfold the MIC (4× MIC, black line), or PBS (vehicle) (green line). Dilutions of aliquots taken at 4, 10, and 24 h were plated on LB agar.
D(KLAKLAK)2 Is Independent of Preexisting Antibiotic Resistance.
Considering the potential therapeutic use of this experimental drug, we next examined whether D(KLAKLAK)2 exerts bactericidal activity against clinical isolates and whether its activity would correlate with preexisting antibiotic resistance. To that end, we determined the MICs of a large panel of clinically relevant bacterial strains (n = 89) obtained from the Clinical Laboratory at the St. Luke’s Episcopal Hospital: 42 strains of E. coli, 25 strains of P. aeruginosa, and 22 strains of K. pneumoniae (Fig. S4). Susceptibility profiles indicated that the MIC ranges from 150 µg/mL for E. coli (Fig. S4A) and P. aeruginosa (Fig. S4B) to 600 µg/mL for K. pneumoniae, respectively (Fig. S4C). Comparative analysis of routinely tested antibiotics (Table S2) and D(KLAKLAK)2 activity revealed no correlation between multidrug resistant (MDR) phenotype and susceptibility to the peptidomimetic. For instance, five E. coli clinical isolates sensitive to all antibiotics displayed average sensitivity to D(KLAKLAK)2 at 150 μg/mL. Similarly, a single E. coli clinical isolate resistant to virtually all antibiotics also exhibited comparable sensitivity to D(KLAKLAK)2 at 150 μg/mL (Table S2, bold numbers). Among the P. aeruginosa strains tested (n = 25), four were resistant to several antibiotics (median, six; range five to eight). However, these strains exhibited varying susceptibility to D(KLAKLAK)2, ranging from the lowest to the second highest concentration tested (Table S2, bold numbers). Perhaps the most striking anecdotal example is represented by a K. pneumoniae clinical isolate found to be resistant to all antibiotics tested, yet sensitive to D(KLAKLAK)2 at 75 μg/mL (Table S2, bold numbers). These results suggest that the peptidomimetic may hold a promise as a last resort antimicrobial for highly MDR Gram-negative bacteria.
D(KLAKLAK)2 Activity Is Growth Phase Independent.
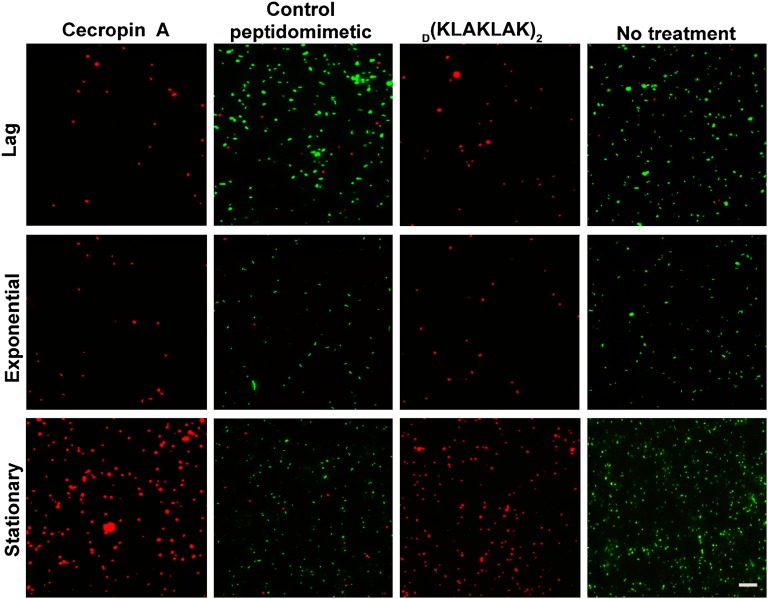
Antibiotic susceptibility is routinely tested on planktonic microorganisms during exponential growth phase (29). However, many severe human infections are caused by quiescent, slow-growing bacteria (31). To determine whether D(KLAKLAK)2 is growth phase dependent, we performed a LIVE/DEAD BacLight viability assay, in which a combination of two nucleic acid stains distinguishes live bacteria with an intact membrane from dead bacteria (32). Viable bacteria stain green with SYTO9, whereas dead bacteria appear red because of staining with propidium iodide. Surprisingly, unlike most antibiotics that are effective only against metabolically active bacteria, D(KLAKLAK)2 equally affected lag, exponential and stationary growth phase P. aeruginosa PAO1 (Fig. 2); in contrast, a control peptidomimetic had no effect on bacterial survival. As expected, cecropin A, an established AMP active against P. aeruginosa (10), was also active independent of the growth phase and served as a positive control (Fig. 2). Taken together, these data suggest that D(KLAKLAK)2 may eliminate dormant bacterial cells, which are usually prone to accumulate antimicrobial resistance (31).
Fig. 2.
D(KLAKLAK)2 activity is growth stage independent. Fluorescence micrographs of P. aeruginosa PAO1 cells at lag, exponential, and stationary growth phase after treatment with D(KLAKLAK)2 (150 µg/mL), control peptidomimetic (150 µg/mL), cecropin A (30 µg/mL), or PBS (vehicle). Viable cells fluoresce in green (due to SYTO9 staining), whereas dead cells fluoresce in red (due to propidium iodide staining). (Scale bar: 10 µm.)
D(KLAKLAK)2 Causes Dose-Dependent Membrane Morphology Damage.
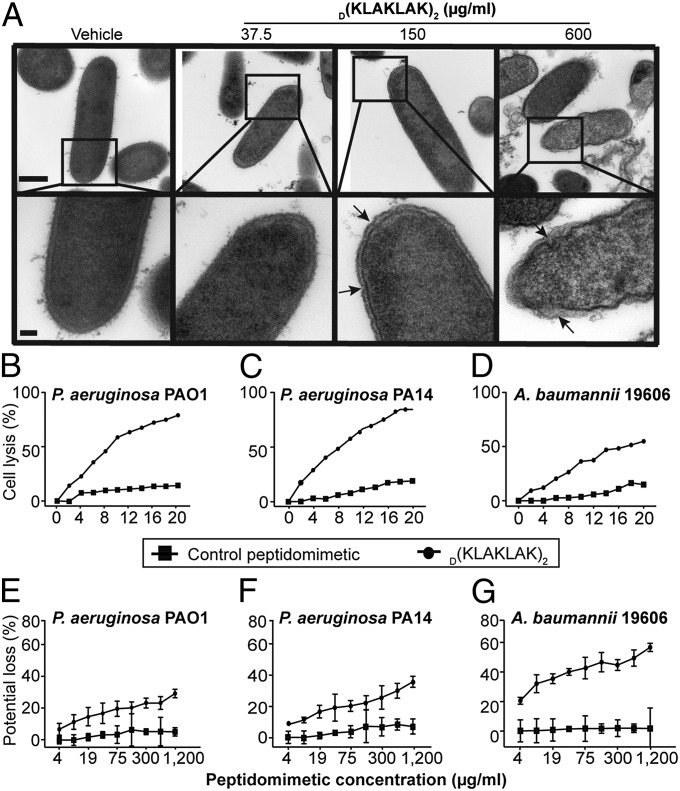
In considering the mechanism of action used by amphipathic-type peptide sequences, we first hypothesized that D(KLAKLAK)2 might induce bacterial membrane damage. Transmission electron micrographs (TEM) of P. aeruginosa PAO1 incubated with increasing concentrations D(KLAKLAK)2 revealed clear morphological evidence of membrane bending and wrinkling (arrows) starting at 150 µg/mL of the peptidomimetic, and severe bilayer damage and bleb formation (arrows) occurring at 600 µg/mL (Fig. 3A). These observations are in agreement with widely accepted, concentration-dependent models of cationic peptide insertion into the lipid bilayer.
Fig. 3.
D(KLAKLAK)2 induces membrane damage. (A) Transmission electron micrographs of P. aeruginosa PAO1 at 50,000× magnification (Upper) and 150,000× magnification (Lower), showing dose-dependent membrane alteration and formation of membrane blebs (arrows) after 25 min of exposure to increasing concentrations of D(KLAKLAK)2. (Scale bars: Upper, 500 nm; Lower, 100 nm.) (B–D) Dose-dependent increase in membrane permeability of P. aeruginosa PAO1 (B), P. aeruginosa PA14 (C), and A. baumannii 19606 (D) (upper lines) in the presence of D(KLAKLAK)2 measured by lysozyme-induced lysis (33, 34). (E–G) Membrane potential loss due to D(KLAKLAK)2 exposure in P. aeruginosa PAO1 (E), P. aeruginosa PA14 (F), and A. baumannii 19606 (G) (Upper lines) measured with the potentiometric dye disk3(5) (34, 35). A control peptidomimetic (Lower lines) did not affect membrane homeostasis.
Having shown that D(KLAKLAK)2 physically alters bacterial surface membranes, we next sought to confirm the lipid bilayer damage mechanism with a more sensitive readout, such as easier access of certain molecules to peptidoglycan. It is well established that when the bacterial surface membrane is damaged, lysozyme gains faster access to its peptidoglycan substrate and induces rapid cell lysis (33). To evaluate this possibility, we exposed P. aeruginosa PAO1, PA14, and A. baumannii 19606 to increasing D(KLAKLAK)2 concentrations in the presence of lysozyme and found a dose-dependent lysis of both P. aeruginosa strains (Fig. 3 B and C). As little as 16 µg/mL D(KLAKLAK)2 induced complete cell culture lysis, whereas only 60% of the A. baumannii 19606 cells were killed at the same peptidomimetic concentration (Fig. 3D). Cell lysis was not observed when bacteria were exposed to a negative control peptidomimetic (Fig. 3 B–D).
Severe membrane injury usually results in membrane potential dissipation and cell death due to loss of lipid bilayer function (34). Thus, we next assessed whether the D(KLAKLAK)2-induced loss of membrane integrity equates with membrane depolarization by using the lipophilic potentiometric dye disk3 (5) assay (35). The addition of D(KLAKLAK)2 triggered an increase in fluorescence intensity, indicating rapid membrane depolarization. We observed a dose-dependent loss of membrane potential in all strains subjected to D(KLAKLAK)2, whereas a control peptidomimetic had no detectable effect (Fig. 3 E–G). We concluded that the lipid bilayer damage induced by D(KLAKLAK)2 is followed by membrane potential dissipation and cell death.
D(KLAKLAK)2 Disrupts the Bacterial Lipid Bilayer.
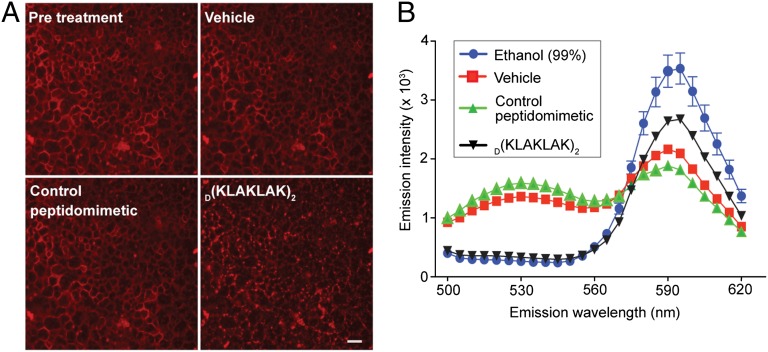
We next sought direct evidence of physical interaction between D(KLAKLAK)2 and bacterial cell membrane lipids. To that end, giant unilamellar vesicles (GUV) derived from E. coli extracts containing rhodamine-labeled phosphatidylethanolamine (Rh-PE) were treated with D(KLAKLAK)2 (34). After 30 s, the GUV membrane appeared punctuated, indicating lipid dissolution and micelle formation as a result of peptidomimetic insertion into the lipid bilayer (Fig. 4A). In the presence of a negative control peptidomimetic, GUV maintained their integrity as demonstrated by continuity in the appearance of lipid membranes (Fig. 4A).
Fig. 4.
D(KLAKLAK)2 induces lipid bilayer disruption. (A) Fluorescence micrographs of giant unilamellar vesicles derived from E. coli extract containing Rh-PE after treatment with 3 µg of D(KLAKLAK)2 or control peptide. The punctuate appearance of lipids after treatment with D(KLAKLAK)2 indicates dissolution of giant unilamellar vesicles. (Scale bar: 100 µm.) (B) FRET between NBD-PE and Rh-PE as a result of D(KLAKLAK)2-induced liposome destruction (black line). No treatment (red line), control peptidomimetic (green line) and 95% ethanol treatment (blue line) served as negative and positive controls.
To evaluate functional membrane damage further, we examined the ability of D(KLAKLAK)2 to disrupt dual-labeled liposomes by using a fluorescence resonance energy transfer (FRET) assay (36). E. coli extract-derived liposomes labeled with either nitro-2-1,3-benzoxadiazol-4-yl (NBD) PE or Rh-PE were incubated with increasing concentrations of D(KLAKLAK)2 or control peptidomimetic. When the liposomes are intact, the two fluorescent lipids are not in close enough proximity to yield an efficient energy transfer. However, when D(KLAKLAK)2 disrupts the liposomal membrane, free labeled lipids are able to come in close contact, leading to FRET. In that setting, energy transfer causes a decrease in emission intensity at 534 nm and a corresponding increase in the emission intensity at 590 nm. In support of our mechanistic hypothesis, D(KLAKLAK)2 disrupted the integrity of liposomes indicated by the energy transfer spectrum (Fig. 4B). Liposome integrity remained unchanged in the presence of a control peptidomimetic, whereas 99% (vol/vol) ethanol (positive control) disrupted both types of liposomes, resulting in an energy transfer similar to the one induced by D(KLAKLAK)2. Our results indicate that the peptidomimetic induces specific lipid bilayer damage similar to the nonspecific damage observed with detergents.
D(KLAKLAK)2 Specifically Disrupts Anionic Phospholipid-Containing Membranes.
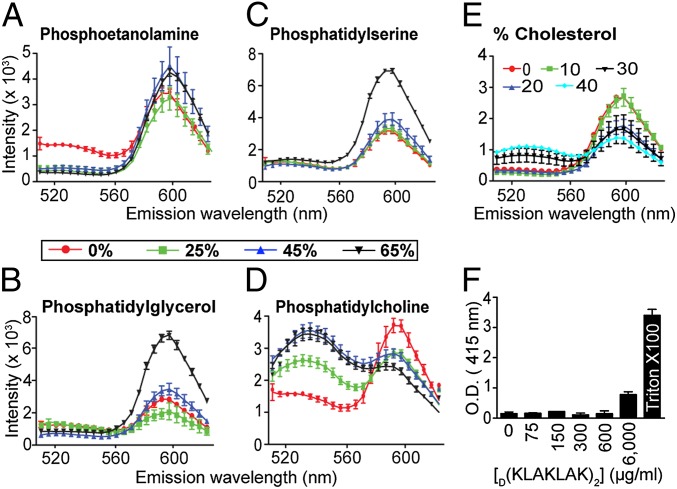
To determine unequivocally a functional mechanism of action for D(KLAKLAK)2, we subsequently examined the effect of lipid specificity on peptidomimetic activity. Most prokaryotic cellular membranes are composed mainly of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and phosphatidylserine (PS), whereas eukaryotic plasma membranes have a more complex lipid content with phosphatidylcholine (PC) and cholesterol as major components (37, 38). In previous work, we clearly showed that only targeted (i.e., ligand-directed and cell-internalized) D(KLAKLAK)2 disrupted mitochondrial membrane (22). Thus, we reasoned that changes in the liposome composition that mimic either type of membrane may alter their susceptibility to D(KLAKLAK)2. Dual-labeled liposomes supplemented with increasing concentrations of each specific lipid were incubated with D(KLAKLAK)2 and assessed for FRET (36, 39). Consistent with our working hypothesis, PE (Fig. 5A), PG (Fig. 5B), and PS (Fig. 5C) increased the susceptibility of liposomes to D(KLAKLAK)2 in a dose-dependent manner. As predicted, the addition of PC (Fig. 5D) and cholesterol (Fig. 5E) decreased the susceptibility of the E. coli extract liposomes to the D(KLAKLAK)2, again supporting the observation (22) that eukaryotic membranes are not affected by the peptidomimetic at similar concentrations.
Fig. 5.
D(KLAKLAK)2 activity is facilitated by anionic phospholipids. (A–D) FRET between NBD-PE and Rh-PE incorporated in liposomes derived from E. coli extract supplemented with increasing percentages of the following: PE (A), PG (B), PS (C), or PC: 0% (red line), 25% (green line), 45% (blue line), or 65% (black line) (D). (E) FRET between NBD-PE and Rh-PE upon lipid bilayer dissolution was carried out with liposomes containing increasing percentages of cholesterol: 0% (red line), 10% (green line), 20% (dark blue line), 30% (black line), or 40% (light blue line). (F) The release of hemoglobin in the supernatant of erythrocytes after treatment with increasing amounts of D(KLAKLAK)2 was measured at 415 nm. Data collected after 48 h of coincubation are presented.
To further corroborate these internally consistent results, we evaluated the ability of D(KLAKLAK)2 to induce erythrocyte hemolysis (34). We measured hemoglobin leakage at 1, 3, and 48 h after exposure to D(KLAKLAK)2 or control peptide. As expected, the D(KLAKLAK)2 peptidomimetic had no hemolytic activity even after 48 h of treatment at MIC, or even higher concentration (Fig. 5F). Together, these data establish that D(KLAKLAK)2 functions through a mechanism of outer membrane bilayer dissolution, which results in nearly complete dissipation of membrane potential and bacterial death. Because D(KLAKLAK)2 acts specifically on the bacterial membrane, disruption of eukaryotic cell membrane homeostasis (i.e., hemolysis) was not observed at the same concentrations, suggesting a favorable drug safety profile.
D(KLAKLAK)2 Has Synergistical Activity in Combination Therapy.
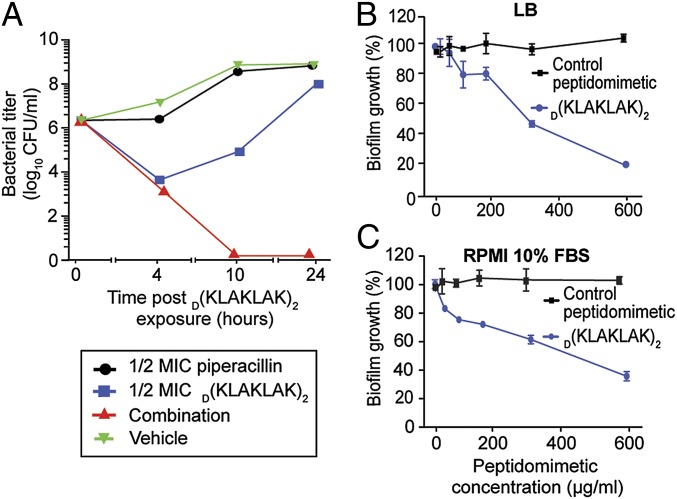
Having shown that D(KLAKLAK)2 activity does not correlate with preexisting antibiotic resistance, we began to examine the potential translational applications for this peptidomimetic. Classic combination therapy enhances antibiotic efficacy and contributes less frequently than monotherapy to selection of drug resistance (40). Thus, we hypothesized that D(KLAKLAK)2 may have synergistic activity with different classes of antibiotics. As an initial exploratory experiment, we performed in vitro time-kill kinetics to determine the activity of D(KLAKLAK)2 in combination with piperacillin, an antibiotic commonly used in therapeutic combinations (i.e., piperacillin/tazobactam) (41). Our results showed great synergistic effect within 4 h after treatment at one-half time the MIC for both the peptidomimetic and the piperacillin (Fig. 6A).
Fig. 6.
D(KLAKLAK)2 acts synergistically with piperacillin and is active against biofilms. (A) The synergistic effect of one-half the MIC D(KLAKLAK)2 and one-half the MIC piperacillin (red line) was compared with one-half the MIC D(KLAKLAK)2 alone (blue line), one-half the MIC piperacillin alone (black line), or untreated bacteria (green line). (B and C) Quantitative assays of biofilm growth inhibition after treatment with D(KLAKLAK)2 (blue line) or control peptidomimetic (black line) in either LB (B) or RPMI medium 1640 containing 10% FBS media (C).
D(KLAKLAK)2 Eliminates Biofilms.
Given that the effect of D(KLAKLAK)2 is not restricted to actively growing microorganisms, we next evaluated its ability to eliminate biofilms, because adherent bacterial growth renders microorganisms 100- to 1,000-fold less susceptible to antibiotics (42). We generated and exposed 24-h-old P. aeruginosa PAO1 biofilm (43) to increasing concentrations of the peptidomimetic. Similar to antibiotics in current clinical use, D(KLAKLAK)2 was less effective against P. aeruginosa PAO1 biofilm (MIC = 600 µg/mL) (Fig. 6 B and C) compared with free-growing cells (MIC = 150 µg/mL) (Table S1). However, there was only a fourfold increase in the MIC value, a result indicating that the peptidomimetic diffuses efficiently through the biofilm and is effective against metabolically active cells as well as the slow-growing bacteria present within the biofilm. Moreover, the activity of D(KLAKLAK)2 was similar in Luria–Bertani (LB) medium (Fig. 6B) and RPMI medium 1640 containing 10% (vol/vol) FBS (Fig. 6C). These results suggest that D(KLAKLAK)2 activity is robust and independent of the culture media used for bacterial growth.
Discussion
Despite the postulated ability of microbes to acquire resistance to any antiinfectives, AMP-conserved efficacy has been proven by their ubiquitous presence among eukaryotes (44). Although such peptides have received great attention over the past decade, additional work is required to translate this emerging class of drugs into preclinical and clinical applications (8, 9). In this report, we describe proof-of-concept experiments designed to evaluate whether the proteolytic enzyme-resistant all-D-enantiomer of a synthetic peptide, L(KLAKLAK)2, would retain its antimicrobial activity. In addition, our study provides a workflow template for the functional assessment of activity in this class of peptidomimetics as membrane-disrupting drugs.
MIC-based susceptibility testing showed that (KLAKLAK)2 stereoisomers are equally efficacious against medically important Gram-negative rods. In contrast, Gram-positive bacteria are resistant to both peptidomimetic forms, most likely because these agents do not diffuse efficiently across the cell wall. Time-kill kinetics revealed marked growth inhibition as soon as 4 h after treatment for all bacterial species studied here. Similar to other AMP, e.g., Pexiganan, killing profiles greatly varied, suggesting that additional factors may affect peptide activity (45). Notably, the starting inoculum for the time-kill experiments was very substantial (108 CFU/mL) compared with the number of bacteria usually recovered from a septic patient’s blood (∼102 CFU/mL).
Initially, we evaluated the efficacy of D(KLAKLAK)2 against standard laboratory strains. However, the major challenge facing antibiotic development is treatment of infections caused by resistant strains. Thus, we first screened a large representative panel of clinical isolates with different antibiotic susceptibilities for their sensitivity to the peptidomimetic. We observed no clear correlation with preexisting antibiotic resistance. These data indicate that the peptidomimetic alone or in combination with a commonly used antibiotic may reduce treatment periods for chronic infections.
To gain insight into the molecular mechanism of D(KLAKLAK)2 action, we performed a series of molecular and cell assays with bacteria or model membranes. Formation of membrane blebs shown by TEM indicated lipid bilayer physical alteration. Further analysis demonstrated that the loss of membrane integrity is accompanied by marked membrane depolarization and bacterial cell death. We found that D(KLAKLAK)2 interacts with lipids found only in the bacterial membrane, an interplay inhibited by addition of PC or cholesterol, components present exclusively in the eukaryotic membrane. We conclude that targeted activity allows D(KLAKLAK)2 to neutralize bacteria without perturbing eukaryotic host plasma membranes. The recent success of drugs such as daptomycin, televancin, oritavancin, and clofazimine, which interfere with membrane integrity and function, encourages one to speculate that the prototype D(KLAKLAK)2 or one of its derivatives is likely to become valuable for treating infections, including those caused by MDR bacteria. Even in situations where the peptidomimetic does not show an improved therapeutic ratio compared with commercially available antimicrobials, combination therapy may hold promise for clinical applications, as suggested by the synergistic effect of piperacillin and D(KLAKLAK)2.
Infections growing as biofilms are commonly treated suboptimally with drugs, a factor potentially contributing to emergence of antimicrobial resistance (44, 46). Notably, live-dead microscopy assays indicated that the peptidomimetic is active regardless of growth phase, so that with the rapid reduction in stationary phase bacterial load, the potential of D(KLAKLAK)2 against biofilms was considered. Our data showed that the peptidomimetic can disrupt a 24-h-old P. aeruginosa biofilm developed on plastic plates irrespectively of the growth medium used, because bacterial growth is affected by quorum sensing-controlling genetic factors that differ among strains (47, 48), whereas the peptidomimetic reacts mainly to bacterial cell membrane lipids. An intriguing possibility is that not only growing bacteria, but also quiescent cells or stationary phase bacteria may be susceptible to D(KLAKLAK)2. Thus, D(KLAKLAK)2 appears to have a desirable therapeutic index against challenging biofilm types of clinical infections.
How our in vitro studies will correlate with therapeutic concentrations remains to be determined, but based on the combined experience of our group and others, it is reasonable to envision that a D(KLAKLAK)2-based drug derivative may allow higher bioavailability, eliminate proteolysis-based instability, and improve cost-effectiveness by supporting treatment with lower amounts of drug. Large preclinical safety studies of ligand-targeted D(KLAKLAK)2 drugs in mice, rats, and nonhuman primates revealed low toxicity at therapeutic concentrations (22–28). In ongoing good laboratory practice (GLP) toxicology studies and even in a first-in-human clinical trial, the toxicity was predictable, dose dependent, and reversible clinically and pathologically. However, we acknowledge that toxicity may differ for cancer and obesity versus bacterial infections. GLP toxicology studies in animals will ultimately determine the drug safety window and toxicity profile. Even if the therapeutic index proves unfavorable, one can perhaps at least exploit this prototype for combination therapy, as a coating component for biomedical devices, or even as a topical antibiotic.
In summary, the prototype D(KLAKLAK)2 represents a stepping stone in the design and development of derivative drugs with broad, yet specific, activity against the bacterial lipid bilayer. On this note, a D(KLAKLAK)2 variant with improved proapoptotic activity has recently been reported (49). In a larger context, these efforts may provide groundwork for rapid design and assessment of prokaryotic membrane-disrupting peptidomimetics for translational development.
Materials and Methods
L(KLAKLAK)2, D(KLAKLAK)2 and D(CVRAC) were synthesized to our specifications by PolyPeptide Laboratories (San Diego, CA). Cecropin A was purchased from AnaSpec, (Fremont, CA). D(CVRAC) served as a negative control peptidomimetic unless otherwise specified. Bacterial and biofilm growth conditions, susceptibility assays and membrane studies, peptidomimetics provenance, and electron and florescence microscopy studies are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Marina Cardó-Vila, Bedrich L. Eckhardt, and Fernanda I. Staquicini for critical reading of the manuscript; Kenneth Dunner, Jr. for help with TEM; and Dr. Magnus Höök for insightful suggestions. P. aeruginosa PAO1 and PA14 were a gift from Dr. Frederick M. Ausubel. This work was supported by awards from AngelWorks, the Gillson-Longenbaugh Foundation, and the Marcus Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221924110/-/DCSupplemental.
See Commentary on page 3212.
References
- 1.Goossens H, Ferech M, Vander Stichele R, Elseviers M. ESAC Project Group Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet. 2005;365(9459):579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N Engl J Med. 2009;360(5):439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 3.Klevens RM, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122(2):160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva ON, et al. Exploring the pharmacological potential of promiscuous host-defense peptides: From natural screenings to biotechnological applications. Front Microbiol. 2011;2:232. doi: 10.3389/fmicb.2011.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci USA. 2009;106(12):4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucher HW, et al. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 7.Poole K. Overcoming multidrug resistance in gram-negative bacteria. Curr Opin Investig Drugs. 2003;4(2):128–139. [PubMed] [Google Scholar]
- 8.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6(5):468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Hancock RE, Lehrer R. Cationic peptides: A new source of antibiotics. Trends Biotechnol. 1998;16(2):82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 10.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 11.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462(1-2):55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Huang J, Chen Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell. 2010;1(2):143–152. doi: 10.1007/s13238-010-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pompilio A, et al. Potential novel therapeutic strategies in cystic fibrosis: Antimicrobial and anti-biofilm activity of natural and designed α-helical peptides against Staphylococcus aureus, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. BMC Microbiol. 2012;12:145. doi: 10.1186/1471-2180-12-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladokhin AS, White SH. ‘Detergent-like’ permeabilization of anionic lipid vesicles by melittin. Biochim Biophys Acta. 2001;1514(2):253–260. doi: 10.1016/s0005-2736(01)00382-0. [DOI] [PubMed] [Google Scholar]
- 15.Peters BM, Shirtliff ME, Jabra-Rizk MA. Antimicrobial peptides: Primeval molecules or future drugs? PLoS Pathog. 2010;6(10):e1001067. doi: 10.1371/journal.ppat.1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maloy WL, Kari UP. Structure-activity studies on magainins and other host defense peptides. Biopolymers. 1995;37(2):105–122. doi: 10.1002/bip.360370206. [DOI] [PubMed] [Google Scholar]
- 17.Bessalle R, Kapitkovsky A, Gorea A, Shalit I, Fridkin M. All-D-magainin: Chirality, antimicrobial activity and proteolytic resistance. FEBS Lett. 1990;274(1-2):151–155. doi: 10.1016/0014-5793(90)81351-n. [DOI] [PubMed] [Google Scholar]
- 18.Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Non-haemolytic beta-amino-acid oligomers. Nature. 2000;404(6778):565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 19.Dathe M, et al. Hydrophobicity, hydrophobic moment and angle subtended by charged residues modulate antibacterial and haemolytic activity of amphipathic helical peptides. FEBS Lett. 1997;403(2):208–212. doi: 10.1016/s0014-5793(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 20.Rothemund S, et al. Recognition of alpha-helical peptide structures using high-performance liquid chromatographic retention data for D-amino acid analogues: Influence of peptide amphipathicity and of stationary phase hydrophobicity. J Chromatogr A. 1995;689(2):219–226. doi: 10.1016/0021-9673(94)00909-s. [DOI] [PubMed] [Google Scholar]
- 21.Javadpour MM, et al. De novo antimicrobial peptides with low mammalian cell toxicity. J Med Chem. 1996;39(16):3107–3113. doi: 10.1021/jm9509410. [DOI] [PubMed] [Google Scholar]
- 22.Ellerby HM, et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med. 1999;5(9):1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 23.Arap W, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8(2):121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 24.Lahdenranta J, Sidman RL, Pasqualini R, Arap W. Treatment of hypoxia-induced retinopathy with targeted proapoptotic peptidomimetic in a mouse model of disease. FASEB J. 2007;21(12):3272–3278. doi: 10.1096/fj.07-8273com. [DOI] [PubMed] [Google Scholar]
- 25.Arap W, et al. Targeting the prostate for destruction through a vascular address. Proc Natl Acad Sci USA. 2002;99(3):1527–1531. doi: 10.1073/pnas.241655998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zurita AJ, et al. Combinatorial screenings in patients: The interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64(2):435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 27.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10(6):625–632. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 28.Barnhart KF, et al. A peptidomimetic targeting white fat causes weight loss and improved insulin resistance in obese monkeys. Sci Transl Med. 2011;3(108):108ra112. doi: 10.1126/scitranslmed.3002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing. Document M100-S23. Available at http://www.clsi.org/. Accessed January 10, 2013.
- 30.Müller M, dela Peña A, Derendorf H. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: Distribution in tissue. Antimicrob Agents Chemother. 2004;48(5):1441–1453. doi: 10.1128/AAC.48.5.1441-1453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 32.Berney M, Hammes F, Bosshard F, Weilenmann HU, Egli T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl Environ Microbiol. 2007;73(10):3283–3290. doi: 10.1128/AEM.02750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock RE, Wong PG. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob Agents Chemother. 1984;26(1):48–52. doi: 10.1128/aac.26.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papo N, Oren Z, Pag U, Sahl HG, Shai Y. The consequence of sequence alteration of an amphipathic alpha-helical antimicrobial peptide and its diastereomers. J Biol Chem. 2002;277(37):33913–33921. doi: 10.1074/jbc.M204928200. [DOI] [PubMed] [Google Scholar]
- 35.Arias CA, et al. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365(10):892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itaka K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Evaluation by fluorescence resonance energy transfer of the stability of nonviral gene delivery vectors under physiological conditions. Biomacromolecules. 2002;3(4):841–845. doi: 10.1021/bm025527d. [DOI] [PubMed] [Google Scholar]
- 37.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cronan JE. Bacterial membrane lipids: Where do we stand? Annu Rev Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 39.Mason AJ, Marquette A, Bechinger B. Zwitterionic phospholipids and sterols modulate antimicrobial peptide-induced membrane destabilization. Biophys J. 2007;93(12):4289–4299. doi: 10.1529/biophysj.107.116681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahal JJ. Novel antibiotic combinations against infections with almost completely resistant Pseudomonas aeruginosa and Acinetobacter species. Clin Infect Dis. 2006;43(Suppl 2):S95–S99. doi: 10.1086/504486. [DOI] [PubMed] [Google Scholar]
- 41.Verbist L, Verhaegen J, Wouters C, Vandenhoven G. Multicentre Study Group Comparative activity of piperacillin/tazobactam against 5625 isolates from hospitalised patients. J Antimicrob Chemother. 1996;37(2):285–293. doi: 10.1093/jac/37.2.285. [DOI] [PubMed] [Google Scholar]
- 42.Costerton JW, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 43.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit1B.1.1–1B.1.18.
- 44.Hurdle JG, O’Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat Rev Microbiol. 2011;9(1):62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge Y, et al. In vitro susceptibility to pexiganan of bacteria isolated from infected diabetic foot ulcers. Diagn Microbiol Infect Dis. 1999;35(1):45–53. doi: 10.1016/s0732-8893(99)00056-5. [DOI] [PubMed] [Google Scholar]
- 46.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406(6797):775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 47.Chugani S, et al. Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc Natl Acad Sci USA. 2012;109(41):E2823–E2831. doi: 10.1073/pnas.1214128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz D, Onuchic JN, Ben-Jacob E. Turning death into creative force during biofilm engineering. Proc Natl Acad Sci USA. 2012;109(46):18633–18634. doi: 10.1073/pnas.1215227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horton KL, Kelley SO. Engineered apoptosis-inducing peptides with enhanced mitochondrial localization and potency. J Med Chem. 2009;52(10):3293–3299. doi: 10.1021/jm900178n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.