Abstract
The packaging of DNA into nucleosomal structures limits access for templated processes such as transcription and DNA repair. The repositioning or ejection of nucleosomes is therefore critically important for regulated events, including gene expression. This activity is provided by chromatin remodeling complexes, or remodelers, which are typically large, multisubunit complexes that use an ATPase subunit to translocate the DNA. Many remodelers contain pairs or multimers of actin-related proteins (ARPs) that contact the helicase-SANT–associated (HSA) domain within the catalytic ATPase subunit and are thought to regulate ATPase activity. Here, we determined the structure of a four-protein subcomplex within the SWI/SNF remodeler that comprises the Snf2 HSA domain, Arp7, Arp9, and repressor of Ty1 transposition, gene 102 (Rtt102). Surprisingly, unlike characterized actin–actin associations, the two ARPs pack like spoons and straddle the HSA domain, which forms a 92-Å-long helix. The ARP–HSA interactions are reminiscent of contacts between actin and many binding partners and are quite different from those in the Arp2/3 complex. Rtt102 wraps around one side of the complex in a highly extended conformation that contacts both ARPs and therefore stabilizes the complex, yet functions to reduce by ∼2.4-fold the remodeling and ATPase activity of complexes containing the Snf2 ATPase domain. Thus, our structure provides a foundation for developing models of remodeler function, including mechanisms of coupling between ARPs and the ATPase translocation activity.
Keywords: RSC, X-ray crystallography, protein complex, nucleosome remodeler
Chromatin remodeling complexes (remodelers) perform functions such as nucleosome repositioning, disassembly, and histone-variant exchange to facilitate processes such as transcriptional regulation, DNA repair, and genome stability (1). Although they are diverse, including at least five different families in eukaryotes, each complex contains a large ATPase subunit that translocates DNA and 2–15 additional subunits that provide functions that include targeting to modified histones and/or regulation of the ATPase activity. The SWI/SNF family of remodelers is broadly conserved among eukaryotes but is best studied in yeast, where two separate complexes are found, SWI/SNF and RSC. Although SWI/SNF is involved in transcriptional activation (2, 3), telomere silencing (4), and DNA repair (5), RSC is more abundant and is essential for cell viability because of its role in regulating transcription mediated by RNA polymerase II and III (6–10).
Actin-related proteins (ARPs) are essential functional subunits of the SWI/SNF, SWR1, and INO80 remodeler families and the NuA4 histone acetyltransferase complex but are absent in ISWI and CHD remodelers (1, 11–15). Dimers or higher-order multimers of these actin/ARP proteins bind to a ∼60-residue helicase-SANT–associated (HSA) domain that is part of the ATPase subunit of remodelers and the Eaf1 subunit of NuA4 and, in remodelers, is adjacent to the N-terminal end of the ATPase domain (16). Indeed, prior biochemical experiments support roles for ARPs in regulating remodeler ATPase activity (12, 13, 16).
HSA domains display specific binding for particular ARPs: the HSA domains of Ino80, Swr1, and Eaf1 bind actin and Arp4, whereas the HSA domains of the Saccharomyces cerevisiae RSC and SWI/SNF ATPases, Sth1, and Snf2, respectively, bind Arp7 and Arp9. Interestingly, the genomes of Yarrowia lipolytica and Schizosaccharomyces pombe do not encode an Arp7 homolog but, instead, encode an extra Arp more similar to Arp4, thereby suggesting an evolutionary relationship between actin/Arp4 and Arp9/Arp7 (17). Arp6, together with Arp5, which contains the largest Arp sequence insertion, and Arp8, which contains an additional N-terminal domain, are only observed as components of INO80. Arp8 is associated with the Ino80 HSA domain and copurifies with actin and Arp4 (16), whereas Arp5 incorporation requires the ATPase subunits Rvb1/Rvb2 (18).
ARPs are numbered according to their sequence similarity to actin, where Arp2 is most similar and Arp11 least similar. The structure of the Arp2/3 complex, a seven-membered complex involved in actin filament branch-point formation, has been determined (19–21). Within the complex, neither Arp2 nor Arp3 make significant interactions with either their D-loop (the region contacting DNase in an actin-DNase cocrystal (22)) or their hydrophobic clefts, which both mediate contacts in actin filaments, leaving them accessible for interactions with actin directly as part of the filament branch-point. Similar to actin, both Arp2 and Arp3 bind nucleotides, although structural data suggest that nucleotide binding alone is not sufficient to generate the conformational changes required for full activation of the complex (20, 23). In addition to Arp2/3, the monomeric structures of Arp4 and Arp8 bound to ATP were recently determined and reveal how the actin structural fold accommodates large sequence insertions that likely preclude any potential actin-like “pointed end” interactions (24, 25). In contrast, the ATPase “active site” residues within Arp7 and Arp9 are not conserved with actin, and their mutation does not result in observable phenotypes, suggesting they do not bind or hydrolyze ATP (11).
The ARP-remodeler ATPase subcomplex represents a core functional unit of the SWI/SNF and RSC complexes. The SWI/SNF Arp7–Arp9–Snf2(ATPase) and RSC Arp7–Arp9–Sth1(ATPase) subcomplexes are functional in ATPase, nucleosome remodeling, and DNA translocation assays (26, 27). Additional functional connections between the Arp and ATPase proteins were identified through arp7/9 suppressor mutations located in the STH1 ATPase subunit of RSC (16). Despite the wide importance of remodelers and the central role in DNA translocation performed by the ATPase subcomplexes, understanding of their mechanisms is currently limited by a lack of structural information.
In an effort to advance understanding of the remodeler mechanism, we have determined a structure of the SWI/SNF Arp7–Arp9–Snf2HSA–repressor of Ty1 transposition, gene 102 (Rtt102) complex to 2.8-Å resolution. Surprisingly, this structure reveals heterodimeric interactions between the Arps that are distinct from known actin polymerization interfaces. Arp7 and Arp9 both straddle the HSA domain, which forms a long helix, in a manner that resembles actin associations with numerous binding partners through the “hydrophobic cleft.” Rtt102 stabilizes the Arp7–Arp9 association by wrapping around one side of the complex in a highly extended conformation. The association is very different from that initially anticipated from the Arp2/3 complex structure and provides a foundation to develop models of remodeler mechanism that couple the ARPs to the ATPase core.
Results and Discussion
Crystal Structure of the SWI/SNF Arp7–Arp9–Snf2HSA–Rtt102 Complex.
We have determined a crystal structure of the SWI/SNF Arp7–Arp9–Snf2HSA–Rtt102 complex using single-wavelength anomalous scattering data from selenomethionine-substituted (SeMet) protein and refined the model at a resolution of 2.8 Å to Rwork/Rfree values of 18/22% (Table 1). Many combinations of constructs were prepared by coexpression in Escherichia coli, purified as stable complexes, and subjected to crystallization trials. Successful crystallization required use of an Arp9 construct in which a poorly conserved loop, which is predicted to be highly flexible, was shortened by deleting residues 247–274. Removal of this loop does not elicit a detectable phenotypic growth defect in S. cerevisiae when tested under a wide variety of growth conditions (temperature sensitivity, use of galactose or glycerol, or presence of methyl methanesulfonate, caffeine, hydroxyurea, or high NaCl).
Table 1.
Crystallographic data collection, phasing, and refinement statistics
| Native | SeMet | |
| Data collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 227.37, 104.14, 81.32 | 226.00, 102.79, 81.50 |
| α, β, γ (°) | 90.0, 93.78, 90.0 | 90.0, 93.93, 90.0 |
| Wavelength (Å) | 1.1 | 0.979 |
| Resolution (Å) | 30.00–2.8 (2.9–2.8) | 50–3.2 (3.31–3.2) |
| Rsym (%) | 5.1 (35.3) | 6.5 (42.0) |
| I /σ(I) | 17.4 (2.2) | 10.7 (1.6) |
| Completeness (%) | 98.4 (86.7) | 94.3 (64.9) |
| Redundancy | 2.0 (1.5) | 3.4 (1.9) |
| Refinement | ||
| Resolution (Å) | 30.00–2.8 | |
| No. reflections | 45,768 | |
| Rwork/Rfree (%) | 18.4/22.3 | |
| No. atoms | ||
| Protein | 7,381 | |
| Ligand/ion | 188 | |
| Water | 91 | |
| Average B-factors (Å2) | ||
| Protein | 69.1 | |
| Ligand/ion | 92.4 | |
| Water | 56.6 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.18 |
Values in parentheses are for the highest-resolution shell.
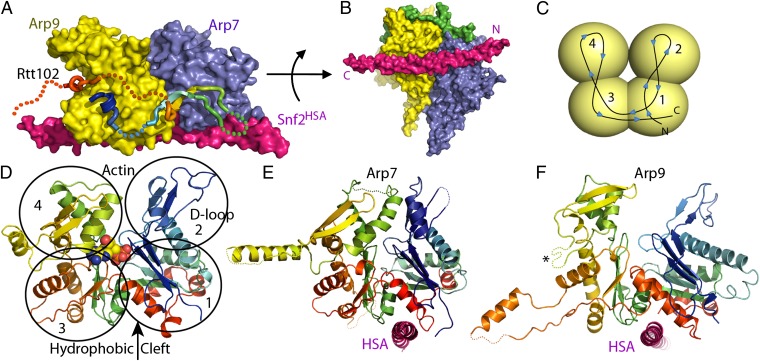
The four-subunit complex forms a compact assembly with overall dimensions of 100 Å × 85 Å × 55 Å (Fig. 1). The architecture is consistent with biochemical studies of subunit associations (16) and provides unexpected information regarding ARP orientations. The HSA domain forms an assembly platform for the ARPs, forming a single 95-residue helix that binds the closely associating heterodimer of Arp7 and Arp9 subunits. In addition to describing a subcomplex of SWI/SNF, the structure also presumably describes the equivalent region of RSC, which contains identical Arp7, Arp9, and Rtt102 proteins and has the Sth1HSA substituted for Snf2HSA (53% sequence identity) (16).
Fig. 1.
Structure of Arp7-Arp9-Snf2HSA-Rtt102. (A) Surface representation. Viewed from the side that binds Rtt102 (rainbow ribbon, disordered residues dotted). (B) Same as A, viewed from below with ordered segments of Rtt102 shown as a green surface. (C) Schematic of actin/ARP fold showing chain trace and domain location viewed from the front, which corresponds to the left side of A for both Arp7 and Arp9. (D) Actin (PDB code 1YAG) (29), shown as a ribbon representation, rainbow-colored N (blue) to C (red), with ATP shown as spheres, (E) Arp7 and (F) Arp9 in the same style and orientation as D. The Arp9 loop that was shortened to facilitate crystallization is indicated with an asterisk.
Structure and Function of Rtt102.
The conformation of Rtt102 is striking. Only 30% of the Rtt102 subunit is visible in the electron density, although the density that is present is clearly defined at a level comparable to other subunits in the structure, indicating that it is present with unit occupancy. The most conserved regions of Rtt102 contact Arp7 and Arp9 by wrapping back and forth in a highly extended conformation across a conserved region of the Arp7–Arp9 interface to form interfaces of 1,455 Å2 with Arp9 and 515 Å2 with Arp7 (28). This interaction is consistent with previous biochemical studies supporting binding of Rtt102 with Arp7–Arp9 rather than direct contacts between Rtt102 and the HSA domain (16). The structure is also consistent with the model that Rtt102 plays a stabilizing role, a view that is supported by the observation that yields are substantially reduced for expression and purification of the ternary Snf1HSA or Sth1HSA complexes lacking Rtt102.
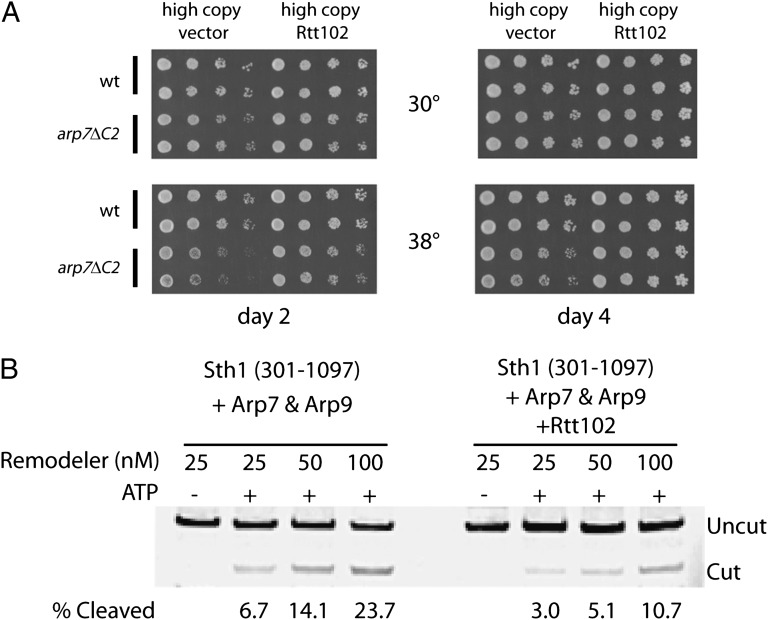
To determine the importance of Rtt102 in S. cerevisiae, we conducted in vivo studies that demonstrated that an rtt102Δ strain in isolation lacks strong phenotypes but that overexpression of Rtt102 gave moderate but clear suppression of certain arp7 temperature-sensitive mutations (Fig. 2A), supporting the notion that it stabilizes ARPs. Interestingly, despite its ability to stabilize the complex, we find that the quaternary complex containing Rtt102 and an Sth1 construct that contains the ATPase domain, Sth1(301–1097), displays ∼2.4-fold less nucleosome remodeling activity, assessed by restriction enzyme accessibility, than the corresponding ternary complex lacking Rtt102 (Fig. 2B). In addition, the corresponding ATPase activity (Vmax) of the complex containing Rtt102 is also diminished ∼2.5-fold. These observations support a model in which Rtt102 stabilizes complex formation while also possibly restricting conformations involved in ATP hydrolysis or coupling to DNA translocation and nucleosome remodeling.
Fig. 2.
Role of Rtt102. (A) Rtt102 overexpression partially suppresses the temperature-sensitive phenotype of a C-terminal truncation of Arp7. A strain (YBC1534) with an Arp7 derivative lacking a small portion of the C terminus of Arp7 (arp7ΔC2) is temperature sensitive compared with a WT strain (YBC1533) (33). Strains were transformed either with an empty vector that is maintained at high copy (pRS423) or with pRS423 expressing RTT102. For each assay, two independent transformants were spotted onto selective plates and incubated at the temperatures indicated for either 2 d (Left) or 4 d (Right). (B) Rtt102 attenuates nucleosome remodeling. Pure complexes of the core ARP/ATPase subcomplex of RSC [Sth1(301-1097) and full-length Arp7 and Arp9], containing or lacking Rtt102, were prepared and quantified as described in the Methods section. Remodeling reactions (Methods) measure the exposure by the remodeler of a restriction enzyme site (AluI), which initially resides within a mononucleosome (174 bp). Cleavage was quantified after DNA extraction and gel electrophoresis. The Cy3-labeled DNA was quantified by scanning on a Typhoon Trio (GE). A representative experiment of two replicates is shown.
Comparison of Arp7 and Arp9 with Actin.
Arp7 and Arp9 share 17% sequence identity, and their structures superimpose with a root mean square deviation of 2.5 Å over 278/396 pairs of Cα atoms. Arp7 and Arp9 share similar (23 and 15%) levels of sequence identity with yeast actin [Protein Data Bank (PDB) code 1YAG] (29), with which they superimpose with root mean square deviation of 2.3 Å over 348/372 pairs of Cα atoms, and root mean square deviation of 2.4 Å over 349/372 pairs of Cα atoms, respectively (Fig. 1 C–F and Fig. S1). Similar levels of structural overlap (2.2–3.9 Å) are seen for Arp7 and Arp9 with Arp2, Arp3, Arp4, and Arp8 (19, 24, 25). Arp7 and Arp9 therefore adopt the canonical actin fold comprising four globular subdomains surrounding a central cleft that, in actin and Arp4, contain the ATPase active site (30). Thus, our structure reveals overall structural similarity of Arp7 and Arp9 to canonical actin.
Despite the overall similarity, Arp7 and Arp9 also deviate from actin in several notable aspects. Unlike actin, and consistent with mutagenesis data (11), neither Arp7 nor Arp9 display conserved residues expected for binding Mg2+ or hydrolyzing ATP, and they are inactive in standard ATPase assays (11). Moreover, electron density maps indicate that nucleotide is not bound in either the native crystal structure or data sets collected after soaks in millimolar concentrations of ATP, although the significance of this observation is potentially limited by the need to maintain crystal lattice contacts and by the crystallization conditions, which include high ammonium phosphate concentrations and result in several phosphate groups being bound in the Arp7 and Arp9 central cavities equivalent to the actin active site. We note that compared with actin, apparent subdomain rotations and deviations in subdomain 4 result in central clefts that are relatively closed in Arp7 and relatively open in Arp9 (Fig. 1 E and F).
Other prominent differences with actin include the D-loop, which in many actin structures and in Arp7 is disordered but in Arp9 is relatively long and forms a crystal contact in the opposite direction to that seen in the actin/DNase I structure (PDB code 1ATN) (22), reinforcing the idea of inherent flexibility at this location. The actin hydrophobic plug, which is a loop in the sequence between subdomains 4 and 3 that has been implicated in interstrand contacts in actin filaments (31), is present in Arp7 but not in Arp9. In comparison with actin, both Arp7 and Arp9 contain extended loop insertions within domains 3 and 4 that are largely disordered except for regions that mediate crystal lattice contacts. One possibility is that these loops mediate contacts to other subunits of the remodeler complexes or to nucleosomes. In comparison, Arp4 contains a large insertion within domain 4 that may play a similar role in its respective complexes (24).
Arp7–Arp9 Heterodimer and HSA Interactions.
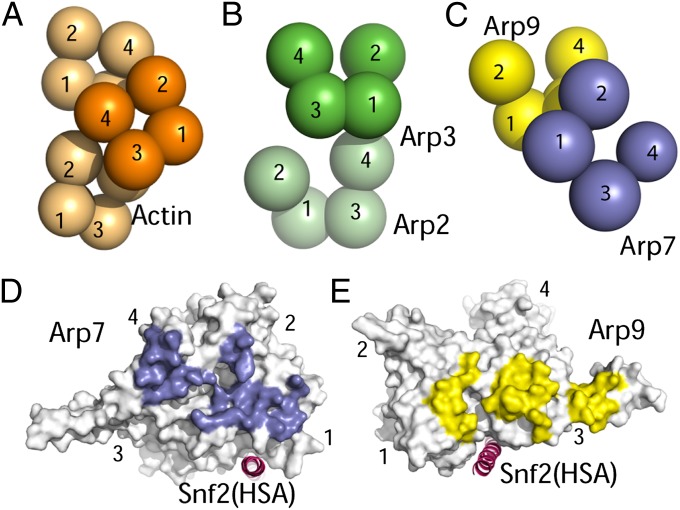
Our most surprising result involves the orientation of the Arp7–Arp9 heterodimer, which assembles very differently from either actin homopolymers (31) or the Arp2/3 branch complex (19) (Fig. 3 A–C). In actin homopolymers, actin monomers pack front-to-front, with an offset that limits individual monomer-monomer interfaces to 494 Å2 but allows repeated twofold helical interactions that extend the filament. The primary interface between Arp2 and Arp3 in the five-protein Arp2/3 complex, which involves domain 4 of Arp2 and domain 1 of Arp3, encompasses 896 Å2. In contrast, Arp7 and Arp9 pack like slightly rotated spoons in a front-to-back arrangement that forms a more extensive 1,330-Å2 interface. Arp7 and Arp9 are related to each other by a translation of ∼40 Å and rotation of ∼20°, with Arp7 residues on the front face of subdomains 1, 2, and 4 contacting Arp9 residues on the back face of subdomains 1 and 3 (Fig. 3 C–E).
Fig. 3.
ARP/actin interactions. (A) Schematic representation of actin/ARP domains within actin filaments (orange) (31). (B) Domain organization within the Arp2/3 complex (green) (19). Arp2 domains 1 and 2 are not visible in the Arp2/3 crystal structure but are shown here in positions inferred from the overall actin fold. (C) Arp7–Arp9 domain organization. Note that residues from Arp7 domain 4 interact with Arp9 residues forming an extended loop from domain 3, although this interaction is not apparent in the stylized representation that depicts spheres centered on domains. (D) Front view of Arp7, with surface contacting Arp9 colored purple. (E) Back view of Arp9, with surface contacting Arp7 colored yellow.
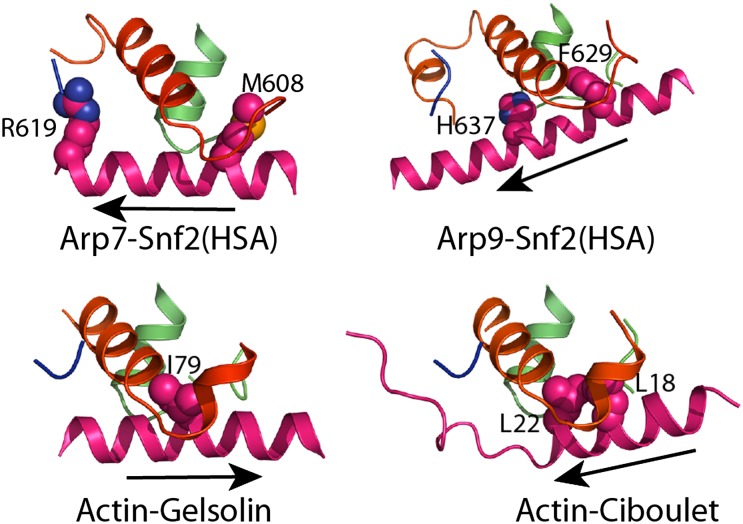
Arp7 and Arp9 bind the Snf2HSA helix through clefts at the base of subdomain 1, near subdomain 3 (Figs. 1 E and F and 3 D and E), to form interfaces of 622 Å2 and 1,002 Å2 with Snf2 residues 604–623 and 623–647, respectively. In actin, this cleft is referred to as the hydrophobic or target-binding cleft (32) and typically mediates contacts with a helix in complexes of actin-binding proteins such as ciboulet, gelsolin, and vitamin D–binding protein (31). The HSA helix runs in the same general orientation as that observed for ciboulet, but in the reverse orientation to the helices of gelsolin or vitamin D–binding protein (Fig. 4). A common feature to these interactions is the insertion of a large hydrophobic group on the binding partner helix into a cleft between the last helix in subdomain 1 before the chain moves to subdomain 3 and the first helix and loop in subdomain 1 after the chain returns from subdomain 3. In the Arp7–Arp9 subcomplex, this role is taken by Snf2HSA Met608, which contacts Arp7, and by Snf2HSA Phe629, which contacts Arp9 (Fig. 4). Arp7 and Arp9 each contain an additional small helix at the front face that extends the cleft and contributes toward making it substantially more hydrophilic than the cleft in actin. The largest surface area contribution to each interface comes from the Snf2HSA hydrophilic residues Arg619 and His637 (Fig. 4).
Fig. 4.
ARP–HSA interactions. Interactions of Arp7 and Arp9 with the Snf2 HSA domain showing prominent interacting side chains from the HSA helix. Structures were aligned on the entire actin/ARP molecule. The helix direction is indicated with an arrow that points in the N to C direction of the helix. Equivalent views are shown for the actin complex with gelsolin (PDB code 1EQY) (43) and with ciboulet (PDB code 1SQK) (44).
Despite the common overall structure of Arp7 and Arp9, there is a clear difference in the structure and importance of residues near their C-termini. Residues near the C-termini of both Arp7 and Arp9 approach the HSA, which explains why truncation of 42 residues from the C terminus of Arp7 or 30 residues from the C terminus of Arp9 results in the selective release of these proteins from preparations of reconstituted S. cerevisiae RSC (33). The C-terminal residue of Arp9 (Phe467) is tucked into the core of the Arp9 structure near both the HSA and Arp7 interface, similar to the position of the C terminus of actin. The Arp7 C-terminal residues, however, are considerably more solvent exposed, which explains why genetic deletion of the Arp9 C-terminal 30 residues is lethal, whereas deletion of the Arp7 42 C-terminal residues elicits only a temperature-sensitive growth phenotype (33).
Models of Other HSA Complexes.
Similar to SWI/SNF, RSC contains an Arp7–Arp9 heterodimer and Rtt102, although, among other differences, the two remodeler complexes have different ATPase subunits, with Sth1 in RSC replacing Snf2 in SWI/SNF. HSA residues that contact Arp7 and Arp9 are only 35% identical in an alignment of S. cerevisiae Snf2 and Sth1 proteins. Although most of the identically conserved HSA residues make contacts at the subunit interface, several residues involved in extensive contacts are not conserved and a few of the identical HSA residues, including Asp643 and Lys646, do not contact Arp7 or Arp9, with one possible explanation for their conservation being a role in binding other SWI/SNF or RSC subunits. Optimal pairwise alignment using pBLAST, and an extensive published alignment (16), both suggest that the RSC Sth1HSA contains an additional four residues near the interface between Arp7 and Arp9 compared with the SWI/SNF Snf2HSA of the structure presented here. We predict that the overall structure of a long α-helix docking into tandem clefts of the ARP heterodimer will be maintained either because the additional residues loop-out (34) or because a shift in the alignment occurs to maintain one long helix with slightly reduced apparent sequence conservation.
The SWI/SNF Arp7–Arp9–Snf2HSA–Rtt102 subcomplex can serve as a model for other HSA complexes. Because overexpression of individual affinity-tagged HSA domains in yeast specifically and solely copurified with the ARP heterodimers for each respective complex (33), the structural similarity likely extends to the Ino80HSA, Swr1HSA, and NuA4 Eaf1HSA ARP subcomplexes, all of which contain an actin–Arp4 heterodimer rather than an Arp7–Arp9 heterodimer. Further, because an Arp7 homolog is missing from two fungi, which instead have a sequence that more closely resembles Arp4 (17), it is attractive to speculate that Arp7 is structurally equivalent to Arp4, whereas Arp9 is structurally equivalent to actin in their respective HSA complexes. In support of this proposal, simple modeling suggests that Arp4 and actin could replace Arp7 and Arp9, respectively, without steric clash, whereas a 54-residue insertion in domain 4 of Arp4, which is disordered in the Arp4 structure (PDB code 3QB0) (24), would lie at the interface and preclude an Arp7–Arp9-like association if Arp4 were placed in the Arp9 position. We therefore favor the model that HSA–actin–Arp4 complexes resemble the structure presented here, with actin in the position of Arp9 and Arp4 in the position of Arp7. This model is consistent with the results of a previous study reporting the crystal structure of isolated Arp4 and its effect on actin filament formation (24), although our complex structure suggests that Arp4 impedes filament elongation by binding to the back surface of actin, thereby blocking interactions with the twofold helically related molecule in a filament rather than directly binding the barbed end of monomeric actin.
The Ino80HSA has the unique distinction of copurifying with Arp8 in addition to actin and Arp4 (16). Arp8 is required for the incorporation of actin and Arp4 in the full complex, suggesting that the heterodimerization of actin–Arp4 is insufficient to sustain binding to the Ino80HSA (12). It is presently unknown whether Arp8 uses its hydrophobic cleft to interact with the HSA specifically or with other subunits in the complex. Large insertions in Arp8 may also be responsible for alternative quaternary interactions (25).
A leading model has been that chromatin remodelers use their ARP heterodimers to interact with other complexes by forming a structure similar to an actin filament (31) in a manner analogous to the Arp2/3 complex, which is thought to undergo a conformational change to mimic an actin filament on activation (19). This model suggested that actin filament–like ARP interactions would bridge complexes to coordinate activities. Importantly, however, this model is inconsistent with the crystal structure because binding of the HSA in the hydrophobic cleft between domains 1 and 3 of Arp7 and Arp9 and the parallel Arp7–Arp9 packing orientation are incompatible with formation of an actin filament, which generally elongates in the direction of the hydrophobic cleft (“barbed” end) and approximates a twofold screw axis (31). Thus, although important questions remain regarding the mechanisms by which ARP associations regulate ATPase and remodeler activities, the possibility of actin filament–like interactions is unlikely.
Nucleosome and DNA Interactions.
To assess the contribution of the Arp7–Arp9–Snf2HSA–Rtt102 subcomplex in nucleosome interactions, the complex was incubated with 174-bp and 147-bp nucleosomes that were assembled with recombinant histones (from S. cerevisiae or Drosophila melanogaster) or with free 174-mer DNA and was subjected to native PAGE (Fig. S2). The subcomplex shifts both free DNA and 174-bp nucleosomes, but not the 147-bp nucleosomes, suggesting that the primary interaction is made through free duplex DNA. Therefore, although it contributes to the nucleosome remodeling process, this subcomplex is not independently capable of nucleosome recognition.
Implications for Future Studies.
Our crystal structure provides a structural model of the Arp7–Arp9–Snf2HSA–Rtt102 module in SWI/SNF-family chromatin remodelers. These ARPs are not required for remodeler assembly because their loss results in a SWI/SNF or RSC complex that simply lacks ARPs (and Rtt102) but retains all other members. Instead, ARPs impart an important regulatory function: The ARP/HSA module is adjacent to the ATPase domain, modulates ATPase activity, and is expected to interact directly with the ATPase domain, as shown through compensatory mutagenesis analysis (16). One speculative model is that the ARP module described here regulates not only ATPase activity but also how efficiently ATPase activity is coupled to DNA translocation. In this model, the ARPs might affect the strength and/or velocity of DNA translocation and affect the outcome of remodeling reactions. An additional possibility is that the ARP module may interact with chromatin factors (presently unknown) and inform the remodeler ATPase of its proper job at the location: sliding or ejection.
Until now, studies of these ARPs have been difficult because they show assembly interdependence and because most mutations obtained in genetic screens simply confer loss of protein and/or assembly at the restrictive temperature. Our structural model provides a foundation for future design of mutation combinations that probe interactions of ARP surfaces with the ATPase domain or other proteins, which should enable mechanistic studies of SWI/SNF and related remodelers. Furthermore, our structure reveals a unique mode of ARP–ARP orientation and interaction (on an HSA platform), which we suggest also applies to remodelers such as INO80 and SWR1 that contain an Arp4/actin (or Baf53/actin) dimer. Notably, INO80 and SWR1 also contain additional ARP proteins, which we speculate may interact with this conserved central ARP dimer module, extending from one end (or both ends) of the central ARP dimer, using the HSA domain as a platform, and forming an ARP polymer. Thus, in addition to guiding mutagenesis studies of mechanism, our structure also guides future structural studies aimed at larger-scale questions of assembly within the remodeler complexes.
Methods
Protein Purification.
Arp7, Arp9(Δ247–274), Snf2(Met–His10–575–667), and Rtt102 were coexpressed in BL21(DE3)RIL from two plasmids, pRSFDuet and pCDFDuet, using the autoinduction method (35). Purification was by Ni2+ chelate and heparin (HiTrap; GE Healthcare) chromatography. Following elution with a NaCl gradient, the protein was concentrated and subjected to sizing chromatography (SD200) in 20 mM Tris, 100 mM NaCl, and 1 mM DTT, pH 7.5. The polyhistidine tag was not removed. The elution profile was consistent with a heterotetramer complex. SeMet protein was expressed and purified in a similar fashion.
Crystallization.
The complex was crystallized by vapor diffusion after mixing 2 µL of complex (OD280 = 10–14; 8.8–10.7 mg/mL) with 2 µL of well solution (1.6–2.0 M ammonium phosphate, 100 mM Hepes pH 7.5, and 1 mM DTT). Crystals grew after 4–7 d at room temperature. Larger crystals for data collection were obtained by streak seeding into an equilibrated (1 d) crystallization drop. Crystals were cryoprotected with well solution made up with 25% glycerol and were vitrified by plunging into liquid nitrogen. Data were collected at 100K at the National Synchrotron Light Source beamline ×25 (SeMet λ = 0.979 Å, Native λ = 1.1 Å). Phases were calculated to a resolution of 3.3 Å and improved by density modification using AutoSol-PHENIX (36). The model was built using COOT (37) and refined in REFMAC5 (38) and PHENIX, (36) using translation, libration, and screw-rotation (39). The following residues are included in the final model: Arp7: 2–39, 44–204, 214–262, 281–344, 380–466; Arp9: 3–223, 275–376, 393–467; Snf2: 592–660; Rtt102: 1–12, 22–34, 54–70, 79–90. The final model has 92.1% core, 6.7% allowed, 1.0% generous, and 0.1% disallowed residues in the Ramachandran plot. Crystallographic statistics are given in Table 1.
Nucleosome Remodeling and ATPase Activity.
Pure complexes containing Sth1(301–1097) and full-length Arp7 and Arp9, either containing or lacking Rtt102, were prepared with the size-exclusion chromatography buffer including 200 mM NaCl and 10% (vol/vol) glycerol. For ATPase determinations, two separate expressions and purifications of these complexes were assayed independently. ATPase activity was measured as described previously (40), using a color (malachite green) absorbance assay that quantitatively measures released free phosphate. Protein levels were quantified by ultraviolet absorbance at 280 nm and by Coomassie-stained SDS/PAGE to ensure equal Sth1 molar equivalents. Control reactions with native RSC ensured that the assays were performed in the linear range. Levels of ATP and DNA were in the saturating range and, therefore, the values reported are at Vmax. Reactions involved incubating 264 ng of Sth1 protein (2.8 μmol of Sth1 molar equivalents) for 30°C for 52 min. Reactions were performed in triplicate. Control reactions on both forms (+/−Rtt102) verified the requirement of DNA to stimulate ATPase activity. For remodeling, mononucleosomes containing 5S rDNA (174 bp, bearing a single AluI site) were prepared as described previously (41), except the DNA was labeled with Cy3. Restriction enzyme accessibility reactions contained nucleosomes (200 nM), ATP (1 mM), AluI (10 U), and increasing concentrations of remodeler complex (indicated) and were incubated at 30°C for 60 min (42). Remodelers were titrated over a linear range. Cleavage was quantified after DNA extraction and gel electrophoresis. The Cy3-labeled DNA was quantified by scanning on a Typhoon Trio (GE).
Supplementary Material
Acknowledgments
We thank Whitney G. Smith for technical assistance and Cedric Clapier for D. melanogaster nucleosomes. Data collection at the National Synchrotron Light Source (NSLS) was funded by the National Center for Research Resources. Operations of the NSLS are supported by the US Department of Energy, Office of Basic Energy Sciences, and by the National Institutes of Health. This work was supported by National Institutes of Health Grants GM076242 (to C.P.H.) and GM60415 (to B.R.C.) and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4I6M).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215379110/-/DCSupplemental.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95(5):717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 3.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97(7):3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dror V, Winston F. The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24(18):8227–8235. doi: 10.1128/MCB.24.18.8227-8235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19(14):1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angus-Hill ML, et al. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7(4):741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- 7.Damelin M, et al. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell. 2002;9(3):563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- 8.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16(7):806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasten M, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23(6):1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soutourina J, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol. 2006;26(13):4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2(5):639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 12.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12(1):147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95(5):625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 14.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 15.Galarneau L, et al. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5(6):927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 16.Szerlong H, et al. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15(5):469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller J, et al. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005;16(12):5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16(3):465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 19.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294(5547):1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 20.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26(3):449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180(5):887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 23.Xu XP, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2012;31(1):236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenn S, et al. Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J. 2011;30(11):2153–2166. doi: 10.1038/emboj.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerhold CB, et al. Structure of Actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res. 2012;40(21):11036–11046. doi: 10.1093/nar/gks842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirinakis G, et al. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011;30(12):2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol. 2007;14(6):540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Vorobiev S, et al. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA. 2003;100(10):5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W, Holmes KC. The actin fold. FASEB J. 1995;9(2):167–174. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- 31.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457(7228):441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22(12):3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz DW, et al. Accommodation of amino acid insertions in an alpha-helix of T4 lysozyme. Structural and thermodynamic analysis. J Mol Biol. 1994;236(3):869–886. doi: 10.1006/jmbi.1994.1195. [DOI] [PubMed] [Google Scholar]
- 35.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 40.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16(16):2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittmeyer J, Saha A, Cairns B. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 2004;377:322–343. doi: 10.1016/S0076-6879(03)77020-7. [DOI] [PubMed] [Google Scholar]
- 42.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12(9):747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364(6439):685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 44.Hertzog M, et al. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117(5):611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.