Abstract
Proper functioning of the mitochondrion requires the orchestrated assembly of respiratory complexes with their cofactors. Cytochrome c, an essential electron carrier in mitochondria and a critical component of the apoptotic pathway, contains a heme cofactor covalently attached to the protein at a conserved CXXCH motif. Although it has been known for more than two decades that heme attachment requires the mitochondrial protein holocytochrome c synthase (HCCS), the mechanism remained unknown. We purified membrane-bound human HCCS with endogenous heme and in complex with its cognate human apocytochrome c. Spectroscopic analyses of HCCS alone and complexes of HCCS with site-directed variants of cytochrome c revealed the fundamental steps of heme attachment and maturation. A conserved histidine in HCCS (His154) provided the key ligand to the heme iron. Formation of the HCCS:heme complex served as the platform for interaction with apocytochrome c. Heme was the central molecule mediating contact between HCCS and apocytochrome c. A conserved histidine in apocytochrome c (His19 of CXXCH) supplied the second axial ligand to heme in the trapped HCCS:heme:cytochrome c complex. We also examined the substrate specificity of human HCCS and converted a bacterial cytochrome c into a robust substrate for the HCCS. The results allow us to describe the molecular mechanisms underlying the HCCS reaction.
Keywords: CCHL, microphthalmia, biogenesis, apoptosis, post-translational modification
Renewed interest in mitochondria stems from recent associations of malfunctioning mitochondria with many cancers (1), neurological diseases (2–4), and even reduced life span (5). The basis for these associations lies in the respiratory chains that power aerobic life. 3D structures for many respiratory complexes and carriers (6–8) have elucidated the detailed mechanisms of electron transport, proton pumping, the reduction of oxygen to water, and ATP formation. Less is known about the biogenesis of these respiratory chain components. The synthesis and insertion of cofactors (e.g., heme and metals) into large, multisubunit membrane complexes represents a new frontier in the study of mitochondrial function. The c-type cytochromes, among the best-studied players in mitochondrial electron transport (9, 10), are redox-active heme proteins whose biosynthesis is only now beginning to be understood. Cytochrome c is a soluble electron carrier in the intermembrane space (IMS) of mitochondria that functions in electron transport between the quinol:cytochrome c oxidoreductase (complex III, or cytochrome bc1) and the cytochrome c oxidase (complex IV, or cytochrome a/a3). In addition to its role in mitochondrial respiration, cytochrome c plays a crucial role in apoptotic signaling (11). A second, membrane-bound c-type cytochrome, cytochrome c1, is an integral part of complex III.
C-type cytochromes differ from other cytochromes in that the heme is attached to the protein covalently via two thioether linkages between the heme vinyls and two cysteine residues of a conserved CysXxxXxxCysHis (CXXCH) heme-binding motif. Two major pathways have been identified for the biogenesis of c-type cytochromes in mitochondria: cytochrome c maturation (CCM) (12–15) and cytochrome c heme lyase, also called “holocytochrome c synthase” (HCCS, the term used in this paper) (16, 17). The CCM system is composed of eight or nine integral membrane proteins and functions in the mitochondrial inner membrane of plants and some protozoa and in the cytoplasmic membranes of alpha- and gamma-proteobacteria (18). Most mitochondria (e.g., those of fungi, invertebrates, vertebrates, and some protozoa) use HCCS for synthesis of cytochrome c. In fungi, two related homologs, HCCS and HCC1S, are dedicated to maturation of cytochrome c and cytochrome c1, respectively (19, 20), whereas in animals a single HCCS enzyme is active toward both cytochrome c and cytochrome c1 (17, 21). Additionally, in yeast and other fungi, the FAD-containing protein Cyc2p is required for heme attachment to apocytochrome c (22–24).
The human HCCS has been increasingly implicated in disease. For example, chromosomal mutations in the gene encoding HCCS can lead to a condition called “microphthalmia with linear skin defects syndrome” (25, 26). Additionally, a role for HCCS in apoptosis (separate from that of cytochrome c) has been described in injured motor neurons (27). Despite the identification of HCCS as the gene product responsible for heme attachment to cytochrome c in Saccharomyces cerevisiae more than 25 y ago (19), the enzyme has never been purified or characterized, and the mechanism of covalent heme attachment is unknown (16, 17). In yeast, HCCS is nuclear encoded and is imported directly into the mitochondrial IMS from the cytosol via the translocase of the outer membrane complex (20, 28, 29). Studies in S. cerevisiae have shown that HCCS is membrane associated in mitochondria and is exposed to the IMS (28–30). The apparent absence of transmembrane helices suggests that membrane association is likely peripheral. Pioneering studies by Sherman and colleagues (30, 31) and by Neupert and colleagues (32, 33) have demonstrated that HCCS also plays an essential role in the import of the apocytochrome c from the cytosol to the mitochondrion. It is unknown how heme enters the IMS from its site of synthesis in the mitochondrial matrix, although early studies showed that reduced heme (Fe2+) is necessary for covalent attachment to cytochrome c (34, 35). Preliminary genetic results suggested that heme binding by HCCS occurred at partially conserved cysteine–proline sequences (36), which serve as heme-regulatory motifs in several other proteins. However, neither of the cysteine–proline sequences in S. cerevisiae HCCS is required for heme attachment to cytochrome c (37), and several HCCS proteins lack cysteine–proline sequences entirely.
Both the mechanism by which HCCS mediates covalent heme attachment to the apocytochrome and the specificity determinants for recognition of heme and the apocytochrome c are poorly understood. Recombinant systems for production of mitochondrial cytochromes c in Escherichia coli, developed by Mauk and colleagues (38), have facilitated some progress in this regard (SI Appendix, Table S1) (39–41). The N-terminal region of the apocytochrome c substrate (including the CXXCH motif) is important for recognition by S. cerevisiae HCCS (reviewed in ref. 16), and a few residues in this region have been identified as important for holocytochrome c maturation (42, 43). However, the features of the cytochrome c substrate that are recognized by the human HCCS have never been examined. Here, we report successful purification and characterization of the human HCCS from recombinant E. coli. The human HCCS is membrane associated and is purified with endogenous heme coordinated by conserved His154. We define the amino acids in the human cytochrome c that are required for holocytochrome c formation by HCCS, and we successfully convert a nonsubstrate cytochrome c, cytochrome c2 from the alpha-proteobacterium Rhodobacter capsulatus, into a robust substrate for the human HCCS by introducing three sequence alterations. Finally, we report purification of trapped heme-containing complexes between cytochrome c and HCCS, with heme ligands coming from His19 (of the CXXCH motif) in cytochrome c and His154 of HCCS. We show that mutation of either cysteine in the conserved CXXCH motif of the cytochrome c leads to accumulation of trapped cytochrome c, with a single covalent attachment to the remaining cysteine, on the HCCS enzyme. Our results suggest mechanisms for heme binding, interaction with apocytochrome c, thioether formation, and a requirement for release of mature holocytochrome c from HCCS.
Results
Purified Human HCCS Contains Heme.
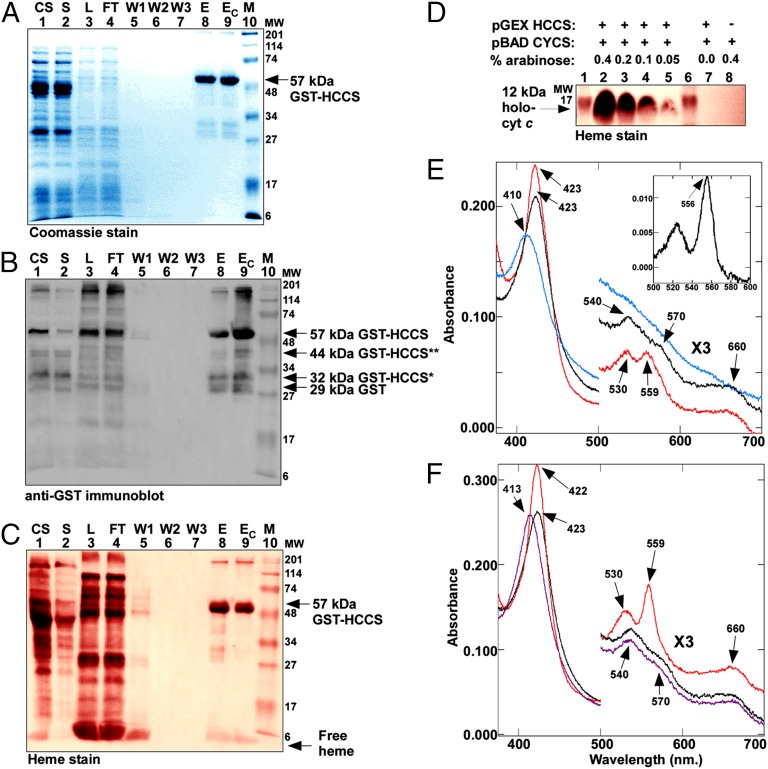
Despite longstanding interest in HCCS, the enzyme has remained refractile to successful purification and biochemical characterization (e.g., refs. 43–45). To address this problem, we engineered the cDNA for the human HCCS in three different vectors (pET Blue-2 with an N-terminal His6 tag, pTXB1 with a C-terminal Intein fusion, and pGEX with an N-terminal GST fusion) for expression and purification in E. coli. Early attempts at purifying HCCS from cytoplasmic fractions were largely unsuccessful for each of these constructs. However, upon fractionation of E. coli expressing GST-HCCS, we observed that the membrane fraction appeared to be enriched for a polypeptide of 57 kDa, the predicted size for the GST-HCCS fusion protein (SI Appendix, Fig. S1). Thus, we directed our efforts toward purifying human HCCS from membranes by solubilization in N-dodecyl β-D-maltopyranoside (DDM) (Fig. 1 A–C) or Triton X-100. Purified full-length GST-HCCS (∼57 kDa) and three minor proteolytic products each reacted with GST antisera (Fig. 1B, lane 9). Note that soluble (cytoplasmic) fractions contained mostly the degraded products and very low levels of full-length GST-HCCS (Fig. 1B, lane 2). Densitometry analysis indicated that more than 98% of the purified, Coomassie-stained protein from the membrane fraction was GST-HCCS. Analysis by LC-MS/MS confirmed that the 57-kDa species was the GST-HCCS fusion protein (SI Appendix, Fig. S2). To test the function of human HCCS, it was coexpressed with an arabinose-inducible gene encoding the human cytochrome c. Maturation of the 12-kDa holocytochrome c (Fig. 1D) confirmed that the GST-HCCS fusion protein was active in heme attachment to cytochrome c (see below). Approximately 3–4 mg of human holocytochrome c was produced per liter of E. coli culture.
Fig. 1.
Purified HCCS is a heme protein. (A) Coomassie blue staining of purified GST-tagged HCCS showing 57-kDa full-length GST-HCCS. (B) Anti-GST immunoblot of purified GST-tagged HCCS showing 57-kDa full-length GST-HCCS, 44-kDa GST-HCCS** and 32-kDa GST-HCCS* degradation products, and 29-kDa GST. (C) Heme staining of purified 57-kDa full-length GST-HCCS. For A–C abbreviations are CS, crude sonicate; E, elution; EC, concentrated elution; FT, flow through; L, load (DDM-solubilized membranes); M, molecular weight standards; S, soluble fraction; W1, wash 1; W2, wash 2; W3, wash3. (D) Heme staining of cytoplasmic extracts (B-PER) showing synthesis of the human holocytochrome c as a function of HCCS expression and arabinose concentration. The 12-kDa holocytochrome c is indicated by an arrow. One hundred micrograms of total protein was loaded in each lane. In C and D, prestained molecular weight standards were overlaid in red onto the heme stains. (E) UV-Vis absorption spectra of purified GST-HCCS (black line), reduced with sodium dithionite (red line), or oxidized with ammonium persulfate (blue line). (Inset) Sodium dithionite-reduced pyridine hemochrome spectrum of purified GST-HCCS from 500–600 nm. (F) UV-Vis absorption spectra of purified GST-HCCS (black line) in the presence of imidazole (purple line) and in the presence of imidazole reduced with sodium dithionite (red line). In E and F, the region from 500–700 nm has been multiplied by a factor of three. Absorption maxima are indicated by arrows.
Preparations of purified GST-HCCS were tinted red, and heme staining revealed that full-length GST-HCCS contained heme (Fig. 1C, lane 9). Heme proteins exhibit characteristic absorptions, referred to as the “α,” “β,” and “Soret” bands, in the UV and visible (UV-Vis) regions of the spectrum. These spectral features can provide information about the identity of the axial ligands and the spin and oxidation states of the heme iron (46). Purified human HCCS showed a Soret peak at 423 nm and three broad shoulders at 540, 570, and 660 nm (Fig. 1E, black line). Longer-wavelength Soret absorptions (e.g., 423 nm) often are characteristic of hexacoordinate, reduced heme (Fe2+) (46). Addition of the reducing agent sodium dithionite to purified GST-HCCS caused no shift in the Soret maximum. However, the Soret band sharpened (because of the loss of its lower-wavelength shoulder), absorbance increased, and small peaks appeared at 559 and 530 nm (Fig. 1E, red line). These spectral features, in combination with the 556-nm absorption in the reduced pyridine hemochrome spectrum (Fig. 1E, Inset) (47), are consistent with noncovalent, or b-type, heme. Because some heme remains with HCCS upon SDS/PAGE (Fig. 1C), we cannot absolutely rule out the possibility that some heme may be covalently bound. However, our spectral results and the detection of variable amounts of free heme in heme stains lead us to favor the idea that heme in HCCS is b-type. Addition of the oxidizing agent ammonium persulfate to purified GST-HCCS caused the Soret band to shift 13 nm to 410 nm, a wavelength typical of hexacoordinate, oxidized heme (Fe3+), and diminished the shoulders at 540, 570, and 660 nm (Fig. 1E, blue line). These observations suggest a mixture of oxidized heme and hexacoordinate, reduced heme in purified GST-HCCS.
To interrogate the heme-binding environment in HCCS further, exogenous imidazole was added to purified GST-HCCS, and UV-Vis absorption spectra were recorded. Imidazole is the side chain of histidine and often is used to probe heme–ligand interactions in vitro (e.g., refs. 48 and 49). Upon addition of 100 mM imidazole, the Soret peak shifted from 423 to 413 nm (Fig. 1F, purple line). Subsequent addition of sodium dithionite to GST-HCCS in the presence of imidazole (Fig. 1F, red line) caused a shift in the Soret peak from 413 to 422 nm and resulted in the appearance of pronounced absorptions at 530 and 559 nm. These spectral features are hallmarks of hexacoordinate-reduced heme with bis-His axial ligation but rarely are observed in hemes with histidine–cysteine, histidine–tyrosine, or histidine–methionine axial ligation (50–52). These observations are consistent with at least one natural histidine axial ligand in HCCS and an unknown axial ligand (possibly a small molecule) that can be replaced by exogenous imidazole.
His154 Is a Heme Ligand in HCCS.
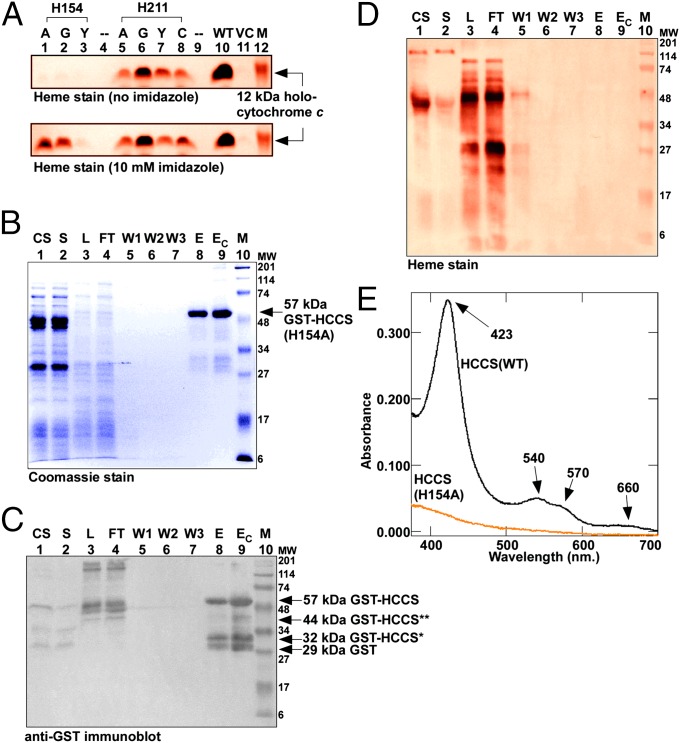
Two histidines, His154 and His211, in human HCCS are conserved in HCCSs from a wide variety of organisms (SI Appendix, Fig. S3). Given the established role of histidine residues as axial heme ligands and our spectroscopic analyses of pure HCCS, we reasoned that one or both of these histidines might coordinate heme in HCCS. We engineered substitutions of each histidine residue and, to test the function of the resulting HCCS variants, coexpressed each with WT human cytochrome c in E. coli ∆ccm (Fig. 2A, Upper). HCCS His154 is essential for heme attachment; mutation to alanine, glycine, or tyrosine completely abolished synthesis of human holocytochrome c (Fig. 2A, Upper, lanes 1–3). For His211, substitutions showed only minor effects on heme attachment (Fig. 2A, Upper, lanes 5–8).
Fig. 2.
HCCS His154 is a heme ligand. (A) Heme staining of cell extracts (B-PER) showing covalent 12-kDa holocytochrome c assembled by WT and site-directed mutants of HCCS in the E. coli cytoplasm in the absence (Upper) or presence (Lower) of 10 mM imidazole added to culture. His154 was changed to alanine (lane 1), glycine (lane 2), or tyrosine (lane 3); His211 was changed to alanine (lane 5), glycine (lane 6), tyrosine (lane 7), or cysteine (lane 8). M, molecular weight standards; VC, vector control. (B) Coomassie blue staining showing purified 57-kDa full-length GST-tagged HCCS(H154A). (C) Anti-GST immunoblot showing 57-kDa full-length GST-tagged HCCS(H154A), 44- and 32-kDa degradation products, and 29-kDa GST. (D) Heme staining of purified 57-kDa full-length GST-HCCS. For B–D abbreviations are as follows: CS, crude sonicate; E, elution; EC, concentrated elution; FT, flow through; L, load (DDM-solubilized membranes); M, molecular weight standards; S, soluble fraction; W1, wash 1; W2, wash 2; W3, wash3. For A and D, prestained molecular weight standards were overlaid in red onto the heme stains. (E) UV-Vis absorption spectra of purified GST-HCCS(H154A) (orange line) shown with the spectrum of purified GST-HCCS(WT) (black line).
To investigate further the function of His154, we tested whether the cytochrome c assembly defect of the His154 mutants could be restored by the addition of exogenous imidazole to E. coli culture. In vivo chemical complementation by imidazole has been used to correct the heme-binding function of histidine mutants for recombinant myoglobin (53) and the cytochrome c synthetases CcsBA (54) and CcmF (55). The addition of 10 mM imidazole restored the cytochrome c assembly defects of the H154A and H154G HCCS mutants to ∼40% of WT levels (Fig. 2A, Lower, lanes 1 and 2) but HCCS H154Y was not corrected (lane 3). This result is consistent with our previous findings on correction of histidine mutants in CcmF (55), which showed that amino acids with bulkier side chains (such as tyrosine) do not accommodate imidazole in the heme-binding cavity and thus are not functionally complemented. For the His211 substitutions in HCCS, imidazole addition had no effect on cytochrome c assembly (Fig. 2A, Lower, lanes 5–8). To test directly whether His154 is required for binding of heme to HCCS, we expressed and purified the nonfunctional GST-HCCS(H154A) (Fig. 2 B and C). GST-HCCS(H154A) was purified as a stable polypeptide with yields similar to those of the WT GST-HCCS, but GST-HCCS(H154A) contained no detectable heme by heme staining (Fig. 2D, lane 9) or UV-Vis absorption spectroscopy (Fig. 2E, orange line). We conclude that His154 is an axial ligand to the heme iron in HCCS.
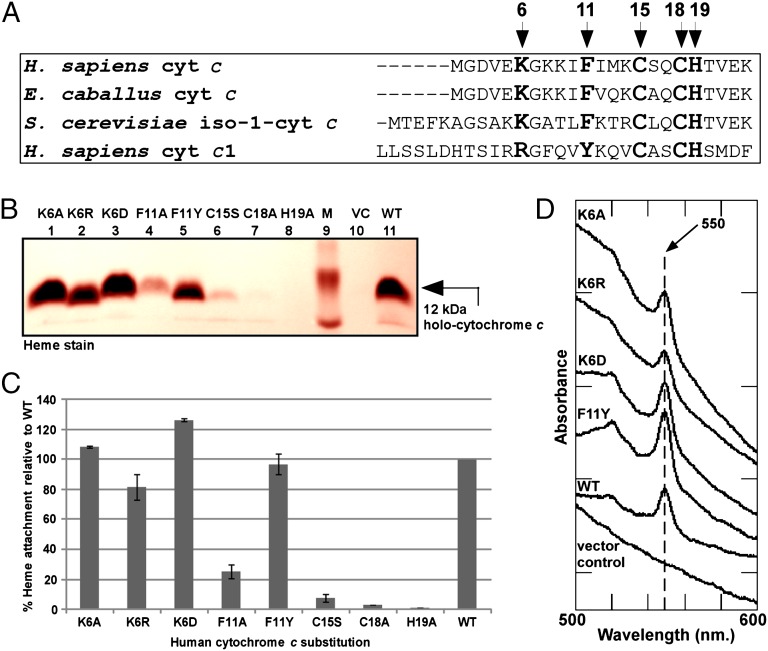
Key Determinants in Human Cytochrome c for Maturation by Human HCCS.
To define the apocytochrome c substrate requirements of human HCCS, we used the isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible human GST-HCCS and a compatible, arabinose-inducible pBAD plasmid carrying the human cytochrome c. Previous studies with S. cerevisiae HCCS (SI Appendix, Table S1) led us to focus on several conserved residues in the amino terminus of the Homo sapiens cytochrome c, namely Lys6, Phe11, Cys15, Cys18, and His19 (residues shown in bold in Fig. 3A). Throughout our report we refer to the initiation methionine of human cytochrome c as “residue 1.” Cell extracts of recombinant E. coli expressing both human HCCS and cytochrome c were assayed for covalent heme attachment (to human cytochrome c) by SDS/PAGE followed by heme staining (Fig. 3 B and C) and UV-Vis absorption spectroscopy (Fig. 3D). In the absence of HCCS, no holocytochrome c was detected by heme staining (Fig. 3B, lane 10) or spectrally (Fig. 3D, vector control). Coexpression of human HCCS and cytochrome c resulted in the production of a 12-kDa holocytochrome c (Fig. 3B, lane 11) that showed a UV-Vis absorption spectrum consistent with covalent heme attachment, based on the typical α maximum at 550 nm (Fig. 3D, WT). Replacement of Lys6 with alanine, arginine, or aspartic acid did not impair maturation of human cytochrome c (Fig. 3 B–D). Substitution of Phe11 with alanine impaired heme attachment to cytochrome c by ∼75% relative to WT under these conditions (Fig. 3 B and C). Human cytochrome c1 contains a tyrosine residue instead of the phenylalanine residue at this position (Fig. 3A); to test whether tyrosine could substitute for phenylalanine in human cytochrome c, we engineered a tyrosine substitution at Phe11. Coexpression of the F11Y variant with human HCCS resulted in levels of heme attachment equal to those in WT cytochrome c (Fig. 3 B and C), and the peak at 550 nm in the UV-Vis absorption spectrum confirmed covalent heme attachment (Fig. 3D). We also engineered substitutions at the two cysteine residues and the single histidine residue of the CXXCH motif (Cys15, Cys18, and His19) in human cytochrome c. The C15S and C18A substitutions were matured at ∼8% and <3% of WT levels, respectively (Fig. 3 B and C). Substitution of alanine for His19 resulted in undetectable levels of cytochrome c (Fig. 3 B and C).
Fig. 3.
Maturation determinants in human cytochrome c. (A) Amino acid sequence alignment of the region encompassing the heme-attachment site (CXXCH) for the indicated cytochromes c and c1. The amino acids mutated in this work are shown in bold and are indicated by arrows; numbering begins at the N-terminal methionine in H. sapiens cytochrome c. (B) Representative heme stainingof B-PER extracts showing synthesis of 12-kDa WT holocytochrome c and the indicated cytochrome c variants by the human HCCS. M, molecular weight standards; VC, vector control. One hundred micrograms of total protein was loaded in each lane. Prestained molecular weight standards were overlaid in red onto the heme stain. (C) Quantification of the results of heme staining of B-PER extracts from three independent experiments. Percent heme attachment for each variant is relative to synthesis of WT cytochrome c, which has been set at 100%. Error bars denote SD. (D) UV-Vis absorption spectra of B-PER extracts from cells expressing HCCS along with the indicated cytochrome c variant.
Our results with regard to the importance of Phe11 differ somewhat from two previous studies, which showed that an F11A variant of cytochrome c was not matured detectably by S. cerevisiae HCCS in the E. coli cytoplasm (42, 43). This difference could result from the broader substrate specificity of the human HCCS relative to the S. cerevisiae HCCS. In S. cerevisiae two related homologs, HCCS and HCC1S, are dedicated to maturation of cytochrome c and cytochrome c1, respectively, whereas in humans (and all animals), a single HCCS matures both the mitochondrial cytochrome c and cytochrome c1.
Requirements for Recognition of Bacterial c-Type Cytochromes by Human HCCS.
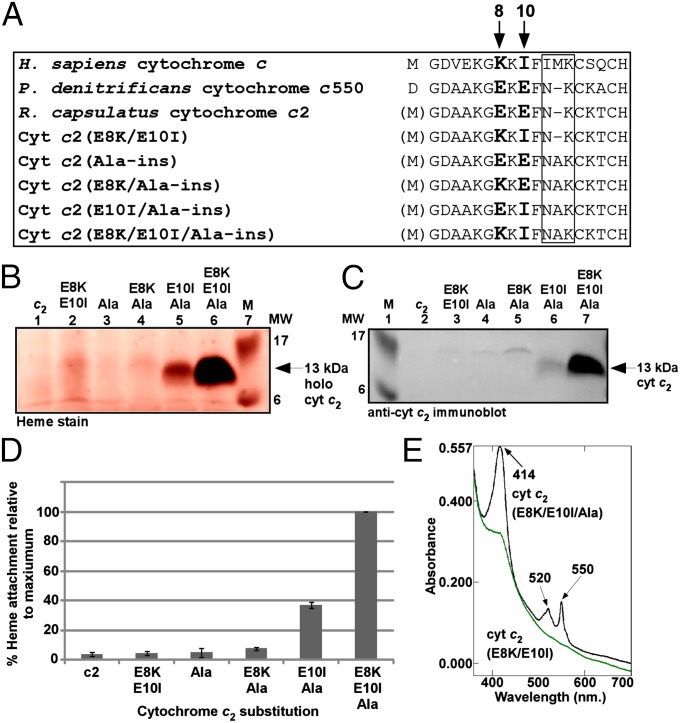
Previous studies have shown that S. cerevisiae HCCS is unable to mature certain bacterial c-type cytochromes (44) despite structural and functional similarities to the mitochondrial cytochrome c (SI Appendix, Fig. S4). Recently, it was shown that a chimeric cytochrome c composed of the N-terminal 18 amino acids of yeast cytochrome c followed by the C-terminal region of cytochrome c550 from Paracoccus denitrificans (including the CXXCH motif from P. denitrificans) could be matured by S. cerevisiae HCCS (43). When the R. capsulatus cytochrome c2, in which the methionine start codon of the human cytochrome c is the engineered initiation codon (Fig. 4A), was coexpressed with the human HCCS, holocytochrome c2 was not detected above background levels (Fig. 4B, lane 1). A notable difference between the N-terminal sequences of R. capsulatus cytochrome c2 and human cytochrome c is the amino acid spacing between the conserved phenylalanine residue (Phe11 in human cytochrome c) and the CXXCH motif (boxed region in Fig. 4A). To test whether a three-residue spacing is a requirement for recognition by human HCCS, we engineered an alanine insertion between Asn11 and Lys12 (human numbering) in R. capsulatus cytochrome c2 and coexpressed it with human HCCS (in E. coli ∆ccm). Analysis of cell extracts by heme staining showed that cytochrome c2(Ala-ins) also was not matured (Fig. 4B, lane 3).
Fig. 4.
Sequence requirements for maturation of a bacterial cytochrome c (cytochrome c2). (A) Amino acid sequence alignment of the N-terminal region (including CXXCH) for the indicated cytochrome c and cytochrome c2 variants. The amino acids mutated in this work are shown in bold and are indicated by arrows. The boxed region corresponds to the location of the alanine insertion; numbering refers to the H. sapiens cytochrome c N-terminal methionine. (B and C) Representative heme staining (B) and anti-cytochrome c2 immunoblot (C) of B-PER extracts showing synthesis of the indicated 12-kDa cytochrome c2 variants; M, molecular weight standards. One hundred micrograms of total protein was loaded in each lane. Prestained molecular weight standards were overlaid in red onto the heme stain. (D) Quantification of the results of heme staining of B-PER extracts from three independent experiments. Percent heme attachment for each indicated variant is relative to synthesis of cytochrome c2(E8K/E10I/Ala-ins), which has been set at 100%. Error bars denote SD. (E) UV-Vis absorption spectra of whole-cell extracts expressing HCCS and the indicated cytochrome c2 variant. Absorption maxima are indicated by arrows.
An additional difference between R. capsulatus cytochrome c2 and human cytochrome c is the local charge in the region preceding the conserved phenylalanine residue (Fig. 4A); specifically, cytochrome c2 contains two negatively charged glutamic acid residues (shown in bold in Fig. 4A). We engineered E8K and E10I substitutions in the WT cytochrome c2 and the cytochrome c2(Ala-ins) variant. Coexpression of cytochrome c2(E8K/E10I/Ala-ins) with the human HCCS led to the production of high levels of a 13-kDa species with covalent heme (Fig. 4 B, lane 6, and D). Immunoblotting with cytochrome c2 antisera confirmed that this covalent heme species is R. capsulatus cytochrome c2 (Fig. 4C, lane 7). UV-Vis absorption spectra confirmed covalent heme attachment to cytochrome c2(E8K/E10I/Ala-ins) (Fig. 4E). Approximately 1–2 mg of holocytochrome c2(E8K/E10I/Ala-ins) was produced per liter of E. coli culture. In contrast, cytochrome c2(E8K/E10I), containing its natural two-residue spacing between Phe11 and CXXCH, was not matured (Fig. 4B, lane 2, and D). We also engineered each single glutamic acid substitution into cytochrome c2(Ala-ins) to generate cytochrome c2(E8K/Ala-ins) and cytochrome c2(E10I/Ala-ins), and although both cytochrome c2(E8K/Ala-ins) and cytochrome c2(E10I/Ala-ins) were matured at higher levels than WT cytochrome c2 (Fig. 4B, lanes 4 and 5, and D), neither was as high as the double substitution.
While our manuscript was in preparation, a report was published on the conversion of R. capsulatus cytochrome c2 into a substrate for S. cerevisiae HCCS (56), with results that are consistent with our findings using the human HCCS. In their study, Verissimo et al. (56) engineered a lysine insertion instead of an alanine insertion and replaced Glu10 with leucine instead of isoleucine. Our results here indicate that positively charged and neutral residues at positions 8 and 10, respectively, and proper spacing between Phe11 and CXXCH constitute the minimal determinants for maturation by human HCCS. We describe the evolutionary implications of these results in SI Appendix.
HCCS Copurifies with Human Cytochrome c.
To investigate further the interactions between HCCS and the substrate cytochrome c, we coexpressed several of the human cytochrome c variants (F11A, C15S, C18A, or H19A) or WT cytochrome c with the human HCCS and analyzed the soluble and membrane fractions. We noticed that the detergent-solubilized membrane fractions from cells expressing the C15S or the C18A cytochrome c variant (along with HCCS) had higher levels of holocytochrome c than the soluble fractions (SI Appendix, Fig S5 C and D, lanes 3, L fractions). Membrane fractions from cells expressing the WT, F11A, or H19A variants of cytochrome c contained low levels of holocytochrome c (SI Appendix, Fig S5 A, B, and E, lanes 3, L fractions).
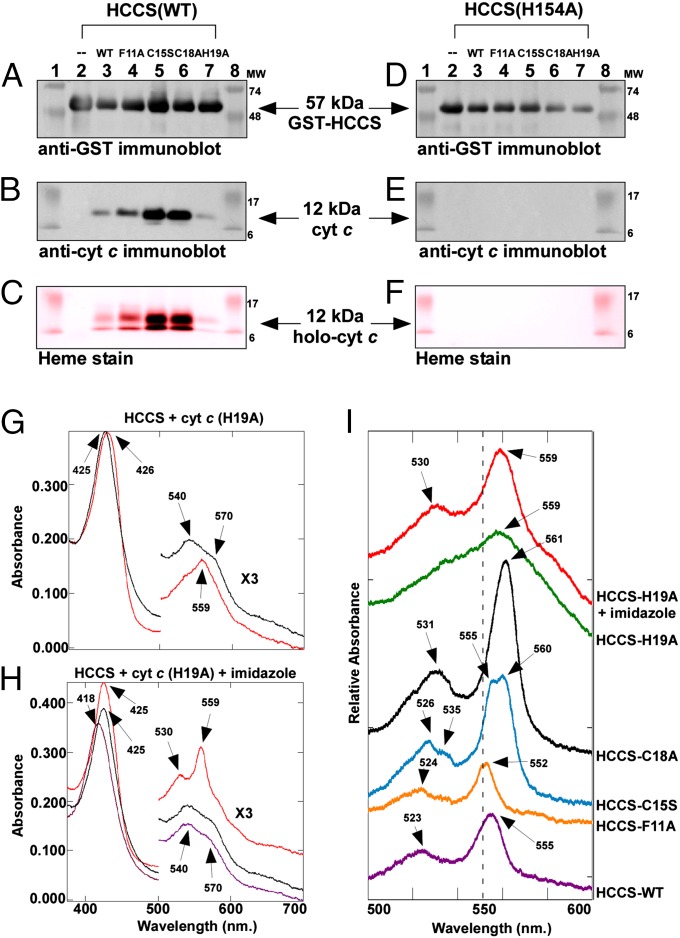
To investigate whether the apparent membrane localization of the cytochrome c variants was related to the association of HCCS with the E. coli membrane, we purified GST-HCCS and analyzed each preparation for the presence of the human cytochrome c. In each case, GST-HCCS was purified from detergent-solubilized membranes as a stable polypeptide with minimal degradation (Fig. 5A). In addition to the full-length GST-HCCS fusion protein (∼57 kDa), we observed in the elution fraction a 12-kDa polypeptide of the molecular mass for cytochrome c (SI Appendix, Fig. S6A). Analysis by LC-MS/MS (SI Appendix, Fig. S7) and immunoblotting with cytochrome c antisera (Fig. 5B) confirmed that cytochrome c copurified with GST-HCCS. Table 1 quantifies the heme and apocytochrome bound in each complex and summarizes the spectral properties of each complex. Heme staining after SDS/PAGE revealed that the WT cytochrome c and each of the variants contained heme (although at very low levels for the H19A variant) (Fig. 5C). Covalent heme attachment to the 12-kDa cytochrome c was confirmed by retention of the heme through SDS/PAGE after boiling and treatment with 8 M urea (SI Appendix, Fig. S8). HCCS complexes with C15S and C18A variants contained approximately eightfold more heme than HCCS alone, and the WT, F11A, and H19A complexes contained approximately fourfold more heme (Table 1, column C). Total heme in each complex was proportional to the amount of cytochrome c polypeptide bound in each complex (Table 1, column B), suggesting that the interaction of HCCS with heme is stabilized by the presence of the cytochrome c acceptor. To determine the role of heme in formation of the complex between HCCS and cytochrome c, we investigated whether HCCS(H154A), which is defective in heme binding (Fig. 2), copurified with any of the cytochrome c variants. Purified HCCS(H154A) (Fig. 5D) contained no detectable cytochrome c (Fig. 5E) or heme (Fig. 5F), just as HCCS(H154A) alone had no heme. The failure of HCCS(H154A) to copurify with cytochrome c indicates that heme binding by HCCS (through His154) is a requirement for interaction with the apocytochrome c.
Fig. 5.
Cytochrome c copurifies with HCCS. (A) Anti-GST immunoblot showing full-length 57-kDa GST-HCCS, (B) anti-cytochrome c immunoblot showing 12-kDa cytochrome c, and (C) heme staining showing 12-kDa holocytochrome c, from elution fractions of copurifications of each cytochrome c variant with GST-HCCS(WT). (D) Anti-GST immunoblot showing full-length 57-kDa GST-HCCS, (E) anti-cytochrome c immunoblot, and (F) heme staining from elution fractions of copurifications of each cytochrome c variant with GST-HCCS(H154A). For A–F, 5 µg of purified total protein was loaded in each lane. See SI Appendix, Fig. S6 for full panels. For C and F, prestained molecular weight standards were overlaid in red onto the heme stains. (G) UV-Vis absorption spectra of GST-HCCS copurified with cytochrome c(H19A) (black line) and reduced with sodium dithionite (red line). (H) UV-Vis absorption spectra of GST-HCCS copurified with cytochrome c(H19A) (black line), in the presence of imidazole (purple line), and in the presence of imidazole reduced with sodium dithionite (red line). The region from 500–700 nm has been multiplied by a factor of three. (I) The α-β region showing the characteristic sodium dithionite-reduced UV-Vis absorption spectrum for each indicated cytochrome c variant copurified with GST-HCCS. Spectra have been offset for clarity. Absorption maxima are indicated by arrows.
Table 1.
Characterization of the complexes of HCCS with cytochrome c
| HCCS complex | A | B | C | D | E | F | G | H | I |
| GST-HCCS levels1 | Cytochrome c bound (in complex)* | Total heme (in complex) † | Covalent 12-kDa heme (in complex) ‡ | Released holo-cytochrome c§ | Reduced α band absorption§ | First heme axial ligand¶ | Second heme axial ligand¶ | Thioether bond formation¶ | |
| HCCS alone | 1.01 ± 0.04 | – | 0.27 ± 0.09 | – | – | 559 | HCCS His154 | Unknown | None |
| HCCS + cytochrome c (WT) | 1 | 1 | 1 | 1 | 1 | 555 | HCCS His154 | Cytochrome c His19 | Cys15-2-vinyl |
| Cys18-4-vinyl | |||||||||
| HCCS + cytochrome c (F11A) | 1.03 ± 0.14 | 0.94 ± 0.14 | 0.83 ± 0.13 | 1.02 ± 0.02 | 0.25 ± 0.05 | 552 | HCCS His154 | Cytochrome c His19 | Cys15-2-vinyl |
| Cys18-4-vinyl | |||||||||
| HCCS + cytochrome c (C15S) | 1.00 ± 0.11 | 2.07 ± 0.10 | 1.84 ± 0.01 | 2.22 ± 0.24 | 0.08 ± 0.03 | 555/560 split | HCCS His154 | Cytochrome c His19 | Cys18-4-vinyl |
| HCCS + cytochrome c( C18A) | 0.98 ± 0.16 | 1.97 ± 0.07 | 1.93 ± 0.08 | 2.24 ± 0.36 | 0.03 ± 0.01 | 561 | HCCS His154 | cytochrome c His19 | Cys15-2-vinyl |
| HCCS + cytochrome c (H19A) | 1.00 ± 0.15 | 0.83 ± 0.02 | 0.89 ± 0.01 | 0.25 ± 0.10 | Not detected | 559 | HCCS His154 | Unknown | Cys15-2-vinyl |
| Cys18-4-vinyl | |||||||||
| Mature cytochrome c (WT) | – | – | – | – | – | 550 | Met80 | His19 | Cys15-2-vinyl |
| Cys18-4-vinyl |
Each value is relative to the complex of HCCS with WT cytochrome c, which has been set at 1, and is based on at least three separate experiments.
*The amount of GST-HCCS and cytochrome c protein in each complex was calculated by densitometry analysis of the chemiluminescent signal from anti-GST and anti-cytochrome c immunoblots, respectively.
†Total heme in each complex was calculated from the Soret absorption in the UV-Vis absorption spectrum and adjusted for total protein concentration.
‡Covalent 12-kDa heme in each complex and released holocytochrome c in B-PER fractions were calculated by densitometry analysis of the chemiluminescent signal from heme stains.
§The reduced α absorption (nm) for each complex is from the UV-Vis absorption spectrum in the presence of sodium dithionite.
¶The axial ligands to the heme in each complex and thioether formation between the Cys residues in the cytochrome c and the heme vinyls are based on experimental data shown here.
Spectral Analyses of the HCCS:Heme:Cytochrome c Complexes.
UV-Vis spectra were recorded on each of the purified complexes to gain additional insight into the heme–protein interactions within each complex. In the absence of residue His19 of apocytochrome c (i.e., the HCCS:H19A complex), the UV-Vis absorption spectrum is reminiscent of the spectrum of HCCS alone, with a Soret absorption peak at 425 nm and broad absorptions at 540 and 570 nm (Fig. 5G, black line). Addition of the reducing agent sodium dithionite to the purified HCCS:H19A complex caused minor changes in the Soret peak and the α-β region of the spectrum (Fig. 5G, red line). To probe ligand interactions in the HCCS:H19A complex further, we added 100 mM imidazole to purified HCCS-H19A and recorded spectra (Fig. 5H, purple line). The effect of imidazole addition on heme in the HCCS:H19A complex was similar to that observed with HCCS alone, with a blue-shift in the Soret band to 418 nm. Upon addition of sodium dithionite to HCCS:H19A in the presence of imidazole, the Soret band shifted to 425 nm, and pronounced α and β absorptions were observed at 559 and 530 nm (Fig. 5H, red line). These results suggest that in the HCCS:heme:cytochrome c(H19A) complex His154 of HCCS provides one axial ligand and that exogenous imidazole can occupy the second axial coordination site. In contrast, the spectra of each of the other complexes of cytochrome c with HCCS (Fig. 5I; full spectra are shown in SI Appendix, Fig. S9) were markedly different from that of HCCS alone, and imidazole caused no change in the spectra of these complexes (SI Appendix, Fig. S10). We propose that, in the C15S, C18A, F11A, and WT complexes of cytochrome c with HCCS, heme is coordinated by His154 of HCCS and His19 of cytochrome c.
To analyze the heme environment in each complex (Table 1, columns G–I), we focused on the sodium dithionite-reduced absorptions in the α-β region of the spectrum (500–600 nm; see Fig. 5I), which are diagnostic for covalent heme attachment and ligand interactions with the heme iron (46). The reduced spectrum of the HCCS:WT cytochrome c complex exhibited α and β maxima at 555 and 523 nm, respectively (Fig. 5I, purple line). The complex of HCCS with the F11A variant of cytochrome c showed α and β absorptions at 552 and 524 nm (Fig. 5I, orange line). The HCCS:C15S complex exhibited a unique and rare split α maximum (57) at 555–560 nm and a split β peak at 526–535 nm (Fig. 5I, blue line). We note that a split α absorption has been observed at room temperature for heme in the CcmCDE complex (58, 59), where, similarly, there is a single covalent attachment to a heme vinyl and bis-His coordination of the heme iron. The HCCS:C18A complex exhibited a strong α absorption at 561 nm and a β peak at 531 nm (Fig. 5I, black line). For the C18A variant, because the α maximum was atypical for a c-type heme (i.e., 561 nm absorption; Fig. 5I, black line), we confirmed covalent attachment of the heme group to the protein by MS; a trypsinized peptide corresponding to the heme-binding motif of this variant plus covalent heme was detected (SI Appendix, Fig. S11).
Thus, for each complex of cytochrome c with HCCS, there are signature absorptions in the reduced UV-Vis spectrum that are diagnostic for the specific heme–protein interactions of that complex. We note that the spectra of the complexes of HCCS with WT cytochrome c and the C15S variant are markedly different from the spectra that have been reported for released, mature WT holocytochrome c (41, 44) and site-directed variants of Cys15 (60, 61), respectively. These differences support our contention that the spectra of the complexes are indeed reflective of interactions with the heme by both the HCCS and the cytochrome c. We propose that these trapped complexes provide insight into intermediates on the path to full maturation and release of the human cytochrome c by the human HCCS.
Discussion
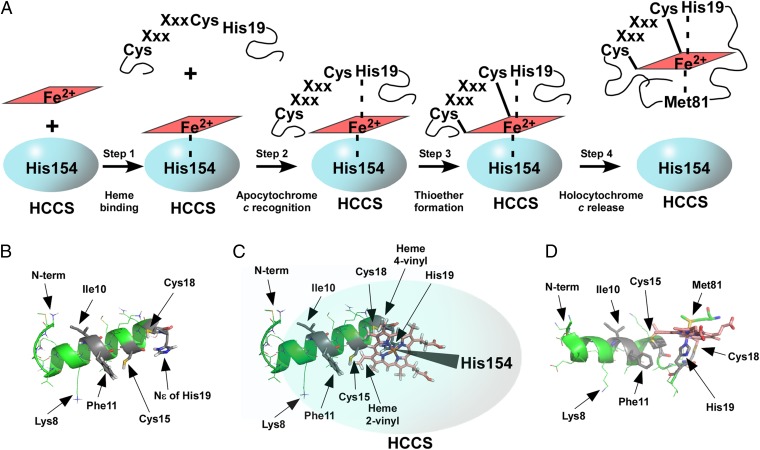
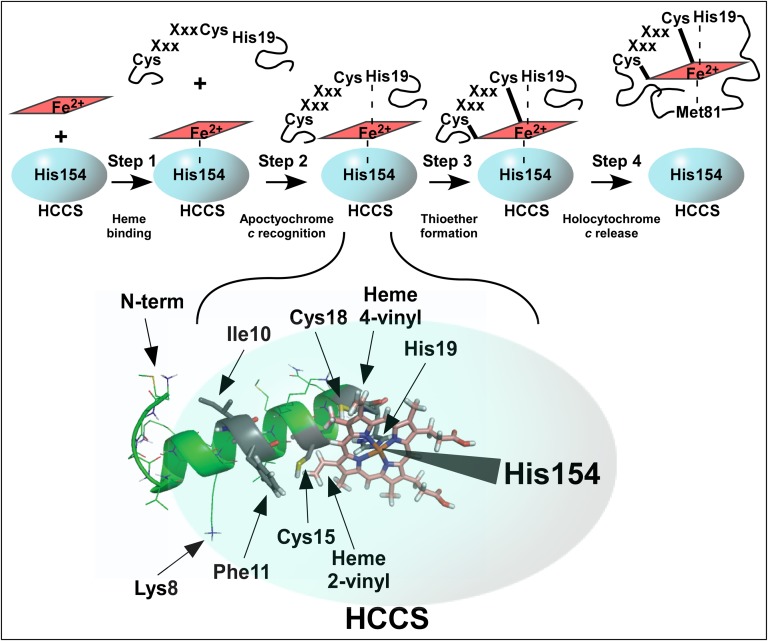
Our results are discussed in the context of four distinct steps in the synthesis of holocytochrome c by the human HCCS: heme binding, apocytochrome c substrate recognition, thioether formation, and release of holocytochrome c (Fig. 6A). We also address long-standing questions about the mechanisms of heme binding by HCCS and the roles of individual residues in both HCCS and the apocytochrome c substrate. A major conclusion is that heme is the central molecule mediating interaction between HCCS and apocytochrome c.
Fig. 6.
Steps in the maturation of cytochrome c by human HCCS. (A) Diagram depicting the four steps in holocytochrome c maturation by HCCS (discussed in text). (B) PEP-FOLD generated model of the N-terminal 20-aa helix in human cytochrome c before interaction with HCCS. Amino acids important for maturation are indicated by arrows. Selected oxygen atoms are shown in red, nitrogen in blue, sulfur in yellow, the heme macrocycle in pink, and the iron atom at the heme center in orange. (C) The HCCS:heme:cytochrome c complex (modeled with heme shown in pink) before thioether formation. Heme is coordinated by His19 of apocytochrome c (PEP-FOLD ribbon diagram, green, as in B) and His154 of HCCS (overlaid cartoon, blue). Amino acids and heme vinyls important for maturation are indicated by arrows. (D) The 20 N-terminal amino acids from the X-ray crystal structure of mature, released E. caballus holocytochrome c (PBD ID HRC1). Amino acids important for maturation are indicated by arrows. Heme is coordinated by His19, as in the complex with HCCS, and cytochrome c Met81, which replaces HCCS His154 in the folded, released holocytochrome c. PyMOL was used for displays in B–D. Amino acid numbering refers to the H. sapiens cytochrome c N-terminal methionine, as shown in Fig 3A (because the initiation methionine is processed off, PBD ID HRC1 refers to Lys8 as Lys7, His19 as His18, Met81 as Met80, and so forth).
Step 1. Heme Binding by HCCS.
Human HCCS is purified from membrane fractions as a full-length GST fusion protein with endogenous heme. Given that the function of HCCS is to attach heme covalently to apocytochrome c, heme binding by this enzyme has long been suspected (34, 36, 62) but never has been shown directly. Spectral studies provided insight into the HCCS heme-binding environment, suggesting that one ligand to the heme iron was a histidine and that the second axial coordination site was occupied by an unknown ligand that could be replaced by exogenous imidazole in vitro. Various approaches were used to demonstrate that residue His154 in HCCS was an axial ligand to the heme in HCCS (Fig. 2). First, HCCS(H154A) did not purify with heme. Additionally, HCCS(H154A) was not functional in holocytochrome c synthesis, but activity was corrected in vivo by addition of exogenous imidazole. Finally, HCCS(H154A) did not form ternary complexes with heme and cytochrome c (Fig. 5). Therefore, we conclude that the heme-bound form of HCCS binds the apocytochrome c (see step 2).
Step 2. Recognition of Apocytochrome c by HCCS:Heme.
We undertook two approaches to define the substrate determinants in the apocytochrome c that human HCCS requires for formation of a mature holocytochrome c. The first approach was to identify key residues in the N terminus of the cognate human apocytochrome c that are required for holocytochrome formation; the second was to convert a nonsubstrate bacterial cytochrome c (R. capsulatus cytochrome c2) into a substrate for human HCCS. For the human apocytochrome c, residues Phe11, Cys15, Cys18, and His19 were important (although to differing degrees) for optimal holocytochrome c maturation (Fig. 3C). Maturation of cytochrome c2 by HCCS required the insertion of an alanine residue between Phe11 and Cys15 and substitution of residues Glu8 and Glu10 (Fig. 4). Predicted structures of the cytochrome c2 N terminus demonstrate that the alanine insertion significantly repositions Phe11 and the two glutamic acid residues (SI Appendix, Fig. S12, compare A and B); therefore, both amino acid spacing and charge (SI Appendix, Fig. S12, compare A and C) are critical for heme attachment by HCCS. Fig. 6B shows the predicted structure of the first 20 residues in the human apocytochrome c, with each of the critical determinants highlighted.
We gained remarkable insight into HCCS substrate recognition when we discovered that each human cytochrome c variant (F11A, C15S, C18A, and H19A) copurified with HCCS (Fig. 5 and Table 1). Each of the HCCS:heme:cytochrome c complexes comprised a unique heme environment (Fig. 5I), suggesting that each variant complex reflects a distinct intermediate in maturation. Thus, these variants are not defective in recognition by and binding to HCCS:heme (step 2) but are defective in a later step in maturation, as described below. Our results are consistent with prior genetic studies in yeast, in which a class of mutant apocytochrome c (including variants at Cys15, Cys18, and His19) was found to be competent for mitochondrial import (in an HCCS-dependent manner) and therefore still was recognized by HCCS but was not matured into holocytochrome c (63).
Fig. 6C diagrams the central role of heme in the formation of the complex of HCCS (blue) with the apocytochrome c N terminus (PEP-FOLD–generated structure, green). Each complex of HCCS with apocytochrome c (including HCCS:H19A) contained more total heme than did HCCS alone (Table 1, column C), indicating that apocytochrome c binding stabilizes heme in the HCCS:heme:cytochrome c ternary complex (Fig. 6A, step 2). His19 of the apocytochrome CXXCH supplies the second axial ligand to heme iron in the complex with HCCS, and HCCS His154 provides the first axial ligand (Fig. 6C). This conformation is suggested by spectral analyses of the HCCS:heme:cytochrome c(H19A) complex compared with the complexes with WT and other cytochrome c variants. That the HCCS:heme:cytochrome c(H19A) complex contained more total heme than HCCS alone further indicates that other residues in apocytochrome c (in addition to His19) likely interact with heme. The spectral features and biochemical characterization of these ternary complexes also inform us of the later steps in holocytochrome c synthesis.
Step 3. Formation of Thioether Bonds.
Each of the complexes of cytochrome c with HCCS contained at least WT levels of covalent 12-kDa heme, with the exception of the H19A variant (Table 1, column D). Although heme in the complex with HCCS still was stabilized by binding of cytochrome c(H19A) (Table 1, column C), thioether formation was very low for this variant. We conclude that His19 of CXXCH in the apocytochrome c is essential for efficient thioether formation. We propose that coordination of the heme iron by His19 in the HCCS:heme:cytochrome c complex positions the two cysteines for stereospecific ligation to reduced (Fe2+) heme, preparing it for thioether bond formation. This proposal is consistent with the effect of exogenous imidazole on the heme environment in the HCCS:heme:cytochrome c(H19A) complex, as described in results (Fig. 5 H and I). The essential role of His19 in step 3 is supported further by the complete absence of released holocytochrome c(His19) (Table 1, column E, and Fig. 3 B and C).
Holocytochrome C15S and C18A were bound to HCCS at steady-state levels twofold higher than the WT holocytochrome c (Table 1, columns B and D). Because the levels of mature holocytochrome c were very low relative to WT (8% for C15S and <3% for C18A), it appears that these variants are trapped in HCCS, unable to undergo release. Thus, the formation of a covalent bond at both cysteines appears to be required for efficient release from HCCS. Interestingly, Wang et al. (63) observed a class of mutant apocytochrome c (including variants in the cysteine residues of CXXCH) whose expression inhibited the maturation of normal cytochrome c when coexpressed in S. cerevisiae. They speculated that this class of variant apocytochrome might form “dead-end” complexes with HCCS. Here, we show that the cysteine variant apocytochromes indeed are bound in nonproductive complexes with HCCS, consistent with the “dominant” cytochrome c variants observed in the aforementioned study by Wang et al. (63). It is striking that the ratio of released to trapped WT cytochrome c is one hundred times higher than in the C18A variant, highlighting the consequences of thioether formation for full release and maturation. Additionally, that both the C15S and C18A single-cysteine complexes contained covalent heme suggests that there may not be a preferred order for thioether formation.
Step 4. Release of Holocytochrome c from the Complex.
In addition to the requirement for formation of two thioether bonds (step 3), optimal production of mature holocytochrome c requires a mechanism for release from HCCS (Fig. 6A). A number of studies on mitochondrial importation of apocytochrome c in whole yeast cells (30, 33, 63) and in isolated yeast mitochondria (31, 64) have concluded that apocytochrome residues important for heme attachment may represent only a subset of the residues involved in mitochondrial import. Thus, in addition to Phe11, Cys15, Cys18, and His19, there are likely multiple residues in the apocytochrome that mediate interaction with HCCS:heme. We propose that a dedicated release mechanism (step 4) is essential for the cognate human holocytochrome c because of multiple, stable interactions between HCCS:heme and the apocytochrome that are related to HCCS-mediated import of the apocytochrome c into the mitochondrion.
Binding of the human HCCS to its cognate cytochrome c in the ternary complex is surprisingly stable, as demonstrated by the fact that the HCCS:heme:cytochrome c complex can be purified (in the oxidized state) and maintained in vitro. We speculate that, in the cell, the proper milieu (e.g., reducing environment) promotes folding of the WT holocytochrome c, which provides the energy to release heme (and the holocytochrome) from the HCCS active site. Although we have not addressed the mechanisms in step 4 directly in our study, a rich literature exists on the analysis of holocytochrome c folding in vitro, which is useful to consider here. For example, reduced cytochrome c (Fe2+) shows a notable difference in folding relative to oxidized cytochrome c (Fe3+), exhibiting greater stability toward denaturation (65). Even the binding of small axial ligands such as carbon monoxide, which can replace the natural methionine axial ligand, has a significant impact on the folding and unfolding processes (66). Thus, the importance of reduced heme (Fe2+) both for thioether formation and for holocytochrome c release might be considered for HCCS-mediated cytochrome c maturation. Possible factors involved in this reduction in mitochondria have been described (24). Release of holocytochrome c from the ternary complex clearly demands that HCCS axial ligand His154 be discharged from the heme iron (Fig. 6C); this discharge may be facilitated by interaction of the heme with other residues in the cytochrome c during folding. Although the second axial coordination site in mature holocytochrome c is occupied by Met81 (Fig. 6D), there is ample evidence that this methionine residue can be replaced and still yield mature cytochrome c (67). Therefore, other residues in cytochrome c may play a role in displacing HCCS His154 from the heme during folding. Besides a molecular understanding of the mechanisms underlying HCCS function, our study provides the foundation for future analyses of other key residues in the human HCCS and the cytochrome c and the framework for delineating the steps in which those residues are involved.
Materials and Methods
Bacterial Growth Conditions.
E. coli strains (SI Appendix, Table S2) were grown at 37 °C in LB broth (Difco) supplemented with the appropriate antibiotics (Sigma-Aldrich) and other medium additives at the following concentrations, unless otherwise noted: carbenicillin, 50 µg⋅mL−1; chloramphenicol, 20 µg⋅mL−1; isopropyl-β-D-thiogalactopyranoside IPTG (Gold Biotechnology), 0.1 mM; arabinose (Gold Biotechnology) 0.2% (wt/vol).
Protein Expression and Purification.
For HCCS expression (with or without coexpression of cytochrome c), E. coli ∆ccm strain RK103 (68) or RK112 (69) was used. Starter cultures were initiated from a single colony and grown overnight at 37 °C with shaking at 200 rpm in 100 mL of LB medium with the appropriate antibiotics. One liter of LB was inoculated to 10% and was grown with shaking at 120 rpm for 1 h (OD600 <1.0), at which point the culture was induced with 0.1 mM IPTG for pET Blue-2–, pTXBI-, or pGEX-based expression for 5 h. For coexpression of pBAD-based cytochrome c, the culture was induced with 0.2% arabinose (wt/vol) 3 h after inoculation (2 h after induction of HCCS expression) and grown for an additional 3 h. Cells were harvested at 5,000 × g for 10 min and frozen at −80 °C overnight. Cell pellets were thawed and resuspended in PBS (100 mM NaCl, 7.5 mM Na2HPO4, 2 mM NaH2PO4) and treated with 1 mM PMSF (Sigma-Aldrich;) and 100 µg⋅mL−1 egg white lysozyme (Sigma-Aldrich ) for 30 min with shaking on ice. Cells were disrupted by repeated sonication for 30-s bursts with a Branson 250 sonicator (50% duty, 60% output) until clearing of the suspension was observed. Crude sonicate was centrifuged at 24,000 × g for 15 min to clear cell debris, and membranes were isolated by centrifugation at 100,000 × g for 45 min. Membrane pellets were solubilized in a modified 1× GST buffer (150 mM NaCl, 50 mM Tris, pH 8) (Pierce) with 1% (wt/vol) DDM (Anatrace) or Triton X-100 (Sigma-Aldrich) on ice for 1 h. Detergent-solubilized membranes were centrifuged at 24,000 × g for 15 min to remove unsolubilized material. Solubilized membranes (L; load) were passed over glutathione agarose (Pierce) per the manufacturer’s recommendations and were washed in 1× modified GST buffer with 0.02% (wt/vol) DDM or Triton X-100. Bound GST-tagged protein was eluted in 1× modified GST buffer containing 0.02% (wt/vol) DDM or Triton X-100 and 20 mM reduced glutathione (E; elution). The purified protein was concentrated and subjected to buffer exchange in an Amicon Ultra Centrifugal Filter 30,000 MWCO (Millipore) after purification (EC; concentrated elution). Total protein concentration was determined using Bradford Reagent (Sigma-Aldrich) per the manufacturer’s instructions, and samples were measured with a PowerWave XS2 Microplate Spectrophotometer (BioTek).
Cytochrome Reporter and Imidazole Complementation Assays.
Human holocytochrome c and R. capsulatus holocytochrome c2 production were assayed in RK103 (68) harboring pGEX:GST-HCCS and a pBAD-based plasmid [human cytochrome c: K6A, K6R, K6D, F11A, C15S, C18A, H19A, WT; R. capsulatus cytochrome c2: c2(WT), c2(Ala-ins), c2(E8K/Ala-ins), c2(E10I/Ala-ins), c2(E8K/E10I), c2(E8K/E10I/Ala-ins)] (SI Appendix, Table S2). Cultures were grown at 37 °C with shaking at 200 rpm in 5 mL LB with appropriate antibiotics for 3 h and were induced for an additional 3 h with 0.8% arabinose (wt/vol) and 1 mM IPTG. Cells were harvested by centrifugation at 10,000 × g, and the cell pellet was resuspended in 200 µL of bacterial protein extraction reagents (B-PER; Thermo Scientific) to lyse cells and extract protein. Total protein concentration was determined using the Nanodrop 1000 spectrophotometer (Thermo Scientific), and 100 µg was analyzed by SDS/PAGE followed by heme staining. Imidazole complementation assays were performed in the same way, except that 10 mM imidazole (pH 7) was added to culture before inoculation.
Heme Stains and Other Methods.
Heme stains and immunoblots were performed as described in ref. 70. B-PER fractions were mixed 1:1 (vol/vol) with loading dye without reducing agents and were boiled for 5 min, separated by 12.5% SDS/PAGE, and transferred to Hybond C nitrocellulose membranes (GE Healthcare). Purified proteins (unboiled) were mixed 1:1 (vol/vol) with loading dye without reducing agents before separation by 12.5% SDS/PAGE and transfer to nitrocellulose. Antiserum to human cytochrome c (Santa Cruz) was used at a dilution of 1:5,000. Anti-GST antibody (Sigma-Aldrich) was used at a dilution of 1:10000. Protein A peroxidase (Sigma-Aldrich) was used as the secondary label. The chemiluminescent signal for heme staining and anti-cytochrome c immunoblots was developed with the SuperSignal Femto kit (Thermo Scientific) or the Immobilon Western kit (Millipore) for anti-GST immunoblots. Chemiluminescent signal was detected with the LAS-1000 Plus detection system (Fujifilm-GE Healthcare) or the ImageQuant LAS-4000 Mini detection system (Fujifilm-GE Healthcare). Heme concentration in purified preparations was determined by pyridine extraction as described in ref. 47, heme staining as described in ref. 71, or quantification of the indicated UV-Vis spectral absorptions. The relative abundances of purified GST-HCCS and copurified human cytochrome c were quantified by densitometry analysis of the chemiluminescent signal from anti-GST and anti-cytochrome c immunoblots, respectively, and were normalized to WT levels. Protein purity was assessed by Coomassie Blue staining of SDS/PAGE.
UV/Vis Absorption Spectroscopy.
UV-Vis absorption spectra were recorded with a Shimadzu UV-2101 PC UV-Vis scanning spectrophotometer at room temperature as described in ref. 54. All spectra were recorded in same buffer in which the proteins were purified: 150 mM NaCl, 50 mM Tris (pH 8), 0.02% (wt/vol) DDM or Triton X-100. Chemical reduction and oxidation of samples were achieved by the addition of several grains of sodium dithionite (sodium hydrosulfite) or ammonium persulfate, respectively. To analyze the effect of imidazole on the electronic spectrum, small quantities of a concentrated imidazole solution (1 M, pH 7) were added to purified protein, and spectra were recorded.
3D Models.
3D models of the N terminus of human cytochrome c and the cytochrome c2 variants from R. capsulatus were generated by PEP-FOLD, an online resource for de novo 3D peptide structure prediction (http://bioserv.rpbs.univ-paris-diderot.fr/PEP-FOLD) (72, 73). Each of the peptides shown represents the lowest energy conformation of the five structures generated by PEP-FOLD. X-ray crystal structures for Equus caballus cytochrome c [Protein Data Bank (PDB) ID code HRC1] and R. capsulatus cytochrome c2 (PDB ID code 1c2r) were obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (www.rcsb.org/pdb/home/home.do).
Supplementary Material
Acknowledgments
We thank Huifen Zhu, John D’Allesandro, Cindy Richard-Fogal, Jing Jiang, Shalon Ledbetter, and the Washington University in St. Louis Bio437 class (Fall 2011) for technical contributions. We thank Kenton Rodgers for advice on the spectra of purified HCCS. This study was supported by National Institutes of Health Grant R01 GM47909 (to R.G.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 3221 (volume 110, number 9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213897109/-/DCSupplemental.
References
- 1.Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutr Metab (Lond) 2010;7:7. doi: 10.1186/1743-7075-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vos M, et al. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science. 2012;336(6086):1306–1310. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su B, et al. Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochim Biophys Acta. 2010;1802(1):135–142. doi: 10.1016/j.bbadis.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunnari J, Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci USA. 2002;99(5):2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun F, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121(7):1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 8.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272(5265):1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 9.Dickerson RE, et al. Ferricytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J Biol Chem. 1971;246(5):1511–1535. [PubMed] [Google Scholar]
- 10.Hampsey DM, Das G, Sherman F. Yeast iso-1-cytochrome c: Genetic analysis of structural requirements. FEBS Lett. 1988;231(2):275–283. doi: 10.1016/0014-5793(88)80834-2. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 12.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 2009;73(3):510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: The Ccm system. Trends Microbiol. 2010;18(6):266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawyer EB, Barker PD. Continued surprises in the cytochrome c biogenesis story. Protein Cell. 2012;3(6):405–409. doi: 10.1007/s13238-012-2912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens JM, et al. Cytochrome c biogenesis System I. FEBS J. 2011;278(22):4170–4178. doi: 10.1111/j.1742-4658.2011.08376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen JW. Cytochrome c biogenesis in mitochondria—Systems III and V. FEBS J. 2011;278(22):4198–4216. doi: 10.1111/j.1742-4658.2011.08231.x. [DOI] [PubMed] [Google Scholar]
- 17.Hamel P, Corvest V, Giegé P, Bonnard G. Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim Biophys Acta. 2009;1793(1):125–138. doi: 10.1016/j.bbamcr.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Beckman DL, Trawick DR, Kranz RG. Bacterial cytochromes c biogenesis. Genes Dev. 1992;6(2):268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- 19.Dumont ME, Ernst JF, Hampsey DM, Sherman F. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 1987;6(1):235–241. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zollner A, Rödel G, Haid A. Molecular cloning and characterization of the Saccharomyces cerevisiae CYT2 gene encoding cytochrome-c1-heme lyase. Eur J Biochem. 1992;207(3):1093–1100. doi: 10.1111/j.1432-1033.1992.tb17146.x. [DOI] [PubMed] [Google Scholar]
- 21.Bernard DG, Gabilly ST, Dujardin G, Merchant S, Hamel PP. Overlapping specificities of the mitochondrial cytochrome c and c1 heme lyases. J Biol Chem. 2003;278(50):49732–49742. doi: 10.1074/jbc.M308881200. [DOI] [PubMed] [Google Scholar]
- 22.Dumont ME, et al. CYC2 encodes a factor involved in mitochondrial import of yeast cytochrome c. Mol Cell Biol. 1993;13(10):6442–6451. doi: 10.1128/mcb.13.10.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernard DG, Quevillon-Cheruel S, Merchant S, Guiard B, Hamel PP. Cyc2p, a membrane-bound flavoprotein involved in the maturation of mitochondrial c-type cytochromes. J Biol Chem. 2005;280(48):39852–39859. doi: 10.1074/jbc.M508574200. [DOI] [PubMed] [Google Scholar]
- 24.Corvest V, et al. The flavoprotein Cyc2p, a mitochondrial cytochrome c assembly factor, is a NAD(P)H-dependent haem reductase. Mol Microbiol. 2012;83(5):968–980. doi: 10.1111/j.1365-2958.2012.07981.x. [DOI] [PubMed] [Google Scholar]
- 25.Prakash SK, et al. Loss of holocytochrome c-type synthetase causes the male lethality of X-linked dominant microphthalmia with linear skin defects (MLS) syndrome. Hum Mol Genet. 2002;11(25):3237–3248. doi: 10.1093/hmg/11.25.3237. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz QP, Cox TC. Complementation of a yeast CYC3 deficiency identifies an X-linked mammalian activator of apocytochrome c. Genomics. 2002;79(1):51–57. doi: 10.1006/geno.2001.6677. [DOI] [PubMed] [Google Scholar]
- 27.Kiryu-Seo S, Gamo K, Tachibana T, Tanaka K, Kiyama H. Unique anti-apoptotic activity of EAAC1 in injured motor neurons. EMBO J. 2006;25(14):3411–3421. doi: 10.1038/sj.emboj.7601225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lill R, Stuart RA, Drygas ME, Nargang FE, Neupert W. Import of cytochrome c heme lyase into mitochondria: A novel pathway into the intermembrane space. EMBO J. 1992;11(2):449–456. doi: 10.1002/j.1460-2075.1992.tb05074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steiner H, Zollner A, Haid A, Neupert W, Lill R. Biogenesis of mitochondrial heme lyases in yeast. Import and folding in the intermembrane space. J Biol Chem. 1995;270(39):22842–22849. doi: 10.1074/jbc.270.39.22842. [DOI] [PubMed] [Google Scholar]
- 30.Dumont ME, Cardillo TS, Hayes MK, Sherman F. Role of cytochrome c heme lyase in mitochondrial import and accumulation of cytochrome c in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(11):5487–5496. doi: 10.1128/mcb.11.11.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumont ME, Ernst JF, Sherman F. Coupling of heme attachment to import of cytochrome c into yeast mitochondria. Studies with heme lyase-deficient mitochondria and altered apocytochromes c. J Biol Chem. 1988;263(31):15928–15937. [PubMed] [Google Scholar]
- 32.Nargang FE, Drygas ME, Kwong PL, Nicholson DW, Neupert W. A mutant of Neurospora crassa deficient in cytochrome c heme lyase activity cannot import cytochrome c into mitochondria. J Biol Chem. 1988;263(19):9388–9394. [PubMed] [Google Scholar]
- 33.Nicholson DW, Hergersberg C, Neupert W. Role of cytochrome c heme lyase in the import of cytochrome c into mitochondria. J Biol Chem. 1988;263(35):19034–19042. [PubMed] [Google Scholar]
- 34.Nicholson DW, Neupert W. Import of cytochrome c into mitochondria: Reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome c. Proc Natl Acad Sci USA. 1989;86(12):4340–4344. doi: 10.1073/pnas.86.12.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barker PD, et al. Transmutation of a heme protein. Proc Natl Acad Sci USA. 1993;90(14):6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner H, et al. Heme binding to a conserved Cys-Pro-Val motif is crucial for the catalytic function of mitochondrial heme lyases. J Biol Chem. 1996;271(51):32605–32611. doi: 10.1074/jbc.271.51.32605. [DOI] [PubMed] [Google Scholar]
- 37.Moore RL, Stevens JM, Ferguson SJ. Mitochondrial cytochrome c synthase: CP motifs are not necessary for heme attachment to apocytochrome c. FEBS Lett. 2011;585(21):3415–3419. doi: 10.1016/j.febslet.2011.08.042. [DOI] [PubMed] [Google Scholar]
- 38.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998;37(17):6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 39.Patel CN, Lind MC, Pielak GJ. Characterization of horse cytochrome c expressed in Escherichia coli. Protein Expr Purif. 2001;22(2):220–224. doi: 10.1006/prep.2001.1438. [DOI] [PubMed] [Google Scholar]
- 40.Rumbley JN, Hoang L, Englander SW. Recombinant equine cytochrome c in Escherichia coli: high-level expression, characterization, and folding and assembly mutants. Biochemistry. 2002;41(47):13894–13901. doi: 10.1021/bi026543y. [DOI] [PubMed] [Google Scholar]
- 41.Jeng WY, Chen CY, Chang HC, Chuang WJ. Expression and characterization of recombinant human cytochrome c in E. coli. J Bioenerg Biomembr. 2002;34(6):423–431. doi: 10.1023/a:1022561924392. [DOI] [PubMed] [Google Scholar]
- 42.Kleingardner JG, Bren KL. Comparing substrate specificity between cytochrome c maturation and cytochrome c heme lyase systems for cytochrome c biogenesis. Metallomics. 2011;3(4):396–403. doi: 10.1039/c0mt00086h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens JM, et al. The mitochondrial cytochrome c N-terminal region is critical for maturation by holocytochrome c synthase. FEBS Lett. 2011;585(12):1891–1896. doi: 10.1016/j.febslet.2011.04.058. [DOI] [PubMed] [Google Scholar]
- 44.Sanders C, Lill H. Expression of prokaryotic and eukaryotic cytochromes c in Escherichia coli. Biochim Biophys Acta. 2000;1459(1):131–138. doi: 10.1016/s0005-2728(00)00122-5. [DOI] [PubMed] [Google Scholar]
- 45.Sanders C, Wethkamp N, Lill H. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol Microbiol. 2001;41(1):241–246. doi: 10.1046/j.1365-2958.2001.02514.x. [DOI] [PubMed] [Google Scholar]
- 46.Falk J. Porphyrins and Metalloporphyrins: Their General, Physical and Coordination Chemistry, and Laboratory Methods. Vol 2. New York: Elsevier; 1964. [Google Scholar]
- 47.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161(1):1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 48.Dawson JH, Andersson LA, Sono M. Spectroscopic investigations of ferric cytochrome P-450-CAM ligand complexes. Identification of the ligand trans to cysteinate in the native enzyme. J Biol Chem. 1982;257(7):3606–3617. [PubMed] [Google Scholar]
- 49.Wang WH, Lu JX, Yao P, Xie Y, Huang ZX. The distinct heme coordination environments and heme-binding stabilities of His39Ser and His39Cys mutants of cytochrome b5. Protein Eng. 2003;16(12):1047–1054. doi: 10.1093/protein/gzg134. [DOI] [PubMed] [Google Scholar]
- 50.Egeberg KD, et al. Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry. 1990;29(42):9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- 51.Adachi S, et al. Roles of proximal ligand in heme proteins: Replacement of proximal histidine of human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry. 1993;32(1):241–252. doi: 10.1021/bi00052a031. [DOI] [PubMed] [Google Scholar]
- 52.Hildebrand DP, Ferrer JC, Tang HL, Smith M, Mauk AG. Trans effects on cysteine ligation in the proximal His93Cys variant of horse heart myoglobin. Biochemistry. 1995;34(36):11598–11605. doi: 10.1021/bi00036a036. [DOI] [PubMed] [Google Scholar]
- 53.Barrick D. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93—>Gly. Biochemistry. 1994;33(21):6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 54.Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci USA. 2009;106(25):10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.San Francisco B, Bretsnyder EC, Rodgers KR, Kranz RG. Heme ligand identification and redox properties of the cytochrome c synthetase, CcmF. Biochemistry. 2011;50(50):10974–10985. doi: 10.1021/bi201508t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verissimo AF, Sanders J, Daldal F, Sanders C. Engineering a prokaryotic apocytochrome c as an efficient substrate for Saccharomyces cerevisiae cytochrome c heme lyase. Biochem Biophys Res Commun. 2012;424(1):130–135. doi: 10.1016/j.bbrc.2012.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy KS, Angiolillo PJ, Wright WW, Laberge M, Vanderkooi JM. Spectral splitting in the alpha (Q0,0) absorption band of ferrous cytochrome c and other heme proteins. Biochemistry. 1996;35(39):12820–12830. doi: 10.1021/bi960895l. [DOI] [PubMed] [Google Scholar]
- 58.Richard-Fogal C, Kranz RG. The CcmC:heme:CcmE complex in heme trafficking and cytochrome c biosynthesis. J Mol Biol. 2010;401(3):350–362. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richard-Fogal CL, et al. A conserved haem redox and trafficking pathway for cofactor attachment. EMBO J. 2009;28(16):2349–2359. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka Y, Kubota I, Amachi T, Yoshizumi H, Matsubara H. Site-directedly mutated human cytochrome c which retains heme c via only one thioether bond. J Biochem. 1990;108(1):7–8. doi: 10.1093/oxfordjournals.jbchem.a123165. [DOI] [PubMed] [Google Scholar]
- 61.Rosell FI, Mauk AG. Spectroscopic properties of a mitochondrial cytochrome C with a single thioether bond to the heme prosthetic group. Biochemistry. 2002;41(24):7811–7818. doi: 10.1021/bi016060e. [DOI] [PubMed] [Google Scholar]
- 62.Tong J, Margoliash E. Cytochrome c heme lyase activity of yeast mitochondria. J Biol Chem. 1998;273(40):25695–25702. doi: 10.1074/jbc.273.40.25695. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Dumont ME, Sherman F. Sequence requirements for mitochondrial import of yeast cytochrome c. J Biol Chem. 1996;271(12):6594–6604. [PubMed] [Google Scholar]
- 64.Sprinkle JR, Hakvoort TB, Koshy TI, Miller DD, Margoliash E. Amino acid sequence requirements for the association of apocytochrome c with mitochondria. Proc Natl Acad Sci USA. 1990;87(15):5729–5733. doi: 10.1073/pnas.87.15.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pascher T, Chesick JP, Winkler JR, Gray HB. Protein folding triggered by electron transfer. Science. 1996;271(5255):1558–1560. doi: 10.1126/science.271.5255.1558. [DOI] [PubMed] [Google Scholar]
- 66.Jones CM, et al. Fast events in protein folding initiated by nanosecond laser photolysis. Proc Natl Acad Sci USA. 1993;90(24):11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu Y, Casimiro DR, Bren KL, Richards JH, Gray HB. Structurally engineered cytochromes with unusual ligand-binding properties: Expression of Saccharomyces cerevisiae Met-80—>Ala iso-1-cytochrome c. Proc Natl Acad Sci USA. 1993;90(24):11456–11459. doi: 10.1073/pnas.90.24.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feissner RE, et al. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol Microbiol. 2006;60(3):563–577. doi: 10.1111/j.1365-2958.2006.05132.x. [DOI] [PubMed] [Google Scholar]
- 69.Richard-Fogal CL, San Francisco B, Frawley ER, Kranz RG. Thiol redox requirements and substrate specificities of recombinant cytochrome c assembly systems II and III. Biochim Biophys Acta. 2012;1817(6):911–919. doi: 10.1016/j.bbabio.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feissner R, Xiang Y, Kranz RG. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal Biochem. 2003;315(1):90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 71.Richard-Fogal CL, Frawley ER, Feissner RE, Kranz RG. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J Bacteriol. 2007;189(2):455–463. doi: 10.1128/JB.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: An online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37(Web Server issue):W498-503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thévenet P, et al. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40(Web Server issue):W288-93. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]