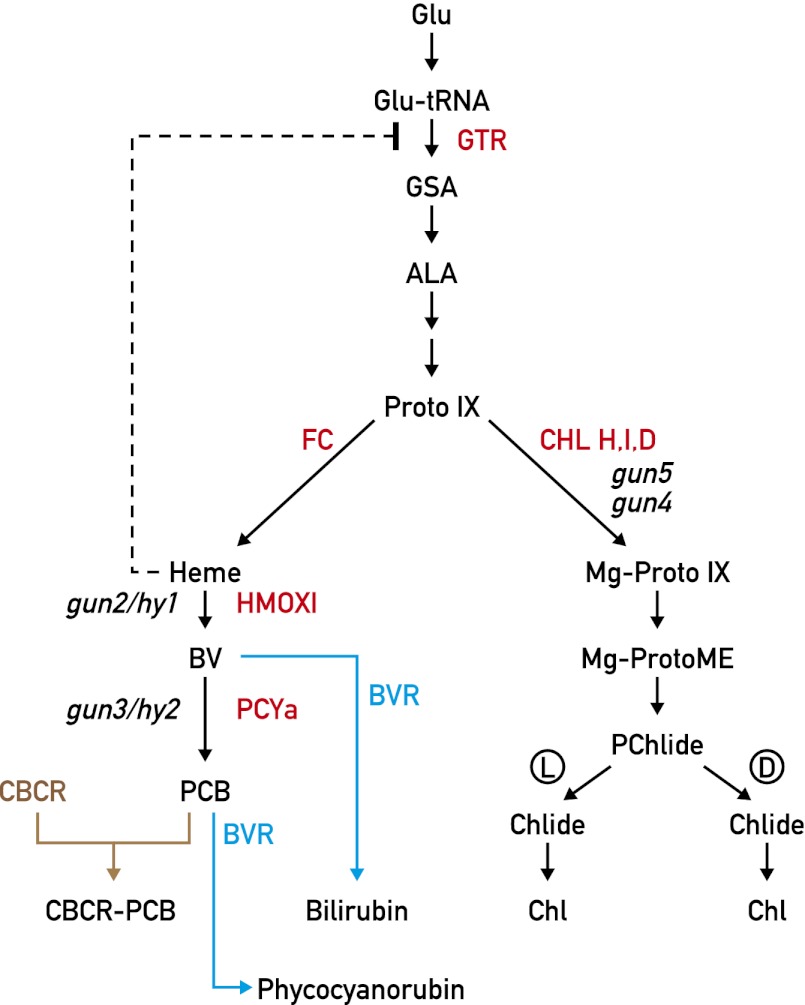
It is well established that the origin of plastids can be traced to an endosymbiotic event in which a free-living photosynthetic prokaryote invaded a eukaryotic cell more than 1 billion years ago. Most genes from the intruder were gradually transferred to the host nucleus whereas a small number of these genes were maintained in the plastid and gave rise to the plastid genome with its associated protein synthesizing system. The products of many of the genes transferred to the nucleus were then retargeted to the plastid to keep it functional. Altogether, approximately 3,000 nuclear genes in plants and algae encode plastid proteins, whereas chloroplast genomes contain between 100 and 120 genes (1). A major challenge for eukaryotic photosynthetic organisms is to coordinate these two genetic systems during chloroplast development and to maintain plastids functional under changing environmental conditions. The situation is even more complex with the existence of a third genetic system in plant and algal mitochondria, which will not be discussed here. Although many nuclear genes have been identified that are involved in chloroplast gene expression, the nucleus is also capable of sensing the state of the chloroplast and to react to maintain chloroplast homeostasis. This process is called retrograde signaling and involves signals originating from the plastid and transmitted to the nucleus, where they elicit a specific transcriptional response. In recent years, several retrograde signaling pathways have been identified mostly through genetic approaches (2, 3). Most of the mutants isolated, called gun mutants, are affected in the tetrapyrrole biosynthetic pathway (4, 5). In plants and algae, this pathway gives rise to heme and chlorophyll, with a branching point at the level of protoporphyrin IX (Fig. 1). This compound is then converted to protoheme and Mg-protoporphyrin IX by Fe- and Mg-chelatase, respectively. In turn, heme acts as prosthetic group for a large number of hemoproteins, but a portion of heme is converted by heme oxygenase (HMOX) to biliverdin IXa and by phytochromobilin synthase (PCYA) to phytochromobilin, which serves as chromophore of phytochromes (Fig. 1). An intriguing feature of all sequenced chlorophyte genomes is that, although they lack phytochromes, their genomes encode two HMOXs, HMOX1 and HMOX2, and PCYA. In PNAS, Duanmu et al. (6) investigate the role of these genes in the green alga Chlamydomonas reinhardtii and made unexpected findings.
Fig. 1.
Tetrapyrrole biosynthetic pathways. The heme and chlorophyll biosynthetic pathways diverge at protoporphyrin IX (ProtoIX). The negative feedback of heme on GTR (Glu-tRNA reductase) is indicated. In C. reinhardtii, conversion of protochlorophyllide (PChlide) to chlorophyllide (Chlide) can occur in a light-dependent (marked as “L”) and a light-independent manner (“D”). In the study of Duanmu et al. (6), the pathway is perturbed by introducing mammalian biliverdin reductase (BVR) for heme depletion. The activity of HMOX1 and PCYA is demonstrated by introducing cyanobacteriochrome (CBCR) reporter and by showing that it covalently binds PCB chromophore. The different steps affected in the gun and hy mutants are indicated. ALA, δ-aminolevulinic acid; Chl, chlorophyll; CHL H/D/I, Mg-chelatase, subunit H/D/I; FC, Fe-chelatase; GSA, glutamate 1-semialdehyde; PCYA, phytochromobilin synthase.
Duanmu et al. first show that HMOX1, HMOX2, and PCYA are catalytically active and produce bilins in vitro (6). They also demonstrate in a very elegant way that these proteins are functional in vivo by expressing a cyanobacteriochrome in the chloroplast of C. reinhardtii, where, remarkably, the photoreceptor is assembled with bound phytochromobilin chromophore. They further show that HMOX1 and PCYA are chloroplast enzymes whereas the animal-type HMOX2 is membrane-bound in the cytosol. The existence of this HMOX associated with cytosolic membranes in C. reinhardtii and other chlorophytes is rather surprising because enzymes of this type are absent from streptophyte algae and land plants.
To assess the role of HMOX1 and HMOX2, the authors isolated mutants unable to produce either of these two enzymes. They show that, under conditions of iron starvation, growth of the WT, but not of the hmox1 or hmox2 mutants, can be rescued by adding hemin to the growth medium, indicating that HMOX1 and HMOX2 are able to degrade exogenously supplied heme for iron acquisition. It is likely that HMOX2 originates from the ancient eukaryotic host that was invaded during endosymbiosis. This gene has been maintained in chlorophyte but not in streptophyte algae, perhaps because the former more often experience iron limitation.
A striking result of this study is that photoautotrophic growth of C. reinhardtii cells is compromised in the absence of HMOX1 whereas heterotrophic growth in the dark is unaffected, and, moreover, the threefold increase of chlorophyll that occurs upon a shift of WT cells from the dark to the light is not observed with the hmox1 mutant. In contrast to land plants, C. reinhardtii is able to produce chlorophyll in the dark, but at a lower level than in the light. These results point to an important role of bilins. Heme is known to exert a negative feedback on chlorophyll synthesis, which explains why chlorophyll deficiency occurs in land plants lacking HMOX. If this explanation is also valid for the C. reinhardtii hmox1 mutant, Duanmu et al. reason that the opposite effect would be expected if the level of heme is decreased (6). This was achieved by expressing, in WT cells, a mammalian biliverdin reductase, an enzyme that converts biliverdin IXα (BV) into bilirubin and phytocyanobilin (PCB) into phycocyanorubin, and is thereby expected to deplete the heme pool (Fig. 1). However, in this case, contrary to the heme feedback hypothesis, the chlorophyll level decreased during a dark-to-light shift, raising the possibility that BV or PCB is responsible for the light-dependent accumulation of chlorophyll. This proposal is in agreement with feeding experiments with biliverdin in which the WT chlorophyll level in hmox1 was restored even though photoautotrophic growth was only partially rescued. However, how the bilins influence chlorophyll accumulation is not yet clear.
To further examine the role of bilins in the regulation of chlorophyll synthesis during a dark/light transition, Duanmu et al. (6) perform a global comparative transcriptomic analysis with WT and hmox1 cells in the presence or absence of biliverdin. Several important results emerged from this study. First, a small set of 76 nuclear genes was identified that are up-regulated by bilins in a light-independent way. These genes include several oxygen-dependent redox enzymes (mono- and dioxygenases, proteins containing redox cofactors and enzymes associated with oxidative amino acid metabolism). A shift of an algal culture from dark to light is accompanied by a sudden increase in oxygen evolution and, as Duanmu et al. suggest, by the release of heme from damaged hemoproteins (6). In contrast, in the light, the plastid-derived bilins appear to have an inhibitory action on nuclear genes involved in photosynthesis. Altogether, these considerations raise the possibility that bilins are part of a retrograde signaling pathway that evolved in C. reinhardtii and more generally in chlorophytes for the detoxification of reactive oxygen species generated during the transition from dark to light. Heme has been proposed previously as positive retrograde signal for the regulation of nuclear gene expression in C. reinhardtii (7) and recently also in Arabidopsis for the coordination of gene expression with chloroplast development (5).
The results of this work raise a number of questions. Do chlorophyte species lacking phytochromes possess an alternative regulatory bilin-based system for regulating chlorophyll synthesis in the light? If so, how does it work? Are there unknown PCB-based photoreceptors in these algae? Was the bilin biosynthetic pathway uniquely maintained for ensuring smooth daily transitions from dark to light with minimal photodamage? How widely distributed is this bilin-mediated retrograde signaling pathway among oxygenic photosynthetic organisms with or without phytochromes? Clearly, future research in this area is likely to provide new answers to these intriguing questions.
Footnotes
The author declares no conflict of interest.
See companion article on page 3621.
References
- 1.Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99(19):12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 3.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74(5):787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 5.Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol. 2011;21(10):897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duanmu D, et al. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc Natl Acad Sci USA. 2013;110:3621–3626. doi: 10.1073/pnas.1222375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell. 2008;20(3):552–567. doi: 10.1105/tpc.107.054650. [DOI] [PMC free article] [PubMed] [Google Scholar]