Abstract
Poxviruses are considered less dependent on host functions than other DNA viruses because of their cytoplasmic site of replication and large genomes, which encode enzymes for DNA and mRNA synthesis. Nevertheless, RNAi screens with two independent human genome-scale libraries have identified more than 500 candidate genes that significantly inhibited and a similar number that enhanced replication and spread of infectious vaccinia virus (VACV). Translational, ubiquitin-proteosome, and endoplasmic reticulum-to-Golgi transport functions, known to be important for VACV, were enriched in the siRNA-inhibiting group, and RNA polymerase II and associated functions were enriched in the siRNA-enhancing group. Additional findings, notably the inhibition of VACV spread by siRNAs to several nuclear pore genes, were unanticipated. Knockdown of nucleoporin 62 strongly inhibited viral morphogenesis, with only a modest effect on viral gene expression, recapitulating and providing insight into previous studies with enucleated cells.
Keywords: nucleoporin 62, poxvirus replication, poxvirus–host interactions, syntaxin 5, vaccinia virus replication
Viruses are dependent on cellular processes for their propagation, and intensive efforts are currently underway to define these pathways, which may provide insight into both host and viral mechanisms and allow the creation of novel therapeutics resistant to the development of virus escape mutations. Poxviruses include species that are highly pathogenic for humans and other animals and are considered potential weapons of bioterrorism. In addition, poxviruses may serve as vaccine vectors to prevent infectious diseases and treat cancers. These DNA viruses carry out their entire replication cycle in the cytoplasm, where they assemble factories for genome replication, transcription, and virion assembly (1). Although lacking translation machinery, poxviruses encode numerous enzymes and factors, including a DNA polymerase, a DNA-dependent RNA polymerase, transcription factors, capping enzyme, and poly(A) polymerase, and thus might be anticipated to require fewer host gene activities for replication compared with other DNA viruses.
Genome-wide RNAi screens have been carried out to assess the roles of cellular genes and pathways on the replication of several RNA viruses, including HIV (2–4), influenza virus (5), West Nile virus (6), hepatitis C virus (7), and vesicular stomatitis virus (8). Although a genome-wide RNAi screen of poxvirus replication had not yet been performed, studies with selected siRNAs demonstrated roles for host proteins involved in vaccinia virus (VACV) entry, DNA replication, gene expression, virion assembly, and intracellular transport in mammalian cells (9–17). Moser et al. (18) determined the effects of siRNAs to 440 genes, composing the “kinome,” on VACV gene expression in Drosophila cells, and found that the AMP-activated protein kinase pathway plays a major role in VACV entry. However, because Drosophila cells abort VACV infection before genome replication (19), the screen provided information only on very early events. Recently, Mercer et al. (20) described a siRNA screen of 7,000 “druggable” gene targets in human cells and identified 188, representing a broad range of host functions, that inhibit VACV gene expression.
Here we describe a human genome-wide RNAi screen on VACV replication. We developed a high-throughput virus spread assay encompassing all steps from virus entry to infectious progeny formation. Using siRNA libraries covering the entire human ORFeome, we confirmed the role of translation factors and the ubiquitin-proteosome pathway and uncovered hitherto unreported interactions, including a role for nuclear pore (Nup) proteins in virion morphogenesis that provides insight into a previous report suggesting a role for the nucleus in VACV replication (21).
Results
Human Genome-Wide siRNA Screen.
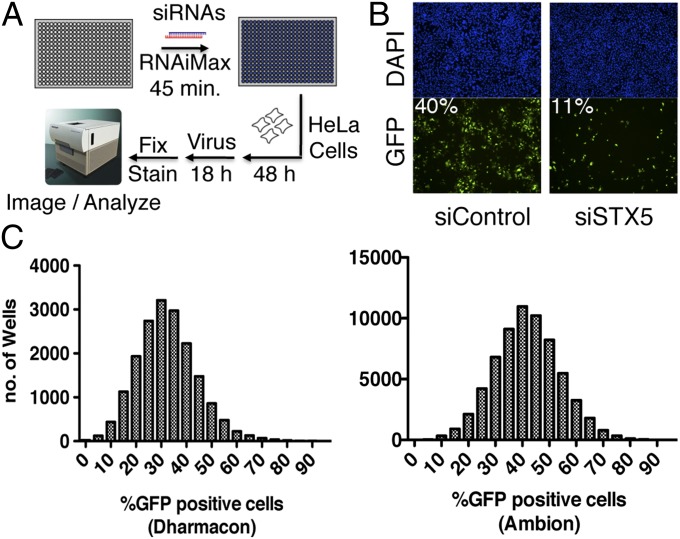
We used a recombinant VACV IHD-J/GFP (22) derived from a virus strain with a point mutation that accelerates the release of progeny from the cell surface (23) and expresses green fluorescent protein (GFP). Pilot experiments indicated that extensive virus spread could be visualized by fluorescence microscopy within 18 h (Fig. S1A). As a control, we treated the cells with ST-246, a drug that has no effect on virus entry or replication but blocks the wrapping of infectious mature virions (MVs), a necessary step for the egress and spread of VACV (24). To optimize the assay for high throughput, we infected HeLa cells in 384-well plates with serial dilutions of VACV IHD-J/GFP and incubated them for 18 h. The GFP-positive cells were scored by automated fluorescence microscopy (Fig. S1B). We determined that a multiplicity of 0.2 plaque-forming unit (PFU) per cell led to ∼40% positive cells in 18 h, which would allow detection of reduced or increased virus spread. Before conducting the large-scale siRNA screen, we examined the effects of siRNAs that would knock down endoplasmic reticulum-to-Golgi transport, a necessary step for the wrapping and egress of virions (25). A previous study reported that knock down of syntaxin 5 (STX5) snare protein inhibited constitutive secretion in mammalian cells (26). We found that STX5 siRNA is a potent inhibitor of VACV spread, and that this occurs in the 384-well high-throughput format without reducing cell numbers (Fig. S1 C and D). The STX5 siRNA was used as a knockdown control in subsequent screens.
For maximal depth of coverage and confirmation of results, we chose two genome-wide libraries: siGenome from Dharmacon (DGW), comprising a single pool of four siRNAs for 18,120 genes, and Ambion Silencer Select Version 4 (AGW), a nonpooled library of three independent siRNAs for 21,566 genes. The DGW and AGW libraries had 17,693 overlapping target genes, although the individual siRNAs differed. We imaged virus-expressed GFP and the nucleus in each well, enabling single-cell determination of an infection event (Fig. 1A). With the STX5 siRNA, 11% of the cells were GFP-positive, compared with ∼40% in control siRNA wells (Fig. 1B). Genome-wide, the screen peaked at around 30–35% GFP-positive cells in DGW and 35–45% GFP-positive cells in AGW (Fig. 1C) and was nearly normally distributed. The coefficient of variation was ∼30%, an acceptable value for siRNA screening. To ensure screening quality, we manually examined each plate for aberrant patterns and calculated the assay z′-factor (27). Plates that failed quality control were redone. The screen performed with a high, robust assay z′-factor score of ∼0.5 per plate. Each plate was normalized to the median of 16 negative control wells per plate, set to 100% virus spread.
Fig. 1.
High-throughput RNAi screen. (A) Diagram of screen setup. HeLa cells were transfected with siRNAs for 48 h and then infected with GFP-expressing VACV for 18 h. After fixation and staining with DAPI, the total number of cells and the fraction expressing GFP were determined. (B) Acquired image. (Upper) DAPI channels. (Lower) GFP channels. The wells were transfected with either siRNA targeting STX5 (Right) or nontargeting control (Left). Percentages of GFP-positive cells are indicated. (C) Signal distribution across wells of the genome-wide screens using the Dharmacon (Left) and Ambion (Right) libraries.
Gene Set Enrichment, Hit Selection, and Network Analysis.
For hit selection, we used the median absolute deviation (MAD) calculated from the distribution of results for a given library of siRNA reagents. This statistical approach is sensitive, yet robust to outliers (28). For hit calling, we used a cutoff of −1.5 MAD, taking the value of the second most active reagent to represent the gene target when multiple reagents per gene were screened individually. We took this stringent approach to balance false-positive selection (off-target effects) and false-negative dismissal (poor cross-library validation). Our high-confidence list contained ∼500 candidate genes that demonstrated a <−1.5 MAD decrease in virus spread with at least two different siRNA reagents from the same or different libraries and a similar number with an increase in virus spread. The siRNAs inducing a >50% reduction in cell count were filtered out as toxic, because they would be expected to reduce virus spread nonspecifically. We followed up the two primary screen libraries by testing siRNAs from a third company, Qiagen, that targeted 237 genes selected on the basis of magnitude of inhibition in a primary screen, pathway analysis, and biological interest. From the primary and secondary screens, we prepared a high-confidence list containing 576 genes associated with a significant decrease in virus spread with at least two different siRNA reagents (Tables S1 and S2). Of these genes, 180 were active in both primary screens; additional hits were active with two or more AGW siRNAs, but not with the DGW and others with the Qiagen siRNAs and either DGW or one AGW. The 530 genes associated with an increase in virus spread after knockdown are summarized in Table S3.
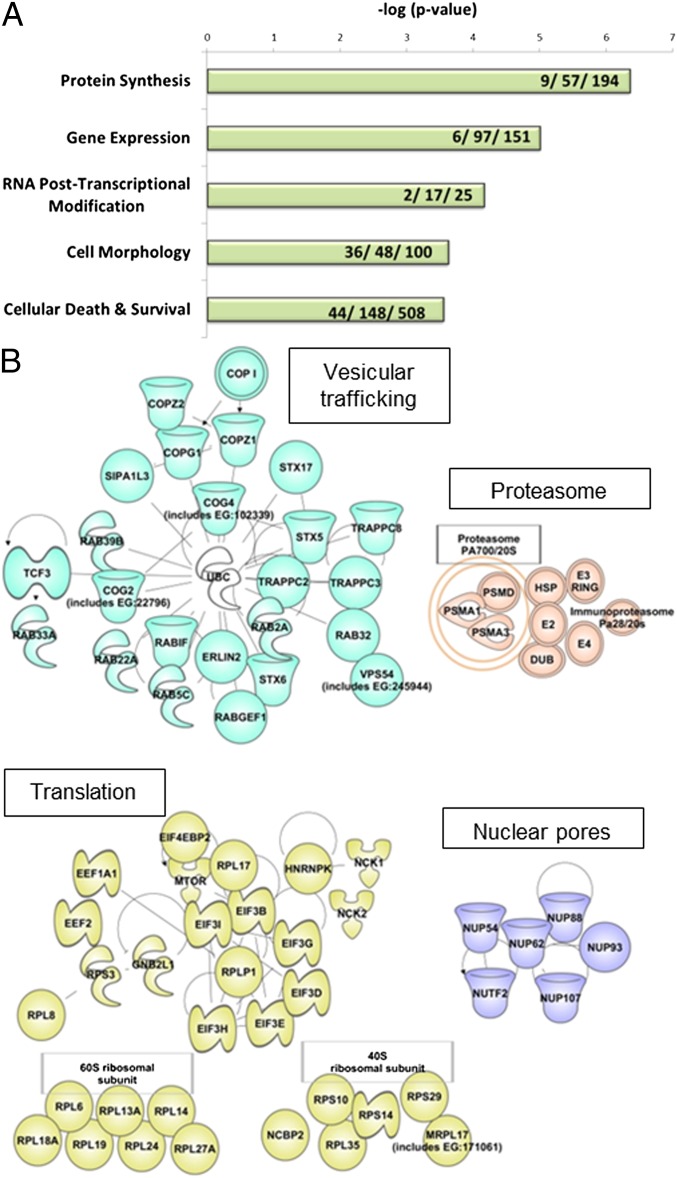
We used Ingenuity Systems Pathway Analysis (IPA) software to classify the genes with multiple active hits into their molecular and cellular functions. The most significantly enriched terms were related to protein synthesis, emphasizing the dependence of VACV on the translational machinery of the cell (Fig. 2A). Additional high scores involved gene expression and RNA posttranscriptional modification, morphology, and cellular death and survival (Fig. 2A). By using curated protein–protein interaction databases, we created interaction maps of several key cellular pathways using the genes listed in the Gene Ontology (GO) annotation and superimposed our hit list on them. The large number of hits for translational control proteins indicated the deep coverage of the siRNA screen (Fig. 2B). In addition, we confirmed the effects of the proteasome and endoplasmic reticulum-to-Golgi vesicular trafficking pathways previously shown to be essential for VACV spread. However, we did not anticipate enrichment of Nup gene targets (Fig. 2B) in view of the cytoplasmic replication of poxviruses.
Fig. 2.
Genome-wide RNAi screen reveals interacting proteins and networks in the high-confidence hits dataset. (A) Negative log (P values) of enriched molecular and cellular functions according to the Ingenuity Knowledge Base. Numbers of identified functions in each ontological category, numbers of unique genes per category, and numbers of all genes associated with the term are indicated. P values were 4.43E-07–1.19E-02 for protein synthesis, 9.87E-08–2.93E-02 for gene expression, 6.92E-05–6.59E-03 for RNA posttranscriptional modification, 2.83E04–2.93E-02 for cell death and survival, and 2.40E-04–2.93E-02 for cell morphology. (B) Interactions among high-confidence hits associated with vesicular trafficking, translation and ribosomal proteins, proteosome and ubiquitination, and Nup proteins.
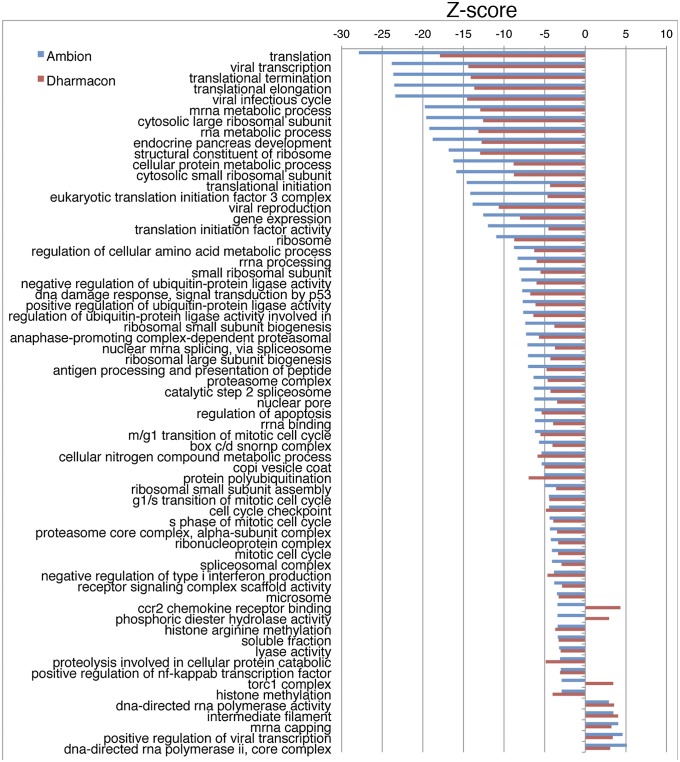
We took a genome-wide analysis approach to better detect subtle changes in global trends, which could be missed when working with rigid thresholds. Toward this end, we used GO annotations to identify associations among RNAi activity of all reagents screened and specific gene function descriptions. Using a parametric analysis of gene set enrichment method (29), we calculated a z-score for each gene set. Only terms with a statistically significant false discovery rate (<0.05) (30) in both DGW and AGW screens are listed (Fig. 3). For siRNAs that inhibited virus spread, translation control factors and ubiquitin-proteasome pathways were highly enriched, reinforcing the importance of these pathways and serving as an internal positive control for our screen and analysis. Again, we found enrichment of Nup components. In contrast, interference with DNA-dependent RNA polymerase II pathways enhanced VACV spread rather than inhibiting it.
Fig. 3.
Gene set enrichment and network analysis. GO annotations were used to identify associations between RNAi activity and specific gene function descriptions. GO terms occurring frequently in the descriptions of gene targets for spread-inhibiting RNAi have a z-score <0, whereas those spread-increasing RNAi have a z-score >0. Only terms with a statistically significant (false discovery rate of 0.05) association in both DGW and AGW screens are listed.
Validation of Nup62 as a True Hit.
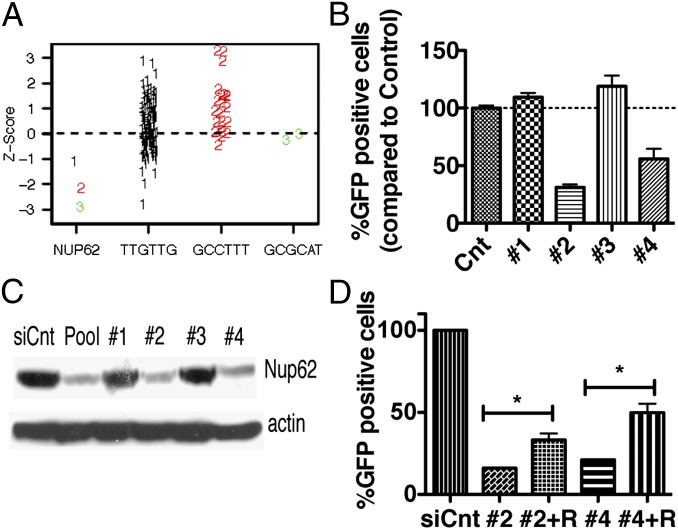
The apparent involvement of Nups prompted us to further investigate the role of the associated proteins in VACV replication. One of the top Nup candidates in the screens was Nup62. To assess whether the activities of siRNAs targeting Nup62 were on target, we applied common seed analysis (31) to the Ambion screen data. This analysis allowed us to empirically distinguish between on-target and off-target effects of siRNAs by comparing the assay results of a siRNA with the assay results of all other siRNAs with the same seed sequence (bases 2–7 of the guide strand). A phenotype increased for the candidate siRNA compared with other siRNAs with the same seed sequence is likely due to knockdown of the intended target. The experiment depicted in Fig. 4A demonstrates that each of the three AGW siRNAs targeting Nup62 inhibited VACV more than other siRNAs with the same seed, suggesting that off-target effects are not the driving force for the phenotype. We also deconvolved the DGW siGenome pool targeting Nup62 and transfected the four different siRNAs separately to test their effect on the spread of virus, using flow cytometry as the readout. Two of the individual siRNAs tested inhibited VACV spread (Fig. 4B). Furthermore, the resulting reduction in Nup62 levels by individual siRNAs was correlated with VACV’s ability to spread, establishing the protein–phenotype linkage (Fig. 4C). In addition, three of four siRNAs from Qiagen targeting Nup62 also induced significant reduction in virus spread (Fig. S2A).
Fig. 4.
Specificity of Nup62. (A) Common seed analysis of three siRNAs targeting Nup62 in the AGW library: 1 (black), 2 (red), and 3 (green). The z-scores of these three siRNAs are shown versus the z-scores for all other siRNAs containing the same seed sequences tested in the assay (indicated on the x-axis), targeted to different genes. (B) Evaluation of four different Dharmacon siRNAs complementary to Nup62 (1–4) or control siRNA (siCnt). At 48 h after transfection, HeLa cells were infected with 0.1 PFU/cell of VACV IHDJ/GFP for 18 h. GFP-positive cells were scored by flow cytometry. The percentage of GFP-positive cells in the control wells was set to 100%. (C) Knockdown of Nup62. HeLa cells were transfected with the siRNAs used in B and 48 h later analyzed by Western blot analysis with antibodies specific for Nup62 and an actin loading control. (D) Rescue of siRNA phenotype. HeLa cells were transfected with siRNA 2 or 4 targeting Nup62 with (+) or without a vector expressing the rat Nup62 (R) gene under the CMV promoter. At 48 h after transfection, the cells were infected with 0.1 PFU/cell of VACV IHDJ/GFP for 18 h, and GFP-positive cells were identified. *Pv (P value calculated with one-way ANOVA and Dunnet post test) < 0.05.
To further reject an off-target effect, we conducted rescue experiments in which we expressed the rat Nup62 (the sequence of which renders it naturally resistant to our siRNA) along with siRNA to the human Nup62, and found a statistically significant increase in virus spread (Fig. 4D). The incomplete effect of rat Nup62 in HeLa cells may reflect species differences in protein–protein interactions.
We carried out experiments to determine whether knockdown of Nup62 induced global effects on the integrity of the nuclear membrane and nuclear export. Mammalian pores exclude molecules that are larger then 40 kDa but allow the diffusion of smaller molecules (32). We tested the functionality of the pores by determining the ability of fluorescently labeled dextrans to diffuse into the nucleus (33). HeLa cells were transfected with siRNA to Nup62 or control siRNA and permeabilized with digitonin after 48 h. Fluorescently labeled 10-kDa or 70-kDa dextran was added and allowed to diffuse for 15 min at room temperature. In both the control and Nup62 siRNA-treated cells, 70-kDa dextran was excluded from the nucleus, whereas 10-kDa dextran was readily detected in the nucleus (Fig. S3).
Cellular topoisomerase II, which is present predominantly in the nucleus, associates with the VACV DNA ligase in cytoplasmic viral factories (34). Localization of topisomerase II with viral factories was not prevented by knockdown of Nup62 (Fig. S4).
Knockdown of Nup62 Arrests Virion Morphogenesis.
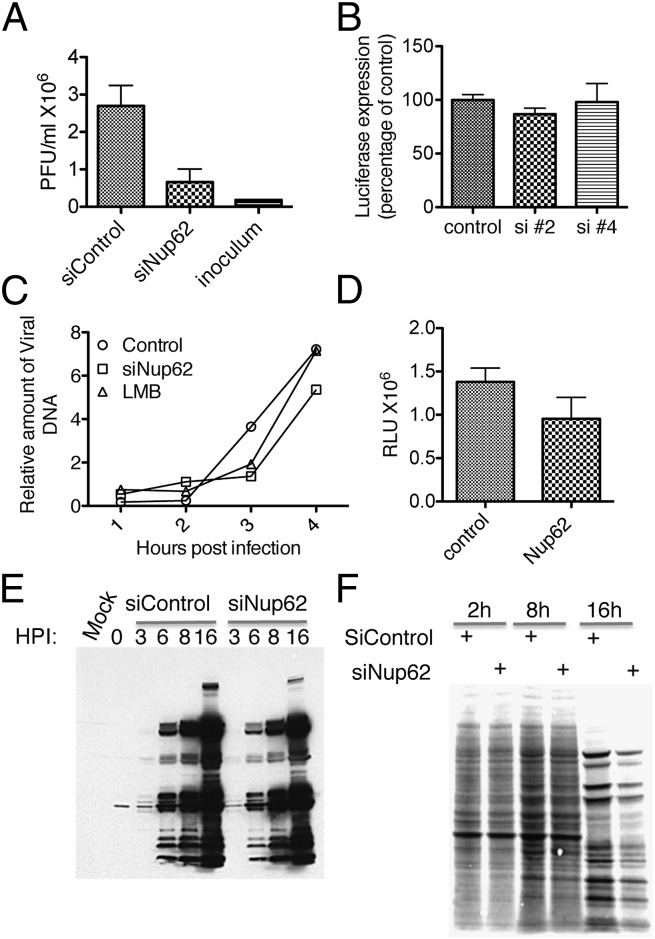
VACV produces two types of infectious virus, the more abundant form of which, the MV, is retained in the cell until lysis. However, a subset of MVs is wrapped with an additional membrane that allows egress from the cell and infection of neighboring cells. Given that our screening assay monitors virus spread, we examined whether Nup62 is required for the formation of infectious virions using a virus inoculum sufficient to infect all cells simultaneously. Nup62 siRNA produced an ∼100-fold reduction in infectious virus yield under one-step growth conditions (Fig. 5A). We next asked whether Nup62 is required for early events in VACV replication, including entry, early gene expression, and DNA replication. We infected HeLa cells with a recombinant VACV that expresses firefly luciferase and measured activity after 90 min, before late gene expression. There was no significant change in early gene expression with Nup62 knockdown (Fig. 5B), or much of an effect on viral DNA synthesis as measured by real-time PCR of total DNA extracted at successive times postinfection (Fig. 5C). A modest decrease of late gene expression by Nup62 siRNA was demonstrated when cells were infected with a recombinant VACV encoding the luciferase gene under a late promoter (Fig. 5D) by Western blot analysis with hyperimmune serum obtained from VACV-infected rabbits (Fig. 5E) and by measurement of incorporation of 35S-amino acid into the major viral proteins (Fig. 5F).
Fig. 5.
Effects of Nup62 on VACV propagation, genome replication, and gene expression. (A) Virus yield. HeLa cells were transfected with control or Nup62 Dharmacon OnTargetPlus siRNAs. After 48 h, the cells were infected with 3 PFU/cell of VACV and harvested at 18 h. The cells were lysed, and infectious virus yields were determined by a plaque assay. (B) Effect of Nup62 knockdown on early expression of firefly luciferase. Control and Nup62 siRNAs (2 and 4, corresponding to Fig. 4 B and C) were transfected into HeLa cells for 48 h, after which the cells were infected with 3 PFU/cell of VACV IHDJ expressing firefly luciferase regulated by an early/late promoter. At 90 min after infection, luciferase activity was determined as a measure of virus entry and early gene expression. (C) Effect of Nup62 knockdown on viral DNA replication. Control and Nup62 siRNA-treated cells were infected with 3 PFU/cell of VACV IHDJ/GFP for the indicated times postinfection. Viral DNA was quantified by real-time PCR and plotted as relative amounts. Leptomycin B (LMB; 20 nM) was added 1 h before infection. (D) Effect of Nup62 knockdown on late expression of firefly luciferase. VACV encoding the firefly luciferase gene under the VACV late F17 promoter was used to infect control and Nup62 siRNA-treated cells at 5 PFU/cell. Luciferase activity was assayed at 6 h postinfection. (E) Effect of Nup62 on viral gene expression determined by Western blot analysis. Cells transfected with control or Nup62 siRNA were infected with 5 PFU/cell of VACV IHDJ/GFP for the indicated times. Western blots were probed with antiserum from VACV-infected rabbits and analyzed by chemiluminescence. (F) Effect of Nup62 knockdown on metabolic labeling of viral proteins. Nup62 and control siRNA-treated cells were infected with 5 PFU/cell of VACV IHDJ/GFP and pulse-labeled with [35S]methionine and [35S]cysteine at 2, 8, and 16 h postinfection (HPI), and the proteins were resolved by SDS-PAGE. An autoradiograph image is shown.
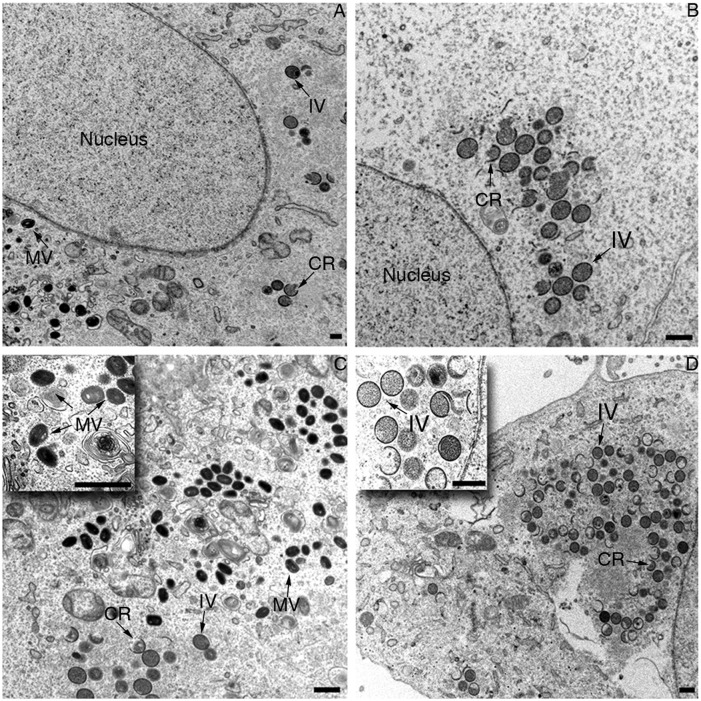
Considering that the biochemical studies indicated only a modest effect of Nup62 siRNA on viral gene expression, we used transmission electron microscopy (TEM) to evaluate the effect of Nup62 siRNA on virion morphogenesis. VACV assembly takes place in cytoplasmic factories in which membrane crescents and spherical immature virions (IVs) form. The latter condense into electron-dense MVs. We carried out two independent experiments to compare and quantify the effects of Nup62 and control siRNAs on morphogenesis. The percentages of control cells with crescents, IVs, and MVs were 86%, 78%, and 41% at 6 h and 90%, 93%, and 62% at 8 h, respectively. In contrast, the percentages of siNup62-treated cells with crescents, IVs and MVs were 66%, 56%, and 3% at 6 h and 100%, 92%, and 17% at 8 h, respectively. The difference in the percentage of cells with MVs was statistically significant (P < 0.01 at 8 h). Thus, the conversion of IVs to MVs was delayed and reduced in siNup62-treated cells compared with control cells. Representative TEM images of cells obtained at 8 h and 20 h are shown in Fig. 6.
Fig. 6.
Effect of Nup62 siRNA on virion morphogenesis. HeLa cells were grown on plastic coverslips and transfected with OnTargetPlus pooled siRNA targeting Nup62 (B and D) or nontargeting siRNA (A and C). After 48 h, cells were infected with 5 PFU/cell of VACV. At 8 h (A and B) and 20 h (C and D) postinfection, the cells were processed for TEM. (Insets) Higher magnification of specific areas. Representative crescents (CR), IVs, and MVs are labeled. (Scale bars: 0.5 μm.)
Discussion
To systematically determine host functions supporting the unique cytoplasmic poxvirus life cycle, we designed a high-throughput assay that measures the cumulative process from entry to spread of infectious progeny virus to other cells. We used two genome-wide siRNA libraries in different formats (one with three individual siRNAs and the other with a pool of four siRNAs), as well as more than 200 additional selected individual siRNAs, making this the most comprehensive virus-host screen reported to date. Of the more than 20,000 genes interrogated, 576 were found to have at least two siRNAs that reduced VACV spread and 180 of them drew from both primary screens. In addition, targeting of 530 genes increased spread, and 251 of these genes were from both primary screens. We attribute the relatively high overlap between the two libraries to the use of the same virus, cells, siRNA protocol, and infectious spread assay.
The recent report of Mercer et al. (20) allowed us to compare datasets in two independent VACV studies. That primary screen was carried out with a library of 6,979 siRNAs from Qiagen, HeLa cells infected 72 h after transfection with thymidine kinase-deficient VACV strain Western Reserve, and an endpoint that measured combined early and late viral protein synthesis. For the present study, primary screens were carried out with siRNAs to 18,120 genes from Dharmacon and 21,566 genes from Ambion, HeLa cells infected with the VACV IHD-J strain at 48 h after siRNA transfection, and an endpoint that measured the entire replication cycle, including spread of infectious virus to other cells.
Because of the different siRNA screen sizes, more than half of our 576 confident hits were not tested in the study of Mercer et al. (20), whereas 186 of their 188 confident hits are represented in the present study. Comparison of those 186 hits with our top candidates revealed only 13 matches; however, removing our toxicity filter yielded 708 candidate genes with two or more reagents scoring <−1.5 MAD, 23 of which overlap with the 186 genes reported by Mercer et al. Although intuitively small, this overlap represents a significant enrichment over what would be expected by chance (approximately six genes). This small overlap may be attributed in part to the experimental differences noted above, as well as to the high rates of false positives and false negatives in RNAi screens. Supporting a high rate of false negatives is the observation that all but 4 of the 76 enriched functional annotation clusters of Mercer et al. (20) overlapped with siRNA hits in our study. Even less correspondence was noted for individual siRNAs targeting host genes required for HIV replication, although enrichment was found at the functional pathway level (35). Nonetheless, the low overlap emphasizes that lists of RNAi screen hits are not to be considered validated without additional evidence (e.g., rescue experiments).
We compared the top molecular and cellular functions that were affected by siRNAs during our genome-wide screen of VACV with screens of several other viruses (Table S4). Twelve common functions were revealed by IPA core analysis to identify the top five molecular and cellular functions for individual viruses. Although each virus queried showed enrichment for all 12 functions, the hierarchy varied, with influenza virus being most similar to VACV (four matches in the top five). Whether this reflects similarities in virus functions or in the siRNA screening methodology is uncertain.
The largest number of hits that decreased VACV spread was seen for host genes involved in translation of mRNA, consistent with the absence of any viral genes dedicated to this process. Moreover, the large number of translation hits, which included translation factors and ribosomal subunits, established the depth of coverage of the screen. The proteasome/ubiquitin pathway and endoplasmic reticulum-to-Golgi transport, which had been identified as important for VACV DNA replication and formation of enveloped virions, respectively, using chemical and dominant negative inhibitors, were enriched in our siRNA screen as well. Mercer et al. (20) focused on the ubiquitin-mediated proteosome pathway for uncoating of the VACV core and genome replication. In our study, the numbers of hits that decreased and increased VACV spread were similar. Examples of the latter are genes encoding subunits of RNA polymerase II and proteins associated with host mRNA formation. It seems plausible that shutting down host mRNA synthesis in the nucleus would attenuate host defenses and conserve cellular resources, thereby benefitting VACV; however, two older studies based on the use of α-aminitin had suggested a positive role for RNA polymerase II in VACV replication (36, 37). One of these studies isolated an α-aminitin VACV-resistant mutant, and the other posited that RNA polymerase II transcribes certain unidentified VACV genes. These studies were performed without the benefit of current molecular tools, however, and further work is needed to identify the putative positive and negative effects of RNA polymerase II on VACV replication.
From among the many host functions that have been implicated in VACV replication, we chose one class to explore in depth. In view of the cytoplasmic site of replication of poxviruses, the finding that siRNAs to nuclear pore genes inhibit VACV spread was intriguing. The nuclear pore complex comprises ∼30 different polypeptides and functions to facilitate or restrict entry of molecules to and from the nucleus (32, 38). Of these, our siRNA screens identified six that knock down VACV spread. Many viruses, including vesicular stomatitis virus, influenza virus, poliovirus, rhinoviruses, and herpesviruses, have developed mechanisms to control or inactivate transport through the pore (39). Disruption of the nuclear-cytoplasmic pathway may diminish the cellular antiviral response, and modifications may allow preferential export of viral mRNAs of nuclear viruses. Knockdown of Nup genes might have been expected to enhance VACV spread; however, the reverse was found.
To explore the basis for the effect of Nup gene siRNAs on VACV, we carried out studies with Nup62, a core component of the nuclear pore complex. We chose this target because robust inhibition was achieved with multiple siRNAs and additional evidence against an off-target effect was obtained. Subsequently, we learned that an annotation cluster-labeled nuclear pore that included Nup62 ranked fourth out of 76 enriched functional groups in the siRNA screen of Mercer et al. (20). Moreover, Nup62 has been shown to interact directly with herpes simplex virus ICP27 (40) and Epstein Barr virus BGLF-4 (41) and to be degraded by poliovirus and rhinovirus proteases (42). We found that knockdown of Nup62 inhibited the production of infectious VACV in a one-step growth experiment, but had no effect on entry and only modest effects on DNA replication and early and late viral gene expression. In addition, Nup62 knockdown did not prevent the previously reported association of cellular topoisomerase II with viral factories (34). However, TEM revealed a severe block in VACV morphogenesis beyond the IV stage. Interestingly, a block in morphogenesis beyond the IV stage and only small decreases in viral gene expression was reported in VACV-infected enucleated cells (21). Although it is possible that Nup62 has a direct role in VACV morphogenesis, it seems more likely that the effect is related to perturbations in nuclear transport, considering that siRNAs targeting Nups in different subcomplexes also decreased VACV spread. When several Nups, such as Nup93 or Nup98, were targeted simultaneously with Nup62, no additive effect was noted beyond the arrest conferred by the individual siRNAs (Fig. S2B), suggesting that they all function in the same nuclear transport pathway. We also found that leptomycin B, a drug that blocks chromosome maintenance region 1 (Crm1)-dependant export from the nucleus, inhibits VACV spread (Fig. S2B). Whether interference with nuclear transport prevents necessary cellular molecules from exiting the nucleus or prevents viral defense proteins from entering cannot be ascertained at this time, although the former possibility seems more likely. So far, only one VACV protein, E3, has been shown to enter the nucleus (43), but its ability to explain the effects seen here is in doubt.
Experimental Procedures
Screening was conducted with the Ambion Silencer Select Human Genome siRNA Library version 4 of nonpooled siRNAs and the Dharmacon siGENOME SMARTpool siRNA, consisting of four unique siRNA duplexes per gene in a single well. The wells of assay plates were imaged with a Molecular Devices ImageXpress Micro high-content platform integrated into an Agilent BioCel robotic system. The screen, statistical analysis, hit identification, and other methods are described in detail in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Elizabeth Fischer [National Institute of Allergy and Infectious Diseases (NIAID) Rocky Mountain Electron Microscopy Laboratory] for providing EM images and Catherine Cotter (NIAID Laboratory of Viral Diseases) for assisting with tissue cultures. This research was supported by the Division of Intramural Research, NIAID, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300708110/-/DCSupplemental.
References
- 1.Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- 2.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 3.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4(5):495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.König R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463(7282):818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 6.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455(7210):242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA. 2009;106(38):16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda D, et al. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc Natl Acad Sci USA. 2011;108(47):19036–19041. doi: 10.1073/pnas.1113643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320(5875):531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 10.Silva PNG, et al. Differential role played by the MEK/ERK/EGR-1 pathway in orthopoxviruses vaccinia and cowpox biology. Biochem J. 2006;398(1):83–95. doi: 10.1042/BJ20060509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahbar R, Rogers E, Murooka T, Kislinger T, Fish EN. Glomulin: A permissivity factor for vaccinia virus infection. J Interferon Cytokine Res. 2012;32(3):127–137. doi: 10.1089/jir.2011.0100. [DOI] [PubMed] [Google Scholar]
- 12.Guerra S, Cáceres A, Knobeloch KP, Horak I, Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4(7):e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paran N, De Silva FS, Senkevich TG, Moss B. Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe. 2009;6(6):563–569. doi: 10.1016/j.chom.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knutson BA, Liu X, Oh J, Broyles SS. Vaccinia virus intermediate and late promoter elements are targeted by the TATA-binding protein. J Virol. 2006;80(14):6784–6793. doi: 10.1128/JVI.02705-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang LL, et al. A role for the host coatomer and KDEL receptor in early vaccinia biogenesis. Proc Natl Acad Sci USA. 2009;106(1):163–168. doi: 10.1073/pnas.0811631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alzhanova D, Hruby DE. A host cell membrane protein, golgin-97, is essential for poxvirus morphogenesis. Virology. 2007;362(2):421–427. doi: 10.1016/j.virol.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schepis A, Stauber T, Krijnse Locker J. Kinesin-1 plays multiple roles during the vaccinia virus life cycle. Cell Microbiol. 2007;9(8):1960–1973. doi: 10.1111/j.1462-5822.2007.00927.x. [DOI] [PubMed] [Google Scholar]
- 18.Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog. 2010;6(6):e1000954. doi: 10.1371/journal.ppat.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bengali Z, et al. Drosophila S2 cells are non-permissive for vaccinia virus DNA replication following entry via low pH-dependent endocytosis and early transcription. PLoS ONE. 2011;6(2):e17248. doi: 10.1371/journal.pone.0017248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer J, et al. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2012;2(4):1036–1047. doi: 10.1016/j.celrep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Hruby DE, Guarino LA, Kates JR. Vaccinia virus replication, I: Requirement for the host-cell nucleus. J Virol. 1979;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bengali Z, Townsley AC, Moss B. Vaccinia virus strain differences in cell attachment and entry. Virology. 2009;389(1-2):132–140. doi: 10.1016/j.virol.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasco R, Sisler JR, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: Effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67(6):3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang G, et al. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J Virol. 2005;79(20):13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husain M, Moss B. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J Virol. 2003;77(21):11754–11766. doi: 10.1128/JVI.77.21.11754-11766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon DE, Bond LM, Sahlender DA, Peden AA. A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic. 2010;11(9):1191–1204. doi: 10.1111/j.1600-0854.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 27.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung N, et al. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13(2):149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- 29.Kim SY, Volsky DJ. PAGE: Parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 31.Marine S, Bahl A, Ferrer M, Buehler E. Common seed analysis to identify off-target effects in siRNA screens. J Biomol Screen. 2012;17(3):370–378. doi: 10.1177/1087057111427348. [DOI] [PubMed] [Google Scholar]
- 32.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18(10):456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore MS, Schwoebel ED. Curr Protoc Cell Biol. 2001. Nuclear import in digitonin-permeabilized cells. Chapter 11:Unit 11.17. [DOI] [PubMed] [Google Scholar]
- 34.Lin YC, et al. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J Virol. 2008;82(12):5922–5932. doi: 10.1128/JVI.02723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135(3):417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hruby DE, Lynn DL, Kates JR. Vaccinia virus replication requires active participation of the host cell transcriptional apparatus. Proc Natl Acad Sci USA. 1979;76(4):1887–1890. doi: 10.1073/pnas.76.4.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dales S. Reciprocity in the interactions between the poxviruses and their host cells. Annu Rev Microbiol. 1990;44:173–192. doi: 10.1146/annurev.mi.44.100190.001133. [DOI] [PubMed] [Google Scholar]
- 38.Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20(6):998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Fontoura BM, Faria PA, Nussenzveig DR. Viral interactions with the nuclear transport machinery: Discovering and disrupting pathways. IUBMB Life. 2005;57(2):65–72. doi: 10.1080/15216540500078608. [DOI] [PubMed] [Google Scholar]
- 40.Malik P, et al. Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J Biol Chem. 2012;287(15):12277–12292. doi: 10.1074/jbc.M111.331777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CW, et al. Epstein-Barr virus protein kinase BGLF4 targets the nucleus through interaction with nucleoporins. J Virol. 2012;86(15):8072–8085. doi: 10.1128/JVI.01058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park N, Skern T, Gustin KE. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J Biol Chem. 2010;285(37):28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195(2):732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.