Abstract
The circadian clock is constituted by a complex molecular network that integrates a number of regulatory cues needed to maintain organismal homeostasis. To this effect, posttranslational modifications of clock proteins modulate circadian rhythms and are thought to convert physiological signals into changes in protein regulatory function. To explore reversible lysine acetylation that is dependent on the clock, we have characterized the circadian acetylome in WT and Clock-deficient (Clock−/−) mouse liver by quantitative mass spectrometry. Our analysis revealed that a number of mitochondrial proteins involved in metabolic pathways are heavily influenced by clock-driven acetylation. Pathways such as glycolysis/gluconeogenesis, citric acid cycle, amino acid metabolism, and fatty acid metabolism were found to be highly enriched hits. The significant number of metabolic pathways whose protein acetylation profile is altered in Clock−/− mice prompted us to link the acetylome to the circadian metabolome previously characterized in our laboratory. Changes in enzyme acetylation over the circadian cycle and the link to metabolite levels are discussed, revealing biological implications connecting the circadian clock to cellular metabolic state.
Keywords: chromatin, epigenetics
The circadian clock is a hierarchical network of biological pacemakers found in all tissues that direct and maintain proper rhythms in endocrine and metabolic pathways required for organism homeostasis (1, 2). The clock machinery consists of core transcription factors, which drive expression of clock-controlled genes (CCGs). An estimated 10% or more of the genome is believed to oscillate in a circadian and clock-controlled manner (3–5). The clock machinery is tightly regulated by transcriptional/translational events (6); posttranslational modifications of key circadian proteins (7), including phosphorylation (8–11), acetylation (12–15), SUMOylation (16) and ubiquitylation (17–19); as well as enzymatic feedback loops that directly link metabolites such as NAD+ with enzymes involved in circadian function and ultimately regulation of CCG expression (20, 21). Importantly, reversible lysine acetylation of clock proteins involved in clock control has been shown for aryl hydrocarbon receptor nuclear translocator-like (BMAL1) (13), Period 2 (PER2) (12), and the glucocorticoid receptor (GR) (14). Furthermore, subsequent reversible deacetylation of BMAL1 (15) and PER2 (12) by the NAD+-dependent histone deacetylase sirtuin 1 appear to be critical events for maintaining proper biological circadian rhythmicity.
Our current understanding of reversible lysine acetylation extends to both histone and nonhistone proteins. Protein acetylation has been implicated in regulating gene expression programs (22–24), cellular structural integrity maintenance (25, 26), metabolic/energy control (27, 28), and cellular growth/proliferation pathways (29). Furthermore, histone acetyltransferases as well as histone deacetylases target specific lysine residues, and the specific addition or removal of an acetyl group can be linked with modulation of enzymatic activity (30, 31), changes in DNA binding capacity of transcription factors (32, 33), alterations in protein–protein interaction (13, 34, 35), as well as regulation of protein stability (26, 36). A number of studies have provided important advancements in mass spectrometry (MS) used for identifying lysine acetylation sites, revealing a role for not only nuclear and cytoplasmic acetylated proteins but also an extensive number of mitochondrial proteins involved in metabolic pathways required for cellular homoeostasis (37–39).
To further expand on these studies, we investigated protein lysine acetylation that is directed by the circadian clock, using quantitative MS in WT and Clock−/− mouse liver at different zeitgeber times (ZTs). A significant number of mitochondrial enzymes were found to be differentially acetylated, mostly enriched in pathways related to energy production and metabolism. Using computational modeling, we correlated our acetylome dataset with available circadian transcriptome data (40, 41), as well as metabolomics data recently obtained in our laboratory, whereby metabolite levels were quantified over the circadian cycle (42). The present study represents a first look at the global circadian acetylome in a metabolic tissue and establishes a link between protein acetylation and presumed modulation of enzymatic activity with metabolites whose pathways could be influenced by the circadian clock. Our results reveal a previously unappreciated, extensive control that the circadian machinery exerts on global protein acetylation.
Results
Circadian Acetylome Reveals Pathways Implicated in Metabolism.
We sought to investigate the circadian acetylome in a physiological setting and explore the contribution of the clock system to global protein acetylation. To do so, we used an MS-based approach to analyze lysine acetylation sites in mouse liver in WT and Clock−/− littermates. Mice were killed at ZT 3, 9, 15, and 21, and three independent livers from each ZT were used for acetylome analysis. A total of 179 proteins were found to be acetylated, with 306 unique sites of acetylation (Table 1). Of these 179 acetylated proteins, 111 contained a single site of acetylation (Table 1). Based on a P value cutoff of 0.05, 19 acetylation sites were found to oscillate in WT livers only, 15 acetylation sites oscillate in Clock−/− livers only, and two acetylation sites oscillate in both WT and Clock−/− livers (Table 1). Moreover, based on acetylation profiles between WT and Clock−/− livers, 47 proteins displayed a differential acetylation at one ZT, and 30 proteins were differentially acetylated at two or more ZTs between WT and Clock−/− livers (Table 1). The correlation coefficients of acetylation profiles comparing WT and Clock−/− livers are graphed in Fig. S1.
Table 1.
Acetylome data from WT and CLOCK−/− liver
| Classification | Protein-level statistics | Acetylation site statistics |
| Total acetylated protein hits | 179 | |
| Protein hits with a single acetylation site | 111 | |
| Protein hits disrupted at one ZT | 47 | |
| Protein hits disrupted at two ZTs | 22 | |
| Protein hits disrupted at three ZTs | 7 | |
| Protein hits disrupted at four ZTs | 1 | |
| Total acetylation sites | 306 | |
| Circadian acetylation in WT liver only | 19 | |
| Circadian acetylation in CLOCK−/− liver only | 15 | |
| Circadian acetylation in both WT and CLOCK−/− liver | 2 |
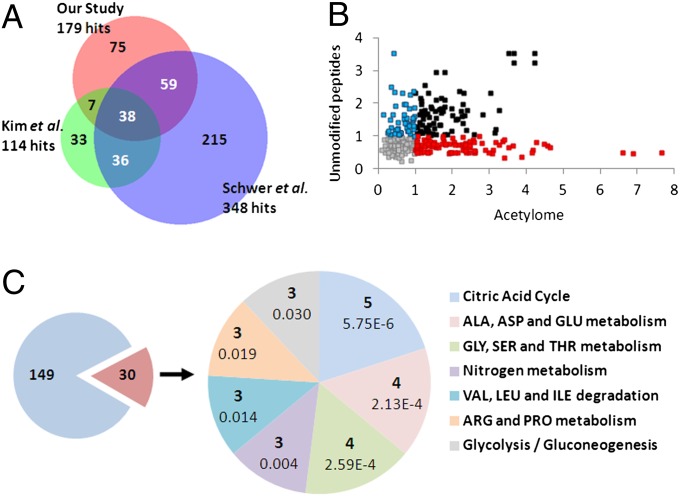
We compared our dataset to other acetylome studies using venn diagram analysis (Fig. 1A). In terms of a mouse liver acetylome, Kim et al. identified 114 acetylated proteins, the majority of which were mitochondrial enzymes (38). Schwer et al. used mouse liver for an acetylome analysis in response to calorie restriction and identified 348 unique acetylated proteins, of which 165 are mitochondrial (43). When these two studies were compared with our hits, 38 proteins were found to be in common among all three datasets, and 45 and 97 proteins were found in common between our dataset and Kim et al. and Schwer et al., respectively (Fig. 1A).
Fig. 1.
Comparison and pathway analysis of the circadian acetylome. (A) Venn diagram comparing protein hits from three different liver acetylomes—our study, Schwer et al. (43), and Kim et al. (38). (B) Scatter plot showing for each acetylation site, the range (max – min) of its acetylation profile from WT liver on the x-axis versus the range of the mean-profile of the three unmodified peptides with the highest average intensity on the y-axis. The mean-profile is used as an estimate of the protein profile (67, 68). Red, acetylation sites with greater than twofold dynamic range in raw acetylation intensities and less than twofold dynamic range in raw unmodified peptide intensities. Blue, acetylation sites with less than twofold dynamic range in raw acetylation intensities and greater than twofold dynamic range in raw unmodified peptide intensities. Black, acetylation sites with greater than twofold dynamic range in both raw acetylation intensities and raw unmodified peptide intensities. (C) The 306 acetylation sites correspond to 179 proteins, of which 30 contain one or more acetylation sites that are differentially acetylated at two or more time points between the WT and Clock−/−, based on a CyberT (64, 65) P value of 0.05. A KEGG pathway enrichment analysis performed using DAVID (48, 49) shows that 24 of these 30 proteins can be mapped to seven KEGG pathways, each containing at least three of these proteins. For each of these pathways, the pie chart on the right shows the corresponding number of proteins and enrichment P value.
The enrichment of acetylated peptides using beads coated with an anti-acetyl lysine antibody results in a higher number of identified lysine sites compared with a direct analysis (44). However, similar to methods used to enrich for phosphorylation sites (45), a substantial number of unmodified peptides were identified using our enrichment procedure (46), as the amount of unmodified protein is expected to exceed acetylated protein abundance. These data enabled us to investigate whether the observed increase in acetylation correlated with an increase in the amount of the corresponding protein as we used the ion intensities of unmodified peptides derived from the identified proteins to measure the total protein abundance. To do so, we plotted the dynamics of unmodified peptide abundance versus variable levels of peptide acetylation in WT liver, with the axes representing fold change of raw intensity. The scatter plot in Fig. 1B reveals the presence of various associations in data population. Data points on the lower far right (red dots) represent dynamic changes in acetylation with minimal changes in unmodified peptide abundance. Conversely, data points in the upper left quadrant (blue dots) represent hits that change in unmodified peptide abundance, while acetylation is less dynamic. Peptides whose levels change both in acetylation and unmodified peptide abundance, not necessarily in phase, are indicated in black (Fig. 1B). The remaining hits shown in gray do not change dynamically. A number of examples of a subset of peptides whose acetylation is oscillatory, while the abundance of unmodified peptides from the same protein is not, are presented in Fig. S2. This analysis directly addresses whether changes in acetylation of a peptide are inherent or independent from its abundance. Our data are further supported by data derived from another study that shows that 12 proteins oscillate in abundance out of the 179 acetylated proteins found here (47).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (48, 49). A subset of significant protein hits that were differentially acetylated between WT and Clock−/− livers at two or more ZTs were used for pathway analysis. Of the 179 protein hits, 30 proteins matched this criteria, 24 of which were matched into pathways defined by DAVID. These pathways include citric acid cycle, amino acid metabolism, nitrogen metabolism, and glycolysis/gluconeogenesis (Fig. 1C). Shown in Fig. S3 is pathway analysis for protein hits differentially acetylated between WT and Clock−/− livers at one ZT, indicating that significant pathways include amino acid metabolism, fatty acid elongation and metabolism, oxidative phosphorylation, and the citric acid cycle.
Global Trends of Circadian Lysine Acetylation.
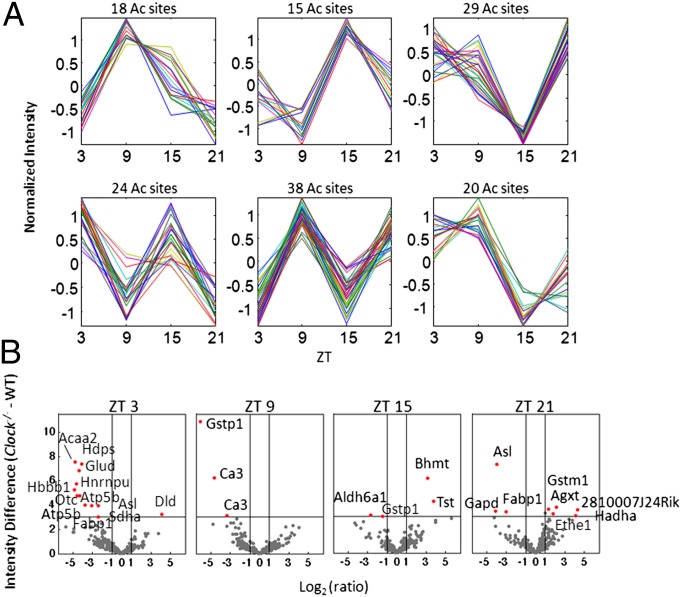
Global trends in cycling acetylation profiles were determined by k-means clustering. The profiles of all acetylation sites were normalized to zero mean and unit variance, and then grouped into 12 clusters using k-means clustering with respect to the Pearson correlation similarity measure. Shown in Fig. 2A are 6 out of 12 clusters with highly oscillating profiles. The largest of these clusters that contained 38 observations or different sites of acetylation demonstrated a robust peak at ZT 9. Interestingly, a ZT 9 peak is also apparent in two other clusters (Fig. 2A). Conversely, two different clusters present the opposite trend with a sharp trough at ZT 9 (Fig. 2A). Furthermore, there could be possible ultradian trends in acetylation based on profiles shown in Fig. 2A, but given the current resolution of the study, it is difficult to decipher ultradian from circadian trends in protein acetylation.
Fig. 2.
Clustering and differential analysis of the circadian acetylome. (A) The k-means clustering of the WT acetylation profiles after normalization of each profile (mean = 0 and variance = 1), using Pearson correlation as the similarity measure with k = 12. Shown are 6 of the 12 clusters with their corresponding number of acetylation sites. (B) Volcano plots show the significance versus fold-change in raw values between the Clock−/− and WT acetylation levels, at each ZT. x-axis, difference in intensity (Clock−/− - WT); Y-axis, –log10 of the CyberT (64, 65) P value comparing Clock−/− and WT acetylation intensities. Protein labels refer to corresponding acetylation sites that are significantly different between Clock−/− and WT liver with a P value less than 0.001 and a raw acetylation intensity fold-change of at least 2.
For each ZT, volcano plots were generated to visualize acetylated protein hits that show a significant change of acetylation levels between the Clock−/− and WT (Materials and Methods) (Fig. 2B). Positive numbers indicate acetylation levels are higher in Clock−/− versus WT livers, whereas negative numbers show lower levels of acetylation in Clock−/− versus WT livers. Significant hits include enzymes involved in mitochondrial fatty acid beta-oxidation such as acetyl-CoA acyltransferase 2 and hydroxyacyl-CoA dehydrogenase alpha (HADHA) and fatty acid binding and transport, such as fatty acid-binding protein 1 (FABP1) (Fig. 2B). Hepatic mitochondrial and cytosolic enzymes involved in the urea cycle were also significant hits, including ornithine transcarbamoylase and argininosuccinate lyase (Fig. 2B). Similarly, detoxifying glutathione-S transferases were significant hits, including glutathione S-transferase pi 1 and glutathione S-transferase mu 1 (GSTM1). Lastly, acetylation levels of the succinate dehydrogenase protein complex subunit A, which is involved in the citric acid cycle, as well as dihydrolipoamide dehydrogenase (DLD), which is part of the pyruvate dehydrogenase and the alpha-ketoglutarate dehydrogenase complexes, were significantly altered in Clock−/− and WT levels (Fig. 2B). With the exception of a few of these enzymes (DLD, HADHA, and GSTM1), relative acetylation of these enzymes/proteins was lower in the Clock−/− with respect to WT livers.
Relating the Circadian Acetylome to the Transcriptome and Proteome.
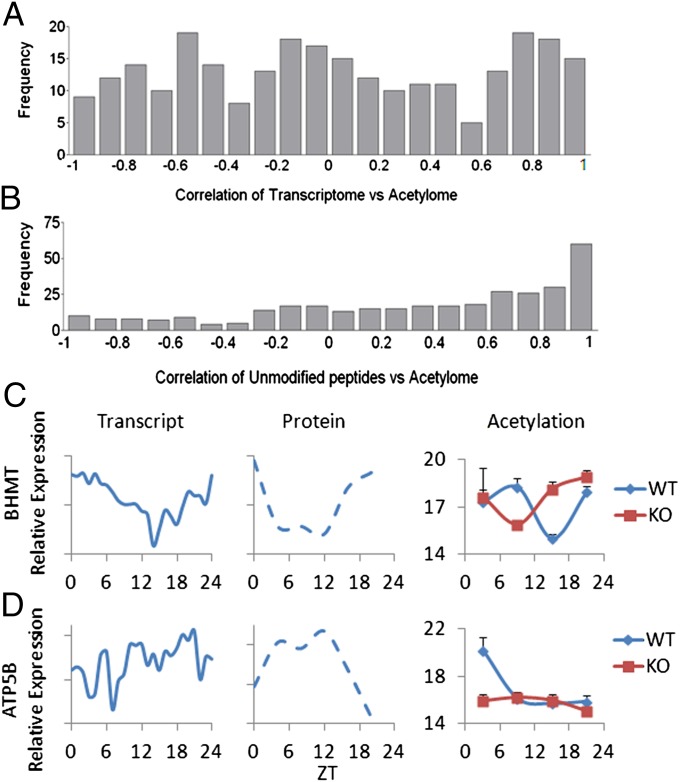
Expression data from previously published liver transcriptome studies (40, 41) were extracted and correlated to the acetylome using Pearson correlation coefficients. The expression level of each mRNA transcript was compared with the acetylation levels of all of the sites on the corresponding protein product. A value of 0 indicates no correlation between the acetylation site and the levels of the transcript; a value close to 1 indicates that these are highly correlated, whereas –1 suggests that an inverted correlation exists (Fig. 3A). This analysis shows that there are different case scenarios, whereby linear correlation, inverse correlation, and no correlation exist between the circadian acetylome and transcriptome (Fig. 3A). Conversely, the histogram in Fig. 3B shows a correlation between the profiles of unmodified peptides versus acetylated peptides from the same protein. Using a correlation cutoff of 0.8, less than one-third of these hits follow the same profile in both acetylation and total peptide abundance (Fig. 3B).
Fig. 3.
Correlation analysis between circadian transcriptome, unmodified peptides, and acetylome. (A) Histogram of Pearson correlations between acetylation profiles and the corresponding gene expression profiles from previously published transcriptome data (40, 41). (B) Histogram of Pearson correlations between acetylation profiles and the estimated protein profile obtained from the unmodified peptide profile (Fig. 1) (67, 68). (C) Previously published gene expression (40, 41) and protein profile (47) for betaine-homocysteine methyltransferase along with the acetylation profile of its acetylation site (JTK cycle P value of 0.0418). (D) Previously published gene expression (40, 41) and protein profile (47) for ATP5B along with the acetylation profile of its most differentially acetylated site (JTK cycle P value of 0.0418).
Specific examples of acetylome and transcriptome correlations are shown in Fig. 3 C and D, using a Jonckheere-Terpstra-Kendall (JTK) cycle P value cutoff of 0.05. Betaine-homocysteine methyltransferase is a highly expressed hepatic methyltransferase involved in methionine synthesis from betaine and homocysteine (50), which subsequently feeds into S-adenosyl-methionine production. The rhythm of transcript (40, 41), total protein levels (47), and unmodified peptides follow a similar circadian profile (Fig. 3C), whereas acetylation at a single lysine is cycling (JTK cycle P value of 0.0418) but antiphasic to both transcript and protein. In this example, a disruption in oscillating acetylation is seen in Clock−/− versus WT livers (Fig. 3C). A slightly different scenario is seen for ATP synthase subunit beta (ATP5B) that encodes a mitochondrial subunit of the enzyme (51). The gene and protein for ATP5B oscillate with different phases (40, 41, 47), whereas the acetylation profile shows a peak at ZT 3 when transcript and protein are low (JTK cycle P value of 0.0418). The oscillation in acetylation is abolished in the Clock−/− liver (Fig. 3D). Conversely, there are examples where transcript rhythm and the circadian oscillation of total protein follow the trend of acetylation. This is the case of carbamoyl phosphate synthetase 1 (CPS1), a mitochondrial enzyme that catalyzes the first step in the urea cycle by producing carbamoyl phosphate (52). Multiple sites of acetylation were detected for CPS1, although only one shows significant oscillation that was changed between WT and Clock−/− livers, with high total protein and acetylation levels during the day that drop at night (Table S1). For information on all protein hits found to be acetylated, the peptide sequence identified by MS, the acetylation intensity in WT and Clock−/− livers, as well as CyberT and JTK_cycle statistics, see Table S1.
Linking the Circadian Acetylome to the Metabolome.
We previously generated “Circadiomics,” a computational resource that connects nodes of oscillating metabolites to transcription networks that may be critical in dictating metabolite levels (53). Using similar computational approaches, the present acetylome data were included with the metabolome data previously published from our laboratory (42) (http://circadiomics.igb.uci.edu/). Importantly, our acetylome and metabolome analyses were performed on the same liver samples, allowing direct comparison of the data. These data can be found at http://circadiomics.igb.uci.edu.
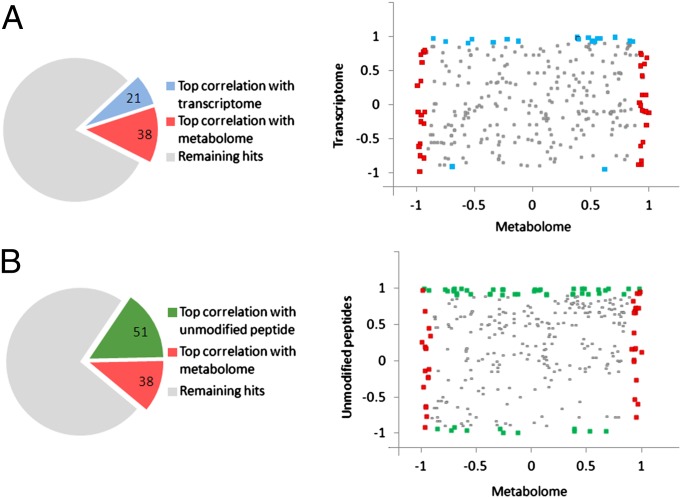
Correlation of the acetylome with the transcriptome, metabolome, and unmodified peptide abundance was performed to visualize distributions in our data by 2D scatter plots. The pie chart shown in Fig. 4A shows top correlated (≥0.9 and ≤–0.9) hits of the acetylome with transcriptome and the acetylome with metabolome datasets. These hits are displayed in the scatter plot in Fig. 4A, which shows global correlation calculated for oscillations in acetylation sites from enzymes versus corresponding metabolites and transcripts, at correlation cutoffs of ≥0.9 and ≤–0.9. Blue dots correspond to highly correlated or highly anticorrelated (|correlation| ≥ 0.9) acetylation site–transcript pairs, whereas red dots correspond to highly correlated or highly anticorrelated (|correlation| ≥ 0.9) acetylation site/metabolite pairs. Likewise, Fig. 4B displays the correlation between enzyme-linked acetylation sites versus unmodified peptides and metabolome datasets. Green dots in this scatter plot correspond to highly correlated or highly anticorrelated (|correlation| ≥ 0.9) acetylation site/mean of unmodified peptide pairs. Based on this analysis, a fairly equal distribution is seen between acetylome and metabolome positively correlated and inversely correlated hits. This is not the case for correlation distribution between acetylome versus unmodified peptides and especially transcriptome data, whereby only three hits were found to be inversely correlated between the acetylome and transcriptome. Correlation coefficients for Fig. 4 A and B are shown in Table S2.
Fig. 4.
Connecting the circadian metabolome and acetylome. (A) Scatter plot corresponding to acetylation sites associated with enzymes. Each point corresponds to an acetylation site on an enzyme paired with a metabolite regulated by that enzyme. (Note: an enzyme can regulate multiple metabolites and thus be associated with multiple dots in the scatter plot.) x-axis, Pearson correlation between the profile of the acetylation site and the profile of the metabolite from the same WT liver sample (42); y-axis, Pearson correlation between the profile of the acetylation site and the gene expression profile of the corresponding enzyme from previously published data (40, 41). The 21 blue dots correspond to highly correlated or highly anticorrelated (|correlation| ≥ 0.9) acetylation site/transcript pairs. The 38 red dots correspond to highly correlated or highly anticorrelated (|correlation| ≥ 0.9) acetylation site/metabolite pairs. (B) Scatter plot and x-axis similar to Fig. 4A. y-axis, Pearson correlation between the acetylation site profile and the abundance of unmodified peptide (Fig. 1) (67, 68). The 51 green dots correspond to highly correlated or highly anticorrelated (|correlation| ≥ 0.9) acetylation site/mean of unmodified peptide pairs. Correlation coefficients for Fig. 4 A and B are shown in Table S2.
Discussion
The remarkable influence played by the circadian clock on cellular and organismal physiology is illustrated by a number of transcriptome (3, 4, 40) and metabolome studies (42, 54). The question remains open, however, on the extent and specificity of circadian enzymatic activities. Indeed, it is not established how much and what types of posttranslational modification reactions are driven by the clock and/or serve circadian regulatory networks. Recent studies showing that mammalian circadian control can be exerted in the absence of transcription underscore the critical role played by posttranslational modifications of proteins (55). Furthermore, the intimate connections between circadian rhythms and cellular metabolism (1, 2, 6) place the mitochondrion in a strategic position as the interface between the clock machinery and energy control (56). Finally, the recent completion of a comprehensive proteome for the circadian liver shows that only 6% of proteins oscillate,* highlighting the importance that posttranslational modifications are likely to have in circadian control. Furthermore, the extent to which clock-dependent histone acetyltransferase activity may play a role in directly regulating protein acetylation will implicate further investigation. This is an important issue as multiple levels of complexity exist: CLOCK-dependent histone acetyltransferase activity may indeed have been abolished in the Clock−/− mice, yet these animals were fed ad libitum, possibly implicating a food-entrainable oscillator that may dictate circadian control in a highly metabolic tissue such as the liver. A compensation mechanism may also be in place to account for the loss of CLOCK protein in the livers of these animals, potentially implicating liver-specific complexes that could direct acetylation.
As increasing evidence links the clock to cellular metabolism (1, 57), we wondered whether there are substantial changes of the acetylome in the liver along the circadian cycle, especially given that posttranslational modifications do play an important role in regulating the clock. The choice of the liver was dictated by a number of considerations. First, the liver comprises a powerful pacemaker, whose function is tightly linked to metabolic pathways. Moreover, we have recently reported the circadian metabolome for the liver, in which the profiles for ∼600 metabolites were determined. These have been presented in a coherent manner within the CircadiOmics resource (http://circadiomics.igb.uci.edu/) (53). This computational resource was recently established to present the regulatory connections between specific metabolic nodes and transcriptome networks. Importantly, as the samples used here for the acetylome were the same as the ones used for the metabolome (42), we have been able to establish parallels within metabolic pathways and the circadian acetylome. Our findings underscore the presence of links between specific metabolites and distinct enzymatic pathways. For example, NAD+, for which oscillation has already been reported (20, 21), feeds into a number of enzymes for which we observe oscillatory acetylation.
An intrinsic limitation of acetylome studies is that they do not allow for comprehensive identification of proteins below a certain abundance threshold (58). In addition, other acetylome studies have been performed in cell cultures that have been treated with histone deacetylase inhibitors to increase the number of acetylated hits (37). Our choice of studying the liver acetylome was based on the interest of identifying physiologically meaningful targets whose acetylation might change along the circadian cycle. It was thereby predictable that a number of known acetylated proteins would not be found in our study. Specifically, nuclear proteins whose acetylation has been shown to be related to clock control, such as BMAL1, PER2, and glucocorticoid receptor (12–14), were not detected. Importantly, because of the metabolic function of the liver, and therefore the high abundance of mitochondrial proteins, we expected to find in this group a large majority of our acetylated hits.
Our analysis demonstrates the presence of all types of case scenarios for protein acetylation along the circadian cycle and in relation to the abundance of unmodified peptides. Based on the notion that acetylation has been linked to stability for a number or proteins (26, 36), our findings indicate that changes in acetylation may be linked to the oscillation of distinct protein levels for one group, or not at all for another group. Our correlation analyses indicate that direct or inverted correlation exists between the acetylome and the circadian metabolome, whereby the transcriptome and the acetylome oscillate exclusively with the same phase.
Disruption of the clock may result in a number of changes in the circadian profile of both transcripts (59) and metabolites (42), which could manifest in dampened rhythms, phase shifts, and also elevated levels of transcripts or metabolites in a clock mutant or clock-deficient background. Our acetylome dataset contains similar examples. First, a significant number of circadian acetylation events are abolished in the Clock-deficient mice (Table 1 and Fig. S1). Also, there is a presence of oscillatory acetylation in proteins identified exclusively in Clock−/− livers (Table 1). As the clock mechanism is disrupted in these livers, the most likely explanation is a dominant effect of food intake in these mice versus their WT littermates. This observation is in keeping with the notion that the presence of food-entrainable oscillators is readily revealed in the absence of a functional clock (60).
Our study constitutes a unique profiling of the circadian acetylome in the mouse liver. The significance of our results lays on the identification of specific links between acetylated proteins and respective metabolites, demonstrating the remarkable extent by which the clock exerts its function in a nontranscriptional manner.
Materials and Methods
Animal Housing and Experimental Design.
WT and Clock−/− animals were a generous gift of S. Reppert (University of Massachusetts Medical School, Worcester, MA) (61). All experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines at the University of California at Irvine. Animals were housed in a 12 h light/dark paradigm and fed ad libitum. Age-matched adult male mice were killed at 6 h intervals at ZT 3, 9, 15, and 21 for metabolomic and acetylomic profiling analysis. Livers were quickly removed from the animals and flash-frozen in liquid nitrogen until use. Livers from three animals were used per experimental condition for the circadian acetylome.
Acetylome Experimental Details.
The acetylome protocol was adapted from Guan et al. (44), with a number of modifications. Frozen liver was minced and homogenized in cold homogenizing buffer [50 mM Tris⋅HCl, pH 7.5, 500 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40 and 20% (wt/vol) glycerol] with the addition of 1× protease inhibitor mixture (Roche), 10 mM trichostatin A, and 6 M Urea. Samples were rocked at 4 °C for 1 h, followed by a brief sonication at 30% power (3×, 10 s). Protein concentration was determined by Bradford, and equal amounts of protein were used for subsequent steps. Samples were reduced with 1 mM DTT for 45 min at room temperature, followed by iodoacetamide treatment for 30 min at room temperature, in the dark. Urea was diluted to a final concentration of 1M with the addition of 40 mM ammonium bicarbonate, and trypsin (Worthington) was added at a concentration of 1:100 and incubated overnight at room temperature. Samples were acidified and diluted with trifluoroacetic acid (TFA) and loaded onto Sep-Pak Plus C18 cartridges (Waters). Columns were washed with 0.1% TFA and subsequently eluted with 60% (wt/vol) acetonitrile/0.1% TFA and speed vacuumed until dry. Dried pellets were resuspended in immunoprecipitation buffer (50 mM Hepes, pH 8, and 40 mM NaCl), peptide concentration was determined, and equal concentrations of peptide were subjected to immunoprecipitation with an agarose-conjugated acetyl-lysine antibody (ImmuneChem). Beads were subsequently washed three times in 1× PBST (0.1%) and three additional times in 1× phosphate buffered saline (PBS). Tryptic peptides were eluted from beads with 0.1% TFA.
MS Analysis.
Tryptic peptides were separated in an Ultimate 3000 high-performance liquid chromatography (HPLC) system (LC Packings) as described elsewhere (62), with minor modifications. The effluent from the HPLC was directly electrosprayed into a linear trap quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The MS instrument was operated in data-dependent mode. Survey full-scan MS spectra (from m/z 300–2,000) were acquired in the Orbitrap with resolution R = 60,000 at m/z 400 (after accumulation to a “target value” of 500,000 in the linear ion trap). The six most intense peptide ions with charge states between two and four were sequentially isolated to a target value of 10,000 and fragmented by collision-induced dissociation and recorded in the linear ion trap. For all measurements with the Orbitrap detector, three lock-mass ions were used for internal calibration (63). Typical MS conditions were spray voltage, 1.5 kV; no sheath and auxiliary gas flow; heated capillary temperature, 200 °C; normalized collision-induced dissociation energy 35%; activation q = 0.25; and activation time = 30 ms. Proteins were identified using Maxquant 1.2.2.5 [Database, Swissprot 57.10; taxonomy, Mus musculus; MS tol, 10 ppm; MS/MS tol, 0.5 Da; peptide false discovery rate (FDR), 0.01; Protein FDR, 0.01; min. peptide length, 6; variable modifications, oxidation (M), acetylation (K); fixed modifications, Carbamidomethyl (C)]. Conditions for Maxquant quantification of acetylation sites were Site FDR, 0.01; peptides for protein quantitation, unique and razor; min. peptides, 1; less modified peptides. “Intensity” in this paper refers to log2 normalized MS values, whereas “profile” refers to the intensity at all four ZTs.
Statistical Analysis of Acetylome Data.
For each of the four ZT time points, a regularized paired T-test was performed to compare the mean intensity between Clock−/− and WT liver samples using CyberT (64, 65). The sliding window size for Bayesian SD estimation was set to 7. P values of less than 0.05 were considered significant, and all acetylation sites were further classified based on the number of time points at which each site showed a significant difference between Clock−/− and WT liver samples. These P values were also used for the Volcano plots.
A nonparametric algorithm, JTK_CYCLE, was used to detect acetylation sites that displayed 24-h rhythmicity (66). P values less than 0.05 were considered significant, and the corresponding time series was classified as rhythmic.
Supplementary Material
Acknowledgments
We thank all members of the P.S.-C., A.I., and P.B. laboratories for helpful discussion and technical assistance. Jordan Hayes helped with Web site design (http://circadiomics.igb.uci.edu). We also thank M. Mann, M. Robles, and F. Gachon for providing unpublished information on circadian proteome analysis. Funding was provided by National Institutes of Health (NIH) postdoctoral fellowship GM097899 (to S.M.). K.L.E.-M. was supported by NIH postdoctoral fellowship DK083881. Financial support was provided by NIH Grant AG041504, Institut National de la Santé et de la Recherche Médicale Grant 44790, and Sirtris Pharmaceuticals Grant SP-48984 (all to P.S.-C.). The work of V.R.P. and P.B. was supported by NSF Grant IIS-0513376, NIH Grant LM010235, and NIH Grant T15 LM07443 (to P.B.). Support was also provided by the Bavarian-Californian Technology Center (BaCaTeC 3/2011-1) and by a grant from European Union EpiGeneSys (both to A.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.A.B. is a guest editor invited by the Editorial Board.
Data deposition: The acetylome data reported in this paper are available at http://circadiomics.igb.uci.edu.
*Robles MS, Mann M, 2012 Federation of European Biochemical Societies Conference (September 4–9, 2012, Sevilla, Spain), P28-11 (abstr.).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217632110/-/DCSupplemental.
See Commentary on page 3210.
References
- 1.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111(7):919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 3.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 4.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12(7):540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 5.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12(7):551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 6.Sahar S, Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer. 2009;9(12):886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 7.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8(2):139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 8.Sanada K, Okano T, Fukada Y. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. J Biol Chem. 2002;277(1):267–271. doi: 10.1074/jbc.M107850200. [DOI] [PubMed] [Google Scholar]
- 9.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shim HS, et al. Rapid activation of CLOCK by Ca2+-dependent protein kinase C mediates resetting of the mammalian circadian clock. EMBO Rep. 2007;8(4):366–371. doi: 10.1038/sj.embor.7400920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002;22(6):1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 13.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 14.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: Potential physiological implications. FASEB J. 2009;23(5):1572–1583. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 17.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316(5826):900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 18.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316(5826):897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 19.Siepka SM, et al. Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129(5):1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 24.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 25.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24(2):473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 26.Takemura R, et al. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J Cell Sci. 1992;103(Pt 4):953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- 27.Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36(2):108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wellen KE, Thompson CB. A two-way street: Reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13(4):270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 29.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42(4):426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103(27):10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 33.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396(6711):594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 34.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307(5707):269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 35.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 36.Grönroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10(3):483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 37.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 38.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes M, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8(5):604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan KL, Yu W, Lin Y, Xiong Y, Zhao S. Generation of acetyllysine antibodies and affinity enrichment of acetylated peptides. Nat Protoc. 2010;5(9):1583–1595. doi: 10.1038/nprot.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soderblom EJ, Philipp M, Thompson JW, Caron MG, Moseley MA. Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal Chem. 2011;83(10):3758–3764. doi: 10.1021/ac200213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, et al. The fasted/fed mouse metabolic acetylome: N6-acetylation differences suggest acetylation coordinates organ-specific fuel switching. J Proteome Res. 2011;10(9):4134–4149. doi: 10.1021/pr200313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans JC, et al. Betaine-homocysteine methyltransferase: Zinc in a distorted barrel. Structure. 2002;10(9):1159–1171. doi: 10.1016/s0969-2126(02)00796-7. [DOI] [PubMed] [Google Scholar]
- 51.Neckelmann N, et al. The human ATP synthase beta subunit gene: Sequence analysis, chromosome assignment, and differential expression. Genomics. 1989;5(4):829–843. doi: 10.1016/0888-7543(89)90125-0. [DOI] [PubMed] [Google Scholar]
- 52.Anderson PM, Meister A. Bicarbonate-dependent cleavage of adenosine triphosphate and other reactions catalyzed by Escherichia coli carbamyl phosphate synthetase. Biochemistry. 1966;5(10):3157–3163. doi: 10.1021/bi00874a012. [DOI] [PubMed] [Google Scholar]
- 53.Patel VR, Eckel-Mahan K, Sassone-Corsi P, Baldi P. CircadiOmics: Integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat Methods. 2012;9(8):772–773. doi: 10.1038/nmeth.2111. [DOI] [PubMed] [Google Scholar]
- 54.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469(7331):554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallace DC. Mitochondria, bioenergetics, and the epigenome in eukaryotic and human evolution. Cold Spring Harb Symp Quant Biol. 2009;74:383–393. doi: 10.1101/sqb.2009.74.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism—The epigenetic link. J Cell Sci. 2010;123(Pt 22):3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10(4):1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 59.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA. 2007;104(9):3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mieda M, Williams SC, Richardson JA, Tanaka K, Yanagisawa M. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci USA. 2006;103(32):12150–12155. doi: 10.1073/pnas.0604189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50(3):465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 62.Forne I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics. 2012;11(4):M111.012088. doi: 10.1074/mcp.M111.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17(6):509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 65.Kayala MA, Baldi P. Cyber-T web server: Differential analysis of high-throughput data. Nucleic Acids Res. 2012;40(Web Server issue):W553–W559. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arike L, et al. Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J Proteomics. 2012;75(17):5437–5448. doi: 10.1016/j.jprot.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 68.Cox J, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4(5):698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.