Abstract
Miniaturized laboratory-on-chip systems promise rapid, sensitive, and multiplexed detection of biological samples for medical diagnostics, drug discovery, and high-throughput screening. Within miniaturized laboratory-on-chips, static and dynamic droplets of fluids in different immiscible media have been used as individual vessels to perform biochemical reactions and confine the products. Approaches to perform localized heating of these individual subnanoliter droplets can allow for new applications that require parallel, time-, and space-multiplex reactions on a single integrated circuit. Our method positions droplets on an array of individual silicon microwave heaters on chip to precisely control the temperature of droplets-in-air, allowing us to perform biochemical reactions, including DNA melting and detection of single base mismatches. We also demonstrate that ssDNA probe molecules can be placed on heaters in solution, dried, and then rehydrated by ssDNA target molecules in droplets for hybridization and detection. This platform enables many applications in droplets including hybridization of low copy number DNA molecules, lysing of single cells, interrogation of ligand–receptor interactions, and rapid temperature cycling for amplification of DNA molecules.
Keywords: nanowire, on-chip heating, evaporation
Recent developments in high-throughput screening technologies have made it possible to process thousands of individual reaction volumes at a time (1). Previous subnanoliter screening techniques used droplets-in-oil, micromachined chambers, and other strategies (2–4). Encapsulating droplets with mineral oil, capping them with polydimethylsiloxane (PDMS), or covering and sealing microchambers with glass and a nail polish solution have all been used to minimize evaporation (3, 5–7). Similarly, biologically compatible solvents with low volatility have been used for limiting evaporation in parallel reactions for screening applications (8). Further integrating a miniaturized heating element with droplet screening technologies can enable many temperature-mediated biochemical reactions such as high-throughput melting curve analyses of individually generated subnanoliter droplets. Such individually addressable heating elements at the microscale can allow for greater spatial and temporal control of temperature profiles.
Previous on-chip, localized heating designs focused on peltier heaters, resistive heaters, or other methods (9–13). A variation on the resistive heater uses a transistor as a heater whereby adjusting the source-drain current via modulation of the gate voltage can result in heating of the fluid above the device (14). This approach, however, required a very wide gate region (∼700 μm) and is incompatible with the use of picoliter scale droplets. A second approach involving microwave heating of picoliter droplets in a microfluidic device has been studied (15), but this method does not allow for individualized heating of droplets and also requires mineral oil as an encapsulation layer to minimize evaporation. Finally, optical heating methods have also been used (16, 17), but suffer from setup complexity for individually heating multiple droplets and similarly require an encapsulation layer to minimize evaporation.
Earlier, we demonstrated heating via use of individual transistors by applying an alternating current (AC) voltage at 10 MHz and 10–25 Vrms between the transistor’s leads and the bulk substrate of a silicon-on-insulator (SOI) microribbon transistor structure (18, 19, 20). This technique focused on characterization of the temperature profile at the surface of a device in a bulk fluid and did not offer control of thermal cross-talk and by-product diffusion between heating elements. To allow for: individual reaction volumes, simple placement of droplets on heating units, minimization of evaporation without an encapsulation layer, reduction of thermal cross-talk, and elimination of by-product diffusion between heating elements, we have developed a droplet-in-air method using a low evaporation, biocompatible solvent (Protein Carrier Solution, NanoInk, Inc.) (Fig. 1). In the past, techniques using silicon wells with 0.4 nL volumes with ethylene glycol to limit evaporation when exposed to air have been reported (21). Similar techniques using glycerol as the low volatility solvent showed longer droplet stability, but with temperature limited to 37 °C (8). In our system, individually addressable, spatially multiplexed droplets were heated to above 80 °C over the course of a few minutes without any noticeable evaporation.
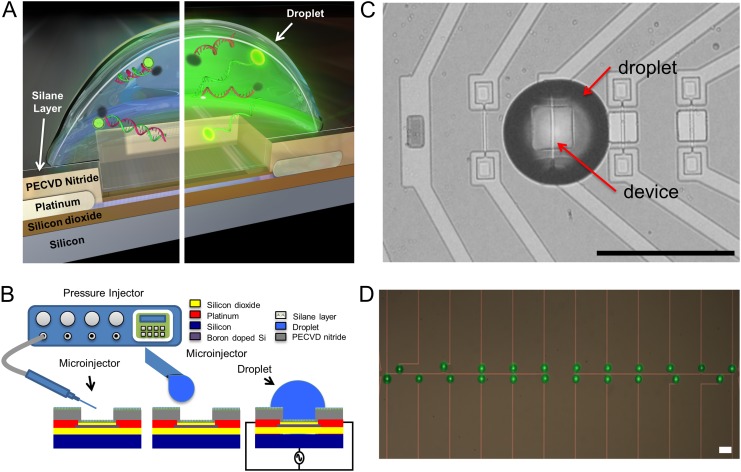
Fig. 1.
Device and methodology schematic. (A) Cross-section of device with a droplet is shown. The left side shows an unheated droplet with the DNA FRET construct in the double-stranded form. The right side shows a heated droplet where the FRET construct has denatured, resulting in an increase in fluorescence. (B) A microcapillary pressure injection system is used to spot droplets on individual devices. An AC signal between the shorted source drain and the back gate of a device is used to heat the droplet. (C) A top view of ∼225 pL droplet placed on a heating element. The heating element is 2 µm wide in a 20 µm × 20 µm release window. Scale bar, 100 µm. (D) An array of droplets is spotted on linked devices. Eleven linked on left module and 11 linked on right module. Scale bar, 100 µm.
Results and Discussion
Control of Droplet Evaporation.
The control of droplet evaporation, even at high temperature, is related to the spatial heating profile within the droplet. Simulations show that the unique heating configuration allows for highly localized and well-controlled AC heating above the device at the core of the droplet (SI Appendix, Fig. S1). Examination of the droplet’s thermal profile shows the temperature at its perimeter has returned close to room temperature. This forms a room-temperature encapsulating shell of fluid around the droplet’s heated core, which helps minimize evaporation. By comparison, extreme evaporation of droplets in bulk heating experiments compared with AC heating provides further justification for use of this localized heating technique for subnanoliter droplet-in-air heating (SI Appendix, Fig. S2).
Single Droplet Heating.
To demonstrate the feasibility of this methodology, we first focused on temperature-mediated DNA denaturation in individual droplets. A droplet of solution with double-stranded DNA (dsDNA) was placed onto a device using microinjection (Fig. 1B and Table 1). For this assay, the 5′ end of the DNA strand and the 3′ end of a cDNA strand were modified with fluorescein (FAM) and a black hole quencher (BHQ), respectively (22). The double-stranded conformation of the DNA sequence results in energy transfer between the FAM and the BHQ, producing a low level of observed fluorescence from the FAM molecule. When the dsDNA denatures, the fluorophores separate, FRET efficiency decreases, resulting in an increase in observed fluorescence from the FAM molecule. By modulating the applied voltage, we can control the temperature profile within the droplet (SI Appendix, Fig. S1). Once a threshold voltage is exceeded, one expects that the dsDNA FRET construct will denature and observed fluorescence will increase (Fig. 1A).
Table 1.
Sequence and melting temperature of the FRET constructs
| Construct | Sequence | Measured melting temperature (Tm), °C |
| 1 | 5′-/6-FAM/TGGATCCATAGTAG-3′ | 50 |
| 3′-/IABkFQ/TTTTTTTTTTACCTAGGTATCATC-5′ | ||
| 2 | 5′-/6-FAM/TGGATCCATAGTAGCGT-3′ | 61 |
| 3′-/IABkFQ/TTTTTTTTTTACCTAGGTATCATCGCA-5′ | ||
| 3 | 3′-/6-FAM/TTTTTTTTTTCGGAGCGACGGCAGCGGT-5′ | 80 |
| 5′-/IABkFQ/GCCTCGCTGCCGTCGCCA-3′ | ||
| 4 | 5′-/IABkFQ/GTTGATGTAGCGTGTCCATTA-3′ | 72 |
| 3′-/6-FAM/TTTTTTTTTTCAACTACATCGCACAGGTAAT-5′ | ||
| 5 | 5′-/IABkFQ/GTTGATTTAGCGTGTCCATTA-3′ | 70 |
| 3′-/6-FAM/TTTTTTTTTTCAACTAAATCGCACAGGTAAT-5′ | ||
| (4–5) heteroduplex | 5′-/IABkFQ/GTTGATGTAGCGTGTCCATTA-3′ | 64 |
| 3′-/6-FAM/TTTTTTTTTTCAACTAAATCGCACAGGTAAT-5′ |
This table provides the sequence information and dye modification for each of the FRET constructs used. The melting temperatures were determined using a commercial real-time PCR system’s melting curve protocol. The single base mismatch in the (72–70 °C) heteroduplex is highlighted in red.
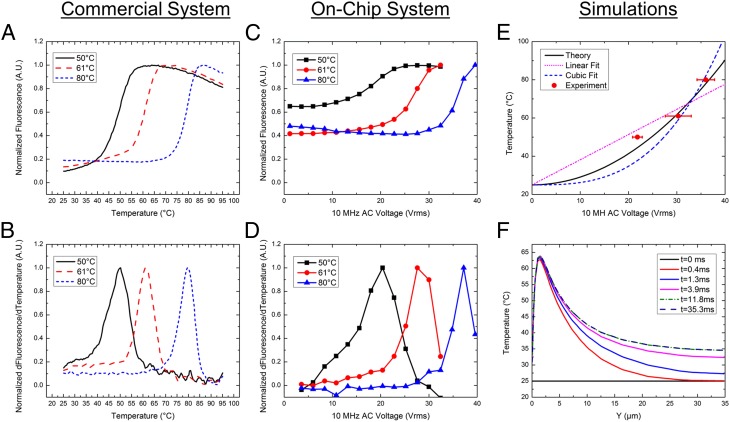
The heating technique was demonstrated using three separate FRET constructs on a single device across multiple chips, as shown in Fig. 2 C and D and SI Appendix, Fig. S3. Similar to the data from a commercial system shown in the Fig. 2 A and B, the on-chip fluorescence data also show a sigmoidal curve. The peak of the sigmoidal curve’s first derivative gives the melting temperature of the dsDNA FRET construct. In our system, the peak of the derivative provides a melting voltage, which can be correlated to the known melting temperature of the dsDNA molecule. Multiple tests provided repeatable melting voltages for the FRET constructs (SI Appendix, Table S5). Replacement of the FRET constructs with nonmodified dsDNA and an observed decrease in fluorescence associated with SYBR Green intercalation corroborated these data (SI Appendix, Fig. S4 and Table S6). Extracting the melting voltage for the three different FRET constructs establishes a calibration curve for melting voltage versus melting temperature.
Fig. 2.
Single droplet melting curves. (A) A melting curve from commercial real-time PCR machine shows an increase in fluorescence as the FRET construct denatures. (B) Derivative of A, the peak of which gives the melting temperature of the FRET construct shown in Table 1. (C) On-chip fluorescence data through a voltage sweep from 0 to 40Vrms. (D) Derivative of plot C showing the melting voltage of the constructs. Averages and SDs across multiple chips are shown in SI Appendix, Table S1. (E) Simulation versus experimental results for temperature-voltage calibration curve. Different fits are shown with R2 values of linear, 0.854; cubic, 0.586; and theory’s fit, 0.935. (F) Simulation of the time it takes for the temperature to stabilize within the droplet.
Mapping Voltage to Temperature for AC Heating.
To understand the physical basis of the calibration curve shown in Fig. 2E, we solved coupled electrical and thermal equations self-consistently through detailed numerical simulations of the device that includes both the transistor as well as the droplet (SI Appendix, Fig. S5). Details of the model are explained in the SI Appendix, and the numerical parameters used are tabulated in SI Appendix, Tables S2 and S4. Fig. 2E shows that the theoretical model anticipates, with no fitting parameters, the temperature rise within the droplet well with an R2 value of 0.935. Three observations related to heating are easily explained: (i) temperature scales roughly as the square of applied bias (i.e., 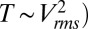 ; (ii) despite the inevitable variation of droplet size, the temperature can be set with excellent precision, and (iii) steady-state temperature is obtained within milliseconds of the onset of AC voltage (Fig. 2F). To explain the first observation, recall that the maximum temperature of the droplet,
; (ii) despite the inevitable variation of droplet size, the temperature can be set with excellent precision, and (iii) steady-state temperature is obtained within milliseconds of the onset of AC voltage (Fig. 2F). To explain the first observation, recall that the maximum temperature of the droplet, 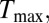 is related to the power dissipated within the droplet approximately as
is related to the power dissipated within the droplet approximately as 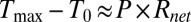 , where
, where 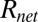 is the net thermal resistance offered to change temperature,
is the net thermal resistance offered to change temperature, 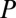 is the power generation due to dielectric heating, and
is the power generation due to dielectric heating, and 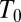 is the temperature of the surroundings. Because the field
is the temperature of the surroundings. Because the field 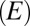 in the device is proportional to voltage
in the device is proportional to voltage 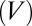 , power scales as,
, power scales as, 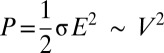 (SI Appendix, Table S3 and Fig. S10), where
(SI Appendix, Table S3 and Fig. S10), where 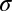 is the electrical conductivity of the dielectric medium (buffer solution/oxide). Therefore, temperature follows the scaling relationship,
is the electrical conductivity of the dielectric medium (buffer solution/oxide). Therefore, temperature follows the scaling relationship, 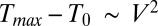 . Second, to understand the size-independent temperature control, note that heat loss can occur through either the substrate stack or through the droplet. Neglecting the thin bottom oxide layer thickness
. Second, to understand the size-independent temperature control, note that heat loss can occur through either the substrate stack or through the droplet. Neglecting the thin bottom oxide layer thickness 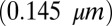 , the ratio of thermal resistance offered by these two processes can be related to the thermal conductivity of the buffer solution within the droplet
, the ratio of thermal resistance offered by these two processes can be related to the thermal conductivity of the buffer solution within the droplet 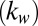 and the substrate region
and the substrate region 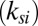 , as
, as 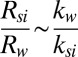 , where
, where 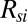 is the thermal resistance of the substrate region and
is the thermal resistance of the substrate region and 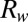 is the thermal resistance of the buffer solution. Because
is the thermal resistance of the buffer solution. Because 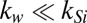 (SI Appendix, Table S4), the substrate region offers a high conduction path for temperature loss to surroundings. Therefore,
(SI Appendix, Table S4), the substrate region offers a high conduction path for temperature loss to surroundings. Therefore, 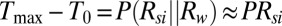 , where
, where 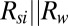 represents the parallel combination of resistances due to the two regions. Hence, the maximum temperature attained is mainly determined by the thermal resistance offered by the substrate region
represents the parallel combination of resistances due to the two regions. Hence, the maximum temperature attained is mainly determined by the thermal resistance offered by the substrate region 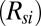 , and the temperature of the droplet can be set regardless of the inevitable variation in the droplet size. This is corroborated by experimental results that show droplets with varying diameters show the same heating characteristics (SI Appendix, Fig. S6). Because the heat source is localized due to fringing fields, uniformity in the temperature profile inside the droplet increases with decreases in the droplet size (SI Appendix, Fig. S7). Finally, a transient analysis of heat conduction (SI Appendix) shows that the temperature saturates quickly to the steady-state value (Fig. 2F) and hence any measurement done after t ∼10 ms is stable. Recently, Issadore et al. have reported a similar saturation time due to dielectric heating of water (15).
, and the temperature of the droplet can be set regardless of the inevitable variation in the droplet size. This is corroborated by experimental results that show droplets with varying diameters show the same heating characteristics (SI Appendix, Fig. S6). Because the heat source is localized due to fringing fields, uniformity in the temperature profile inside the droplet increases with decreases in the droplet size (SI Appendix, Fig. S7). Finally, a transient analysis of heat conduction (SI Appendix) shows that the temperature saturates quickly to the steady-state value (Fig. 2F) and hence any measurement done after t ∼10 ms is stable. Recently, Issadore et al. have reported a similar saturation time due to dielectric heating of water (15).
Application 1: Parallel Melting Curves.
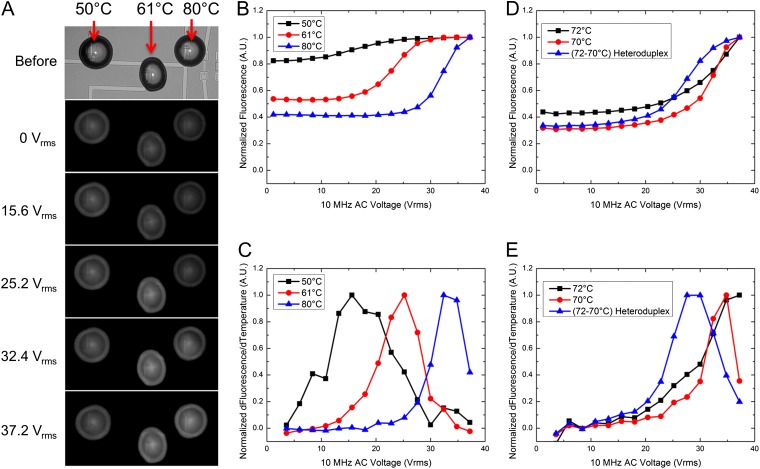
The calibration curve, shown in Fig. 2E, can now be used to achieve specific temperature points required for a variety of biological assays. For example, to further demonstrate the system’s capabilities, we performed a parallel nucleic acid denaturation study. We shorted the source contact of multiple heating elements and placed individual droplets on each device (Figs. 3A and 1D). Using different dsDNA FRET constructs with varying melting temperatures on linked devices allows us to run parallel melting curves on-chip (Fig. 3). In this experiment, a single voltage sweep interrogates three different FRET constructs. Fig. 3A shows the progression of increased fluorescence from each droplet as voltage increases. Fig. 3 B and C provide the measured raw fluorescence and derivative of the raw fluorescence versus voltage for each droplet in the first experiment. To confirm heating uniformity across linked devices (sharing a common source electrode), a single FRET construct was shown to have the same melting voltage across five linked devices (SI Appendix, Fig. S8). This approach provides a simple method of running multiple synchronous DNA melting curves on-chip and, by extracting melting voltages of different FRET constructs, a means of quickly developing a calibration curve for each chip in a single experiment or across multiple chips (SI Appendix, Table S7).
Fig. 3.
Parallel droplet heating of multiple constructs. (A) A sequence of images showing the process of heating of linked devices for plots B and C. Each droplet contains a unique FRET construct with a different melting temperature (50, 61, and 80 °C). (B) A plot of the raw fluorescence data from the droplets during the voltage sweep. (C) The derivative of B provides the melting voltage for each of the constructs. SI Appendix,Table S3 provides averages and SDs for melting curves performed on multiple devices and chips. D and E provide a second example of linked device heating. In this example, it is possible to discern between two fully complementary strands and a heteroduplex that contains a single base mismatch. SI Appendix, Table S4 provides average and SDs for the melting voltage across multiple chips.
Application 2: Single Base Mismatch Detection.
The ability to distinguish shifts in melting temperature associated with single base mismatches can be important in medical diagnostics and genetic applications. A single base mismatch results in a decrease in the overall free energy of the double-stranded complex, which decreases melting temperature (SI Appendix, Fig. S9). Fig. 3 D and E demonstrates a decrease in melting voltage for a heteroduplex of DNA consisting of a single strand from DNA No. 4 and the opposing single strand from DNA No. 5. Wider variation occurred across multiple chips, but the heteroduplex consistently showed a lower melting voltage (SI Appendix, Table S8). Heteroduplexes arise from heterozygous PCR amplifications, which are commonly used to determine donor compatibility for organ transplants (23). This system demonstrates the ability to distinguish a single base mismatch using a DNA melting curve within subnanoliter droplets and could be used to identify a noncompatible donor pair.
Application 3: DNA Microarray Technique.
We also demonstrate the capability of this system to act as a DNA microarray where each pixel also includes a miniaturized heater. In traditional DNA arrays, probe DNA is spotted on the device. The sample target DNA is then modified with a fluorophore, such as cyanine 3 (cy-3), and incubated on the probe DNA. The cy-3 target DNA hybridizes to a specific probe sequence, while nonspecifically bound DNA is washed away. The resulting cy-3 fluorescence intensity of the spot can be correlated to the amount of target DNA in the original solution (24). This system requires strict control of buffers and hybridization/washing temperatures to minimize nonspecific binding associated with false positives. In our version of the system, we use denaturation of the DNA complex to determine whether the strands are complementary, a design similar to the dynamic allele specific hybridization (DASH) assay (25). However, our system compartmentalizes melting curves into individual reaction volumes, easily allowing for large-scale parallel analyses.
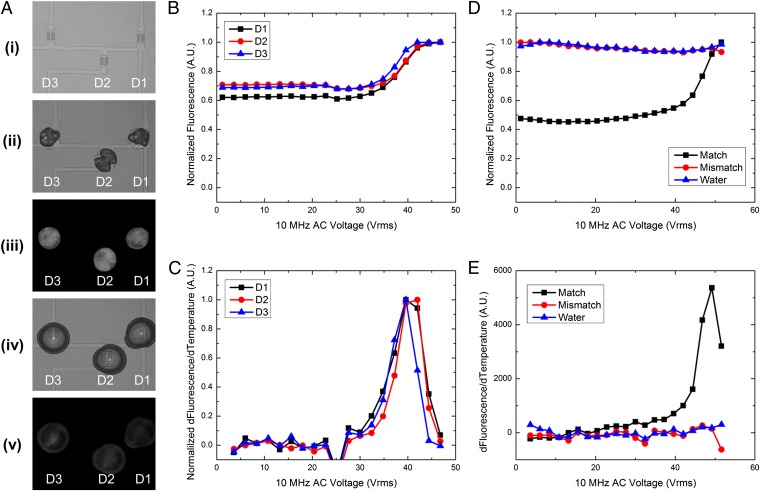
To demonstrate this technique, we dried a solution of probe single-stranded DNA (ssDNA) on the chip’s surface, as shown in Fig. 4A and SI Appendix, Fig. S10A. We then rehydrated the dried probe DNA with the single-stranded probe DNA target suspended in low evaporation solvent. Similar to standard DNA duplexing techniques, the newly rehydrated probe-target droplet was heated and cooled once to ensure proper DNA hybridization (SI Appendix, Fig. S10 B and C). This initial heating curve showed dual peaks in the derivative, which can be attributed to improper hybridization that occurs when DNA is duplexed at room temperature. To confirm that the dual-peak nature of the initial heating/cooling step was due to improper hybridization, we dehydrated a fully duplexed FRET construct on the chip surface and then rehydrated it. As shown in SI Appendix, Fig. S11, the initial melting curve for this complex showed the single peak of a fully hybridized DNA complex. Immediately after the initial heating/cooling step, a second melting curve was run that now showed a single peak in the derivative. This affirms that the probe and target were fully hybridized and complementary (Fig. 4 B and C). To confirm the specificity of this technique, we dehydrated a probe ssDNA strand on three separate devices. We then rehydrated one device with a complementary ssDNA, one device with a noncomplementary sequence, and one device with water as a control. Fig. 4 D and E shows that only the complementary matching sequence shows an increase in fluorescence. The mismatch and water show no increase in fluorescence from DNA denaturation. This methodology is compatible with current DNA microarray technologies and, in addition, promises to extend the capabilities of current DNA microarrays and DASH platforms by including a FRET fluorophore, like cy-3, in the spotted probe DNA; incorporating a heating element under each spot on the array; and using droplets-in-air for individual reaction compartments.
Fig. 4.
Probe DNA dehydration with target DNA rehydration. (A) An example of the process flow is presented. A, i shows the devices before dehydration of the probe ssDNA. After spotting of the probe ssDNA (A, ii), the DNA in solution is allowed to dehydrate, leaving behind residual salts and DNA. A, iii shows a fluorescent image of the dried ssDNA spot. The fluorescence intensity is high without the presence of the FRET quencher. A, iv shows the rehydration of devices 1, 2, and 3. The initial fluorescence before denaturation is show in A, v. The fluorescence intensity is lower than A, iii due to the introduction of the FRET quencher and the DNA hybridization. (B and C) A melting curve of three spots that have been rehydrated with a complementary target sequence. The increase in fluorescence shows a distinct, single peak. (E) This implies that the DNA has hybridized properly without unwanted heterodimer or self-dimer formation. (D and E) A test for specificity in the process. Spots of a dried probe sequence were rehydrated with a complementary sequence, a noncomplementary sequence, or water. A distinct peak in the derivate in E implies a matching sequence.
Conclusions
Integration of various laboratory functions onto microchips has been intensely studied for many years. Lab-on-a-chip technologies are attractive as they require fewer reagents, have lower detection limits, allow for parallel analyses, and can have a smaller footprint. Further advances of these technologies require the ability to integrate additional elements, such as the miniaturized heating element described here, and the ability to integrate heating elements in a massively parallel format compatible with silicon technology (26). Notably, our miniaturized heaters could also function as dual heater/sensor elements, as these SOI nanowire or nanoribbon structures have been used to detect DNA, proteins, pH, and pyrophosphates (27–31).
In summary, by using microfabrication techniques and incorporating the unique design of transistor-based heating with individual reaction volumes, “laboratory-on-a-chip” technologies can be scaled down to “laboratory-on-a-transistor’ technologies that exist as sensor/heater hybrids for point-of-care diagnostics. We elucidate a technique to heat subnanoliter droplets-in-air for visualization of DNA denaturation with resolution down to single base mismatches with application to current DNA microarray technologies. This methodology can be extended to a variety of other high-throughput screening applications such as high-speed PCR, single-cell lysis, single-molecule enzymology, and interrogation of ligand–receptor interactions in protein melting studies.
Methods
Chip Fabrication.
The fabrication flow and preparation of devices as well as techniques for heating and imaging were presented previously (19). A complementary metal-oxide semiconductor (CMOS)-compatible top-down fabrication procedure was followed to create devices in SOI wafers. The wafer’s device layer was thinned down to ∼300 Å by timed dry oxidation followed by buffered oxide etch. Active areas were lithographically defined and the rest of top silicon was etched using deep reactive ion etch. Afterward implant areas were defined with photoresist mask for Boron ion implantation. After doping, around 300 Å of silicon oxide was grown to form the gate oxide. Metal contacts (200 Å titanium/800 Å platinum) were patterned via lift off after wet etch removing of silicon oxide on top of contact regions. Finally, a 5,000 Å nitride-rich plasma enhanced chemical vapor deposition passivation layer was deposited and patterned to expose device channel and probing pads. Resulting devices were 300 Å thick with a channel that was 10 μm long and 2 μm wide.
Chip Preparation.
The chip surface was coated with a hydrophobic silane monolayer. Trichloro-perfluoro-octyl silane (PFOS) was vapor deposited on the chip surface. The chips were first cleaned using an acetone, methanol, deionized (DI) rinse and then oxygen plasma clean for 5 min at 300 W. The chips were then placed in a desiccator inside a Pyrex Petri dish with 20 μL of the PFOS. A vacuum was pulled on the desiccator for 20 min to allow for vapor deposition of the silane on the device surface. After deposition, the devices were cleaned of excess using an acetone, methanol, DI rinse. Any remaining silane excess was removed using a microfiber swab.
FRET Construct Solution Preparation.
Single-stranded FRET constructs were ordered from IDT, Inc. already lyophilized. The DNA was rehydrated to a concentration of 50 μM using nuclease-free DI water. The ssDNA FRET construct was then mixed in equal parts to its complementary ssDNA FRET construct. To ensure proper hybridization, the mixture was heated to 95 °C in a thermocycler for 150 s and then allowed to slowly cool to room temperature over the course of 5–10 min. To make each of the FRET construct solutions for droplet generation, 1 μL of the 25 μM dsDNA FRET solution was added to 10 μL of Protein Carrier Solution from NanoInk and 10 μL of 3× SSC buffer. Macroscale melting temperature readings were taken using an Eppendorf Realplex Thermocycler. These melting temperatures were used for comparing the melting voltage versus the melting temperature for the different FRET constructs.
Microinjection Procedure.
Injection of the subnanoliter droplets was accomplished using a microcapillary pressure injection system typically used for Intracytoplasmic sperm injection. A microcapillary from TransferTip F (ICSI), from Eppendorf, with a 7 μm inner diameter and 15 μm outer diameter tip, was used. The tip was inserted into a holder that connected the tip to a Narishige IM-300 pressure regulation system. The microcapillary tip holder was inserted into a 3D motorized micromanipulator system (Ultraprecise Motorized Micromanipulator from Warner Instruments). This system has 10 nm resolution with a range of 10 mm and allows accurate manipulation of the microcapillary tip for droplet placement. A Leica upright microscope was used for device visualization during droplet placement. The FRET construct, mixed with low evaporation solution, was loaded into the tip by suctioning for 2 s. The tip was positioned above the chosen device active area and the solution injected for 0.01–0.03 s, which resulted in a 50–100 μm diameter droplet with an estimated volume of hundreds of picoliters. The tip was then moved to the next device using the motorized micromanipulator, where the injection procedure was repeated.
Heating Procedure.
Radio frequency (RF) dielectric heating, described previously (18, 19), was induced in each device by applying an AC bias between shorted drain/source (or only source in “common-source” experiments) and silicon substrate. Using double-sided adhesive conductive carbon tape (SPI Supplies), the chips were adhered to a brass plate that acted as chuck to form the back contact. Voltage bias was applied to specific devices by contact with micromanipulator probes to appropriate source/drain pads. Voltages up to 40 Vrms at 10 MHz were applied using a function generator (Agilent 33120A) with a RF power amplifier (EIN – Model 2100L – 50 dB). Earlier studies have indicated this results in joule heating due to the mobile ions responding to the electric field within the double layer at regions of high electric field above the device. Previous studies have also shown that DNA-in-water solutions have a strong relaxation point in the low MHz regime (32, 33). This is typically considered to be due to counter ion movement around the transverse axis of the DNA molecule (34–36). Using 10 MHz and the movement of ions around the DNA molecule allows heating of a DNA-in-water solution at a much lower frequency than a traditional microwave heater. Hence, even though typically microwaves operate at 2.45 GHz as water has a strong dielectric relaxation point at 17 GHz (37), we are able to use a significantly lower frequency of 10 MHz for heating the droplets. A Matlab script was developed to control the function generator output using its embedded general purpose interface bus (GPIB) module. In this way, required voltage amplitudes were set and timed, creating a voltage ramp for melting curve calibrations.
Dehydration/Rehydration of Spotted FRET Construct.
To confirm this system’s ability to be used as a DNA microarray, we dehydrated FAM-modified ssDNA on devices. These DNA solutions did not contain the Protein Carrier Solution to minimize evaporation. These spots were then rehydrated with Iowa BHQ-modified ssDNA. For the match/mismatch test, only one of the spots was rehydrated with the complementary sequence. The other two spots were rehydrated with either water or a noncomplementary Iowa BHQ-modified ssDNA sequence. The droplets were heated using the AC technique and the fluorescence monitored to show an increase in fluorescence from DNA denaturation.
Supplementary Material
Acknowledgments
We acknowledge Lauren Yang for help editing the manuscript and funding support from a cooperative agreement with Purdue University and the Agricultural Research Service of the US Department of Agriculture (Project 1935-42000-035) and a subcontract to the University of Illinois at Urbana–Champaign. We also acknowledge support from National Institutes of Health Grant R01-CA20003, National Science Foundation Nanomanufacturing Science and Engineering Center at Ohio State University, and National Science Foundation Industry-University Collaborative Research Center for Agricultural, Biomedical, and Pharmaceutical Nanotechnology at the University of Illinois at Urbana-Champaign.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219639110/-/DCSupplemental.
References
- 1.Wölcke J, Ullmann D. Miniaturized HTS technologies—uHTS. Drug Discov Today. 2001;6(12):637–646. doi: 10.1016/s1359-6446(01)01807-4. [DOI] [PubMed] [Google Scholar]
- 2.Song H, Chen DL, Ismagilov RF. Reactions in droplets in microfluidic channels. Angew Chem Int Ed Engl. 2006;45(44):7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagai H, Murakami Y, Yokoyama K, Tamiya E. High-throughput PCR in silicon based microchamber array. Biosens Bioelectron. 2001;16(9-12):1015–1019. doi: 10.1016/s0956-5663(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 4.Balon K, Riebesehl BU, Müller BW. Drug liposome partitioning as a tool for the prediction of human passive intestinal absorption. Pharm Res. 1999;16(6):882–888. doi: 10.1023/a:1018882221008. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, et al. A universal multiplex PCR strategy for 100-plex amplification using a hydrophobically patterned microarray. Lab Chip. 2011;11(21):3609–3618. doi: 10.1039/c1lc20526a. [DOI] [PubMed] [Google Scholar]
- 6.Rondelez Y, et al. Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat Biotechnol. 2005;23(3):361–365. doi: 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- 7.Matsubara Y, et al. On-chip nanoliter-volume multiplex TaqMan polymerase chain reaction from a single copy based on counting fluorescence released microchambers. Anal Chem. 2004;76(21):6434–6439. doi: 10.1021/ac0497149. [DOI] [PubMed] [Google Scholar]
- 8.Gosalia DN, Diamond SL. Printing chemical libraries on microarrays for fluid phase nanoliter reactions. Proc Natl Acad Sci USA. 2003;100(15):8721–8726. doi: 10.1073/pnas.1530261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maltezos G, Johnston M, Scherer A. Thermal management in microfluidics using micro-Peltier junctions. Appl Phys Lett. 2005;87(15):1–3. [Google Scholar]
- 10.Park I, Li Z, Pisano AP, Williams RS. Selective surface functionalization of silicon nanowires via nanoscale joule heating. Nano Lett. 2007;7(10):3106–3111. doi: 10.1021/nl071637k. [DOI] [PubMed] [Google Scholar]
- 11.Lee CY, Lee GB, Lin JL, Huang FC, Liao CS. Integrated microfluidic systems for cell lysis, mixing/pumping and DNA amplification. J Micromech Microeng. 2005;15(6):1215–1223. [Google Scholar]
- 12.Giordano BC, Ferrance J, Swedberg S, Hühmer AFR, Landers JP. Polymerase chain reaction in polymeric microchips: DNA amplification in less than 240 seconds. Anal Biochem. 2001;291(1):124–132. doi: 10.1006/abio.2000.4974. [DOI] [PubMed] [Google Scholar]
- 13.Shah JJ, et al. Microwave dielectric heating of fluids in an integrated microfluidic device. J Micromech Microeng. 2007;17(11):2224–2230. [Google Scholar]
- 14.Graf M, Frey U, Taschini S, Hierlemann A. Micro hot plate-based sensor array system for the detection of environmentally relevant gases. Anal Chem. 2006;78(19):6801–6808. doi: 10.1021/ac060467d. [DOI] [PubMed] [Google Scholar]
- 15.Issadore D, et al. Microwave dielectric heating of drops in microfluidic devices. Lab Chip. 2009;9(12):1701–1706. doi: 10.1039/b822357b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, Vishniakou S, Faris GW. Petri dish PCR: Laser-heated reactions in nanoliter droplet arrays. Lab Chip. 2009;9(9):1230–1235. doi: 10.1039/b817288a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terazono H, Hattori A, Takei H, Takeda K, Yasuda K. Development of 1480 nm photothermal high-speed real-time polymerase chain reaction system for rapid nucleotide recognition. Jpn J Appl Phys. 2008;47(6 PART 2):5212–5216. [Google Scholar]
- 18.Elibol OH, et al. Localized heating on silicon field effect transistors: Device fabrication and temperature measurements in fluid. Lab Chip. 2009;9(19):2789–2795. doi: 10.1039/b906048k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy B, Jr, et al. Silicon field effect transistors as dual-use sensor-heater hybrids. Anal Chem. 2011;83(3):888–895. doi: 10.1021/ac102566f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jokilaakso N, et al. Ultra-localized single cell electroporation using silicon nanowires. Lab Chip. 2013;13(3):336–339. doi: 10.1039/c2lc40837f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young IT, et al. Monitoring enzymatic reactions in nanolitre wells. J Microsc. 2003;212(Pt 3):254–263. doi: 10.1111/j.1365-2818.2003.01252.x. [DOI] [PubMed] [Google Scholar]
- 22.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86(4):2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8(6):597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- 24.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21(1) Suppl:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 25.Howell WM, Jobs M, Gyllensten U, Brookes AJ. Dynamic allele-specific hybridization. A new method for scoring single nucleotide polymorphisms. Nat Biotechnol. 1999;17(1):87–88. doi: 10.1038/5270. [DOI] [PubMed] [Google Scholar]
- 26.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475(7356):348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 27.Fritz J, Cooper EB, Gaudet S, Sorger PK, Manalis SR. Electronic detection of DNA by its intrinsic molecular charge. Proc Natl Acad Sci USA. 2002;99(22):14142–14146. doi: 10.1073/pnas.232276699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern E, et al. Label-free biomarker detection from whole blood. Nat Nanotechnol. 2010;5(2):138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elibol OH, Reddy JB, Bashir R. Nanoscale thickness double-gated field effect silicon sensors for sensitive pH detection in fluid. Appl Phys Lett. 2008;92(19):193904–193903. [Google Scholar]
- 30.Reddy B, Jr, et al. High-k dielectric Al2O3 nanowire and nanoplate field effect sensors for improved pH sensing. Biomed Microdevices. 2011;13(2):335–344. doi: 10.1007/s10544-010-9497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu DJ, et al. Surface immobilizable chelator for label-free electrical detection of pyrophosphate. Chem Commun (Camb) 2011;47(29):8310–8312. doi: 10.1039/c1cc12073e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomić S, et al. Dielectric relaxation of DNA aqueous solutions. Phys Rev E. 2007;75(2 Pt 1):021905. doi: 10.1103/PhysRevE.75.021905. [DOI] [PubMed] [Google Scholar]
- 33.Salm E, et al. Electrical detection of dsDNA and polymerase chain reaction amplification. Biomed Microdevices. 2011;13(6):973–982. doi: 10.1007/s10544-011-9567-x. [DOI] [PubMed] [Google Scholar]
- 34.Takashima S, Gabriel C, Sheppard RJ, Grant EH. Dielectric behavior of DNA solution at radio and microwave frequencies (at 20 degrees C) Biophys J. 1984;46(1):29–34. doi: 10.1016/S0006-3495(84)83995-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuwabara S, Umehara T, Mashimo S, Yagihara S. Dynamics and structure of water bound to DNA. J Phys Chem. 1988;92(17):4839–4841. [Google Scholar]
- 36.Saif B, Mohr RK, Montrose CJ, Litovitz TA. On the mechanism of dielectric relaxation in aqueous DNA solutions. Biopolymers. 1991;31(10):1171–1180. doi: 10.1002/bip.360311005. [DOI] [PubMed] [Google Scholar]
- 37.Stuerga D. Microwave-Material Interactions and Dielectric Properties, Key Ingredients for Mastery of Chemical Microwave Processes. Microwaves in Organic Synthesis. Weinheim, Germany: Wiley-VCH; 2008. pp. 1–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.