Abstract
The current epidemic of infections caused by antibiotic-resistant Gram-positive bacteria requires the discovery of new drug targets and the development of new therapeutics. Lipoteichoic acid (LTA), a cell wall polymer of Gram-positive bacteria, consists of 1,3-polyglycerol-phosphate linked to glycolipid. LTA synthase (LtaS) polymerizes polyglycerol-phosphate from phosphatidylglycerol, a reaction that is essential for the growth of Gram-positive bacteria. We screened small molecule libraries for compounds inhibiting growth of Staphylococcus aureus but not of Gram-negative bacteria. Compound 1771 [2-oxo-2-(5-phenyl-1,3,4-oxadiazol-2-ylamino)ethyl 2-naphtho[2,1-b]furan-1-ylacetate] blocked phosphatidylglycerol binding to LtaS and inhibited LTA synthesis in S. aureus and in Escherichia coli expressing ltaS. Compound 1771 inhibited the growth of antibiotic-resistant Gram-positive bacteria and prolonged the survival of mice with lethal S. aureus challenge, validating LtaS as a target for the development of antibiotics.
Methicillin-resistant Staphylococcus aureus (MRSA), glycopeptide-resistant enterococci (GRE), and antibiotic-resistant Clostridium difficile are examples for newly emerging or reemerging drug-resistant Gram-positive pathogens with significant human morbidity (1). As commensals of the human skin or intestine, these microbes are continuously exposed to antibiotics and have evolved resistance traits against commonly used therapeutics (2). To develop novel antibiotics, a suitable molecular target must be identified (3). Ideal targets are found only in bacteria, not in humans, and are positioned in the bacterial envelope, accessible for small molecule inhibitors yet out of reach of multi–drug-resistance transporters protecting cytoplasmic targets (3, 4). Such attributes are found in penicillin-binding proteins and their inhibitors, β-lactam or glycopeptide antibiotics; however, efforts to identify other extracellular targets in Gram-positive bacteria have met with little success (5).
Gram-positive bacteria incorporate lipoteichoic acids (LTAs) into their envelope to scavenge magnesium ions, direct autolysins to subcellular cell wall locations, and enable bacterial cell division (6, 7). LTA from S. aureus and other bacteria is composed of 1,3-polyglycerol-phosphate linked to glycolipid, which provides for LTA anchoring in membranes (8). The glycolipid moiety is composed of β-gentiobiosyldiacylglycerol [glucosyl-(1→6)-glucosyl-(1→3)-diacylglycerol (Glc2-DAG)] (8, 9). PgcA (α-phosphoglucomutase), GtaB (UTP:α-glucose-1-phosphate uridyl transferase), and YpfP (glycosyl-transferase) catalyze the three steps of glycolipid synthesis (10–12), whereas LtaA transports Glc2-DAG across the lipid bilayer (10). S. aureus ltaA, ypfP, pgcA, or gtaB mutants cannot assemble Glc2-DAG but continue to synthesize polyglycerol-phosphate (10, 12). Glycolipid synthesis mutants are viable; however, the variants display an increase in size and aberrant cell shapes (10, 12).
LTA synthesis involves the polymerization of polyglycerol-phosphate and its transfer to Glc2-DAG (13). This reaction is catalyzed by LTA synthase (LtaS), a protein with five transmembrane domains in its N-terminal domain and an extracellular sulfatase-like domain (pfam00884) at the C-terminal end (14). Genetic depletion or loss of the ltaS gene in S. aureus results in severe cell division defects, a phenotype that is exacerbated when staphylococci are grown at >30 °C (14, 15). Characterization of LtaS in Bacillus anthracis, Bacillus subtilis, and Listeria monocytogenes confirmed that membrane proteins with pfam00884 domains are indeed responsible for the synthesis of polyglycerol-phosphate LTA (7). Where examined, ltaS mutants exhibited diminished viability, increased cell size, and altered morphology (16–19). These results suggested that LtaS of Gram-positive bacteria may represent an extracellular target for the development of antibiotics against drug-resistant Gram-positive bacteria. We here provide proof for this hypothesis by isolating a small molecule inhibitor of LTA synthesis.
Results and Discussion
Compound 1771 Inhibits S. aureus Growth and LTA Synthesis.
S. aureus variants that cannot express ltaS are unable to grow at 37 °C (14). We took advantage of the temperature-sensitive phenotype and screened compound libraries at the National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Disease (NSRB) for candidate compounds that inhibit growth of S. aureus at 42 °C. The primary screen identified 73 compounds with greater than 90% growth inhibition (Table S1). Thirty-one compounds were subjected to secondary screening, which included dose–response analyses for growth inhibition of MRSA, as well as the Gram-negative microbe Escherichia coli. Fifteen compounds specifically inhibited the growth of MRSA but not of E. coli and displayed little or no cytotoxicity when added to HL-60 cells, a human promyelocytic leukemia cell (Table S1). One of these molecules, compound 1771 [2-oxo-2-(5-phenyl-1,3,4-oxadiazol-2-ylamino)ethyl 2-naphtho[2,1-b]furan-1-ylacetate], was identified as an inhibitor of LTA synthesis in S. aureus as follows.
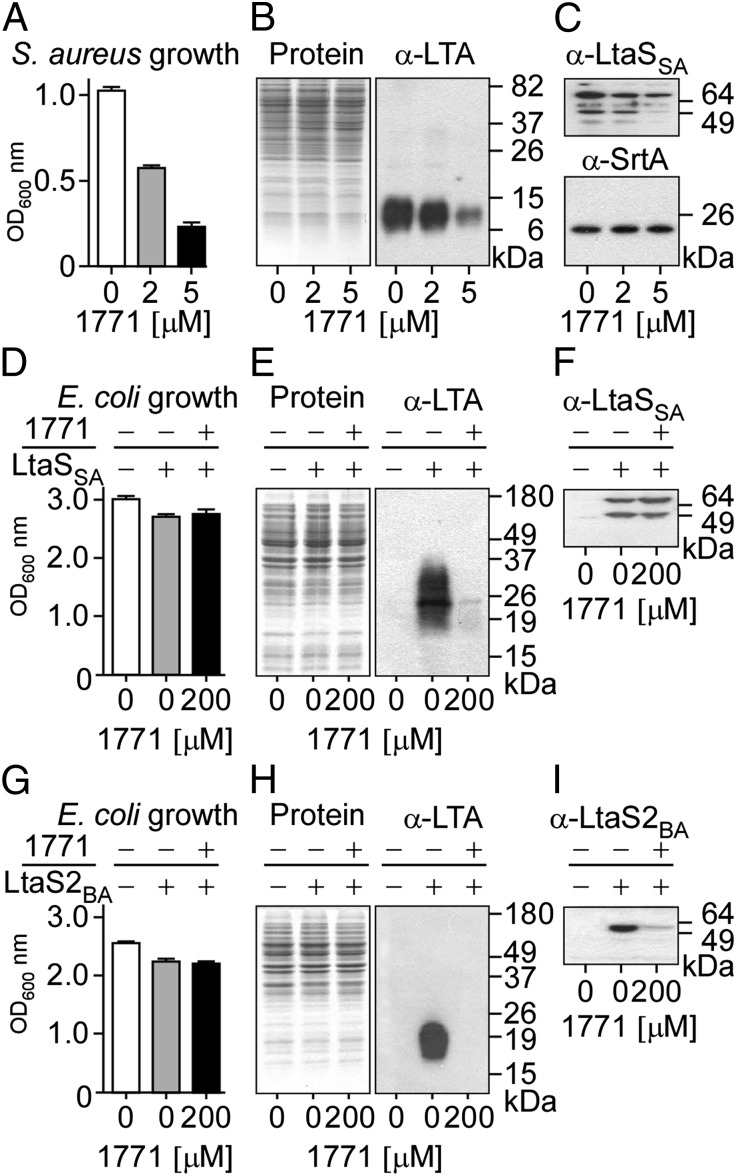
Overnight cultures of S. aureus were diluted and incubated with increasing concentrations of compound 1771 (0, 2, or 5 μM), resulting in increased reduction of growth (Fig. 1A). Extracts prepared from staphylococcal cultures normalized to the same density were analyzed by SDS/PAGE. Coomassie Blue staining of proteins suggested that incubation of staphylococci with compound 1771 did not alter the concentration of bacterial polypeptides (Fig. 1B). However, immunoblotting with 1,3-polyglycerol phosphate-specific monoclonal antibody (α-LTA) revealed that compound 1771 reduced the abundance of LTA (Fig. 1B). At a higher concentration of 1771 (5 μM), LtaS-specific immunoreactive signals were reduced (Fig. 1C). As a control, the abundance of sortase A (SrtA), an enzyme that links proteins to peptidoglycan, was not affected in staphylococcal cultures treated with compound 1771 (Fig. 1C).
Fig. 1.
Compound 1771 inhibits LTA biosynthesis in S. aureus. (A) S. aureus was grown in medium supplemented with either 1% DMSO (control) or two subinhibitory concentrations of 1771 for 1 h, and the optical density at 600 nm (OD600 nm) was recorded. (B and C) Cultures shown in A were normalized to the same density, and cells were lysed to prepare extracts for separation on SDS/PAGE and Coomassie staining (B, Left) or immunoblot analyses with antibodies specific for polyglycerol-phosphate (B, Right; α-LTA) or LtaS (C, Upper; α-LtaS) and SrtA (C, Lower; α-SrtA). Native S. aureus LtaS is detected as both full-length and processed proteins (70 and 49 kDa, respectively). (D–I) Recombinant S. aureus LtaSSA (D–F) or B. anthracis LtaS2BA (G–I) were expressed in E. coli, and synthesis of polyglycerol-phosphate was detected by immunoblot. E. coli cultures induced (+) or noninduced (−) for expression of LtaSSA (D) or LtaS2BA (G) were grown in medium supplemented with either 1% DMSO (−) or 200 µM 1771 (+). Culture density measurements were used to normalize cell lysates that were separated by SDS/PAGE and analyzed by Coomassie staining (E and H, Left) or immunoblotting with antibodies against LTA (E and H, Right) and LtaS (F and I). Recombinant LtaSSA is detected as a double band like the native protein produced in S. aureus, whereas only processed LtaS2BA is detected in E. coli extracts. Molecular weight markers are indicated in kilodaltons.
Expression of LtaS from S. aureus (ltaSSA) or B. anthracis (ltaS2BA) in E. coli leads to the production of 1,3-polyglycerol phosphate, as LTA synthase can use phosphatidylglycerol (PG) substrate even from the membrane of Gram-negative bacteria (Fig. 1, E and H). Unlike its antibiotic activity in S. aureus, compound 1771 did not affect the growth of E. coli even at very high concentrations (200 µM; Fig. 1 D and G). Strikingly, E. coli synthesis of polyglycerol-phosphate via LtaSSA (Fig. 1E) or LtaS2BA (Fig. 1H) was abrogated in the presence of compound 1771. Of note, E. coli expression of LtaS2BA, but not of LtaSSA, was reduced in the presence of 1771 (Fig. 1 F and I).
Compound 1771 Inhibits the Growth of Gram-Positive Bacteria.
To examine the spectrum of antibiotic activity for compound 1771, we analyzed Gram-positive bacteria harboring polyglycerol-phosphate LTA and LtaS homologs (Table S2). Compound 1771 inhibited the growth of antibiotic-resistant MRSA, e.g., the epidemic community-acquired isolate USA300 LAC, and VRE, i.e., vancomycin-resistant Enterococcus faeaclis and Enterococcus faecium whose genomes harbor two ltaS homologs (Table S3). Gram-positive bacteria with three (Clostridium perfringes) or four ltaS homologs (B. cereus and B. anthracis) appeared to be more susceptible to compound 1771–mediated growth inhibition than microbes with only one or two ltaS genes (Table S3).
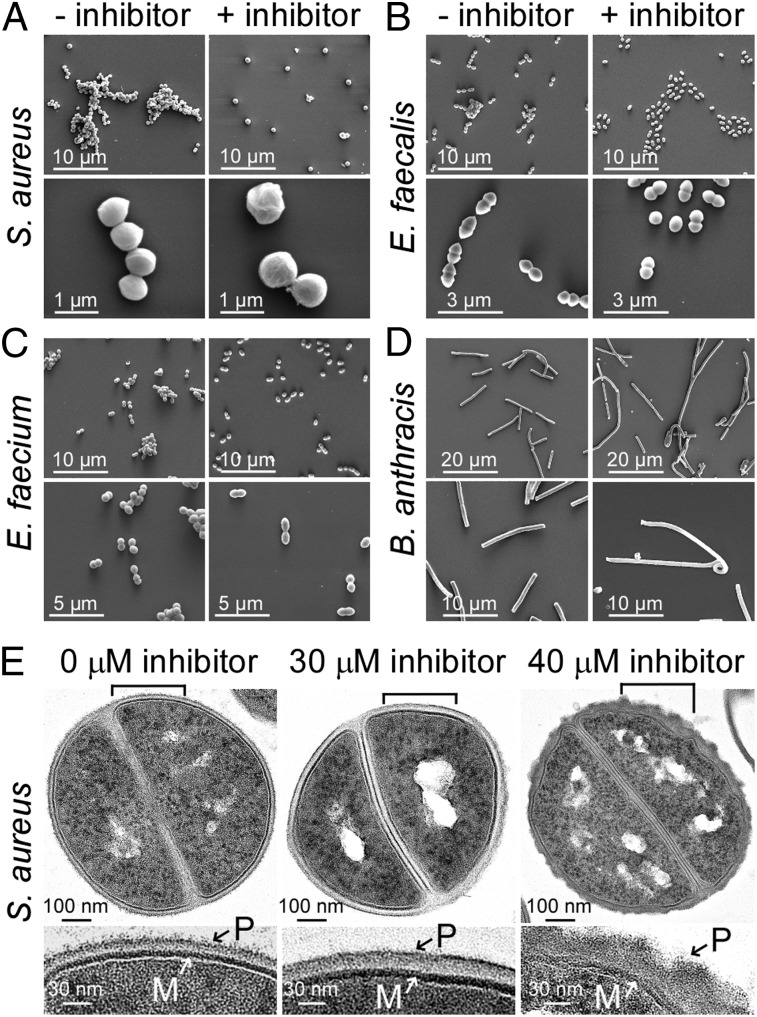
Bacterial cultures incubated with or without sublethal concentrations of compound 1771 were examined by scanning electron microscopy (SEM; Fig. 2). Treatment with compound 1771 dispersed cluster formation in S. aureus or chain formation in E. faecalis and E. faecium (Fig. 2 A–C; Table S4). At higher magnification of SEM images, compound 1771–treated staphylococci and enterococci revealed an increase in cell size, as well as deformations of their cell surface and shape (Fig. 2 A–C). In the presence of compound 1771, B. anthracis formed longer chains of vegetative cells and displayed undulating deformations of its cylindrical cell shape (Fig. 2D; Fig. S1). Compound 1771–induced morphological changes resemble those observed for S. aureus and B. anthracis mutants with defects in LTA synthesis (14, 18). For example, the chain length of the B. anthracis ltaS1/ltaS2 mutant is increased compared with WT (Fig. S1A), and its ability to form colonies is reduced by 1,000-fold (18).
Fig. 2.
Structural changes of Gram-positive bacteria treated with compound 1771. (A–D) Scanning electron micrographs of bacterial cultures grown without or with compound 1771. Bacteria were cultured in BHI medium supplemented with either 1% DMSO (− inhibitor) or subinhibitory concentrations of 1771 (+ inhibitor): (A) S. aureus RN4220 ±30 µM 1771 scanned at 10,000× (Upper) and 80,000× (Lower) magnification; (B) E. faecalis V583 cells ±25 µM 1771 scanned at 10,000× (Upper) and 24,000× (Lower); (C) E. faecium TX0016 cells ±20 µM 1771 scanned at 10,000× (Upper) and 20,000× (Lower); (D) B. anthracis Sterne ±5 µM 1771 scanned at 5,000× (Upper) or 10,000× (Lower). (E) Thin-section transmission electron micrographs of S. aureus reveal a thickening of the envelope with visible deformations in the presence of 30 or 40 μM compound 1771. (Upper) Electron micrographs of staphylococci in the midst of cell division. Brackets in the upper micrographs indicate the positions of enlarged image sections shown below. M, plasma membrane; P, peptidoglycan layer containing teichoic acids. Scale bars are indicated at the bottom left of each micrograph.
Thin-section transmission electron microscopy (TEM) of S. aureus treated with compound 1771 revealed thickening and structural disorganization of the cell wall envelope (Fig. 2E). The smooth surface and structural organization of the envelope were perturbed when staphylococci were grown in the presence of 40 µM compound 1771. Similar results were obtained with TEM images of thin-sectioned enterococci and bacilli, which prompted measurements of envelope thickness. The data revealed increases in envelope diameter for S. aureus, E. faecalis, E. faecium, and B. anthracis grown in the presence of compound 1771 (Fig. S1B; Table S5).
Mechanism of LTA Synthesis Inhibition for Compound 1771.
The 3D structure of the extracellular catalytic domain of LtaS has been determined (17, 20). Overall, extracellular catalytic domain of LtaS (eLtaS) assumes a sulfatase-like fold; however, its active site is distinct from that of sulfatases (20). Threonine (T300) of LtaS together with residues E255, D475, and H476 coordinate a manganese ion and assemble to form a binding pocket for glycerol-phosphate (20). As revealed from the cocrystal structure of eLtaS with glycerol-phosphate, one oxygen atom of the phosphate group is coordinated with Mg2+, whereas the remainder of the phosphate group is stabilized by hydrogen bonding with H416 and W354. Hydroxyl side chains of glycerol-phosphate form hydrogen bonds with H347, D349, and R356 (20). Catalysis has been proposed to involve PG docking in the active site of eLtaS to enable nucleophilic attack from the deprotonated hydroxyl of T300, generating a glycerol-phosphate-threonine intermediate and releasing diacylglycerol. The glycerol-phosphate-threonine intermediate may subsequently be resolved by the nucleophilic attack from the terminal OH group of another PG, thereby extending the LTA chain by one glycerol-phosphate moiety (20).
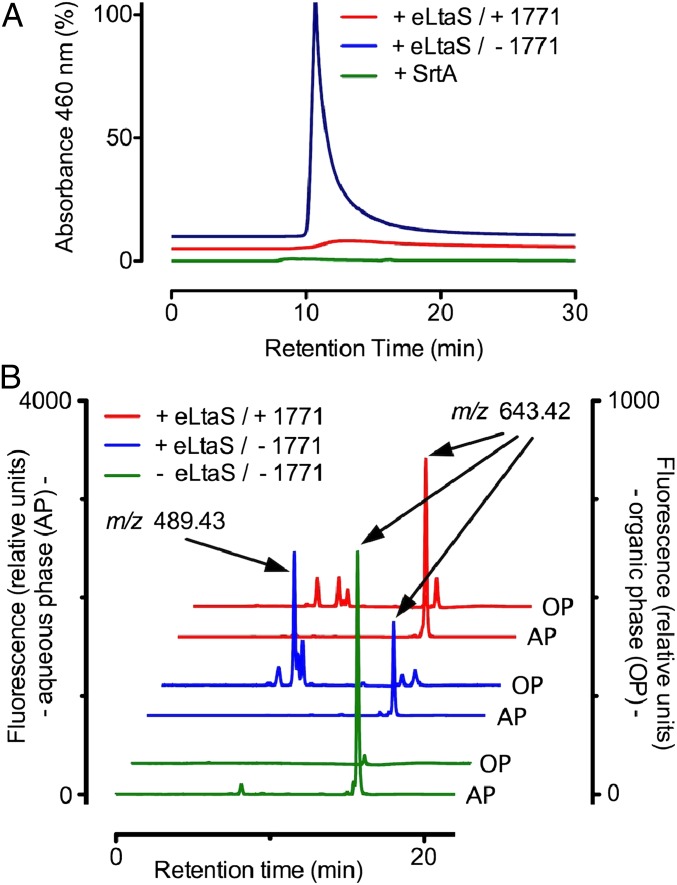
Incubation of purified eLtaS with compound 1771 followed by MS analysis of the enzyme and inhibitor did not reveal the formation of an eLtaS adduct or the cleavage of 1771. We used size-exclusion HPLC with a BioBasic SEC300 column equilibrated with 20 nmol nitro-benzoxadiazole glycerol-phosphate (NBD-GP) to measure binding of enzyme to substrate. Chromatography of 2 nmol eLtaS on the BioBasic SEC300 column led to the elution of an enzyme·NBD-GP complex and absorbance at 460 nm (Fig. 3A). As a control, chromatography of 2 nmol sortase A, a transpeptidase that anchors surface proteins to peptidoglycan and does not bind to glycerol-phosphate (21), did not elute NBD-GP from a preequilibrated BioBasic SEC300 column (Fig. 3A). When the mobile HPLC phase was supplemented with 200 µM LTA synthesis inhibitor 1771, the ability of eLtaS to elute NBD-GP complexes from the preequilibrated column was abolished (Fig. 3A). eLtaS-mediated cleavage of nitrobenzoxadiazole-PG (NBD-PGC6, with six carbon acyl chains) was used to determine whether compound 1771 inhibits LTA synthesis in vitro (22). In the presence of the enzyme (+eLtaS), but not in its absence (−eLtaS), NBD-PGC6 (m/z 643.42) was cleaved to generate NBD-DAGC6 (nitrobenzoxadiazole-diacylglycerol, m/z 489.43), as revealed by HPLC and MS of chloroform-extracted samples, separating NBD-PGC6 substrate in the aqueous phase (AP) from NBD-DAGC6 product in the organic phase (OP; Fig. 3B). Addition of 100 µM compound 1771 inhibited eLtaS-mediated formation of NBD-DAGC6 product from NBD-PGC6, indicating that the molecule inhibited LTA synthesis in vitro (Fig. 3B).
Fig. 3.
Compound 1771 inhibits eLtaS binding to and cleavage of phosphatidylglycerol (PG) in vitro. (A) Size-exclusion HPLC of 2 nmol eLtaS or SrtA on BioBasic SEC300 column preequilibrated with 20 nmol nitro-benzoxadiazole PG containing chains of 16 carbon atoms (NBD-PGC16) reveals elution of the NBD-PGC16·eLtaS complex with absorbance at 460 nm (blue trace), but not formation of a NBD-PGC16·SrtA complex (green trace). Inclusion of 200 µM compound 1771 in the mobile HPLC phase abolished the elution of NBD-PGC16·eLtaS complex (red trace). (B) Using NBD-PGC6 with six carbon acyl chains cleavage of 2 nmol substrate by 2 nmol eLtaS was detectable after 6-h incubation at 37 °C. Chloroform extraction separated nonhydrolyzed NBD-PGC6 [aqueous phase (AP)] from the hydrophobic reaction product nitro-benzoxadiazole diacylglycerol (NBD-DAGC6), which segregated into the organic phase (OP). Both phases were analyzed by normal-phase HPLC on a diol column (blue traces). Elution profiles were monitored by fluorescence (excitation at 460 nm, emission at 534 nm). Identity of peak fractions was confirmed by mass spectrometry. Addition of 100 µM 1771 blocked NBD-DAGC6 production by eLtaS (red traces). A control reaction incubated for 6 h without eLtaS did not contain detectable amounts of NBD-DAGC6 (green traces).
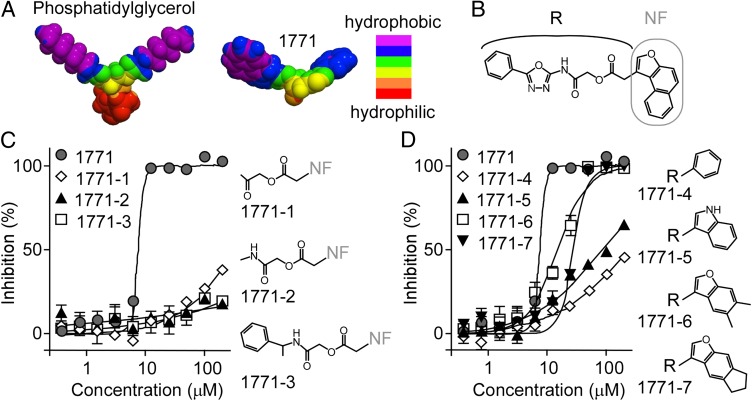
The results in Fig. 3 suggest that compound 1771 may bind to the active site of eLtaS and prevent its association with PG. If so, the distribution of hydrophobic and hydrophilic surface areas may be similar for compound 1771 and PG. This prediction was examined by generating 3D models with the Galaxy 3D Structure Generator v2011.02 (www.molinspiration.com; Fig. 4A). The chemical designation of compound 1771 is 2-oxo-2-(5-phenyl-1,3,4-oxadiazol-2-ylamino)ethyl 2-naphtho[2,1-b]furan-1-ylacetate, and its structural formula can be arbitrarily divided into the R and naphthofuranyl (NF) groups (Fig. 4B). The NF and R groups appear to mimic the polar chains of PG (Fig. 4A). To analyze this possibility, we selected three derivatives with an intact NF group and different R groups for structure–activity relationships (Fig. 4C; 1771-1/-3). Compared with compound 1771, none of these derivatives inhibited S. aureus growth (Fig. 4C; Table S6). Four compounds with intact R group and different NF groups were also identified (Fig. 4D; 1771-4/-7). These derivatives yielded increasing inhibitory activity on acquisition of either naphtho or furan rings (Fig. 4D; Table S6). The relative loss of inhibitory activity by all seven structural derivatives was similar for S. aureus and B. anthracis (Table S6). Together, these findings indicate that compound 1771 is structurally similar to PG and prevents the interaction between PG and eLtaS.
Fig. 4.
Structure-activity relationships of compound 1771. (A) 3D models showing the molecular hydrophobicity of PG (Left) and compound 1771 (Right). Models were generated using Galaxy 3D Structure Generator v2011.02. Color coding for hydrophobic and hydrophilic areas are shown. (B) Structural formula of 1771 with the chemical designation 2-oxo-2-(5-phenyl-1,3,4-oxadiazol-2-ylamino)ethyl 2-naphtho[2,1-b]furan-1-ylacetate. The naphthofuranyl group (NF) is indicated with a gray line, and the remainder of the molecule is referred to as the R group. Growth inhibitory activity and structural formula of substructures with intact naphthofuranyl group (C) or intact R region (D). Growth inhibitory activities were measured by adding compounds to cultures of S. aureus RN4220 and displayed as mean with SDs of three independent experiments. Dose–response graphs were calculated by fitting data with variable slope sigmoidal function using GraphPad Prism 5. Corresponding IC50 values are presented in Table S6.
Compound 1771 Prolongs the Survival of Mice with S. aureus Sepsis.
To examine the therapeutic value of compound 1771, we evaluated its half-life in mice. Animals received a single i.p. injection of 32 mg/kg compound 1771. The blood of three animals was drawn 1, 6, and 12 h following compound 1771 injection, and serum samples were extracted with methanol and chloroform, separated by reversed-phase HPLC, and subjected to MS. This analysis revealed two absorption peaks corresponding to cleavage fragments of compound 1771, none of which retained inhibitory activity. Full conversion of compound 1771 into its two cleavage fragments occurred between 3 and 6 h following injection. The enzyme(s) responsible for compound 1771 cleavage are not yet known, and it is not yet clear whether compound 1771 can be modified to resist cleavage while retaining its antibiotic activity.
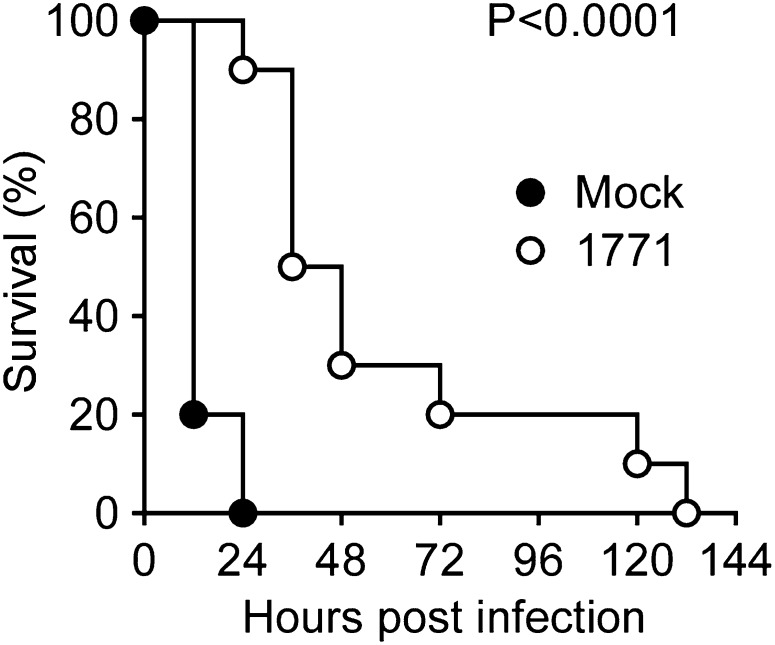
To evaluate the therapeutic efficacy of compound 1771, we used a mouse model of staphylococcal sepsis (23). Animals received i.p. injections with 32 mg/kg of compound 1771 in 12-h intervals. Treatment was initiated 24 h before challenge and terminated at 72 h after infection. Following challenge of mice via bloodstream injection of 1 × 108 CFU S. aureus Newman, mock-treated animals died of sepsis within 24 h (Fig. 5). In contrast, compound 1771–treated animals survived up to 132 h, albeit all animals in this cohort eventually succumbed to the challenge (Fig. 5). Thus, although compound 1771 is unstable in mice with rapid loss of activity, its administration into animals delays time to death following a lethal challenge with S. aureus.
Fig. 5.
Compound 1771 increased the time to death in a mouse model of S. aureus sepsis. Survival of cohorts of BALB/c mice (n = 15) challenged by i.v. injection with 1 × 108 CFU S. aureus Newman. Animals were treated for 4 d starting 1 d before infection in 12-h intervals via i.p. injection with either compound buffer (Mock) or 32 mg/kg weight of compound 1771. Statistical significance was examined with the log-rank test: mock vs. 1771, P < 0.0001. Data are representative of two independent experiments.
Selecting for S. aureus Variants with Increased Resistance to Compound 1771.
The therapeutic value of many antibiotics is limited because bacteria acquire resistance via spontaneous mutations that modify the drug target (5). For example, streptomycin blocks ribosomal protein synthesis (24, 25); however, mutations in rpsL, the structural gene for ribosomal protein S12, alter the polypeptide to prevent antibiotic access to the ribosome (26). In contrast to streptomycin-resistant mutants, which arise at frequencies <10−7 (Fig. S2A), S. aureus RN4220 formed rare small colonies only after 3–4 d of incubation on agar media with a 10–200 µM gradient of compound 1771 (Fig. S2B). These observations suggest that resistant colonies cannot be isolated from S. aureus at frequencies ≤2 × 10−9. When analyzed for their resistance phenotype, none of the three independent colony isolates displayed significant changes in either the minimal inhibitory concentration (MIC) or the IC50 values for compound 1771 (Table S7). Isolated strains produced LtaS and LTA with similar abundance as their S. aureus parent and did not harbor mutational alterations in the ltaS gene (Fig. S2C). Thus, S. aureus selection on agar plates did not lead to variants with a significant increase in resistance to compound 1771. This phenotype is similar to that reported for vancomycin (27), a cell wall active antibiotic, which requires mutations in different genes for staphylococci to acquire an intermediary resistance phenotype (28, 29).
LTA Synthesis Inhibitors as Infectious Disease Therapeutics.
Owing to the frequent use of antibiotics, members of the human microbiome continuously evolve drug resistance (30). For MRSA, drug resistance is associated with therapeutic failure and increased mortality of human infections (31, 32). Glycopeptide (vancomycin) resistance has transferred from enterococci to MRSA (33); the resulting VRSA strains are broadly antibiotic resistant and represent a global infectious threat (34, 35). Daptomycin and linezolid have recently been licensed to address the threat of MRSA and VRSA infections (36, 37). Nevertheless, staphylococci quickly developed daptomycin and linezolid resistance, indicating that additional antibotics are needed to combat S. aureus infections (38). The crisis in antibiotic resistance applies also to other Gram-positive pathogens, including C. difficile, E. faecium, E. faecalis, Staphylococcus epidermidis, and Streptococcus pneumoniae (2, 39).
We explored LTA synthesis as a target for antibiotic therapy. Growth of S. aureus, B. anthracis, L. monocytogenes, or B. subitilis cannot occur without polyglycerol-phosphate LTA synthesis and ltaS expression (14, 16, 17, 40). LtaS, the catalyst of LTA synthesis, harbors five transmembrane domains and a C-terminal catalytic domain that is found in bacteria but not in eukaryotes (14). The unique presence of LTA and LtaS in the envelope of bacterial species, the availability of the catalytic domain of LtaS on the bacterial surface, and the requirement of LTA synthesis for bacterial growth and cell division fulfill key target features for the development of new antibiotics (41). Here we characterized compound 1771 as an LTA synthesis inhibitor and demonstrated its mechanism of action and ability to kill Gram-positive bacteria with polyglycerol-phosphate LTA. We were unable to isolate staphylococcal mutants with resistance against compound 1771, suggesting that LTA synthesis may display target attributes similar to peptidoglycan synthesis. Some Gram-positive bacteria, for example, C. difficile and S. pneumoniae, synthesize LTA with distinct phosphate-polymer structures (42, 43); however, their mechanisms of synthesis and possible inhibition by compound 1771 are not yet known. Future work must develop compound 1771 further to generate molecules that are stable in mammalian tissues and display antibiotic activity against many different bacteria. Such compounds may be useful therapeutics for human infectious diseases caused by drug-resistant Gram-positive bacteria.
Experimental Procedures
High-Throughput Screen.
The NSRB library of 167,405 compounds was screened for molecules that inhibited the >90% growth of S. aureus RN4220 in Mueller-Hinton broth II supplemented with 0.005% Tween-80 in a 384-well format by measuring the optical density at 600 nm (Z′ = 0.72–0.84). A 98.9% (wt/wt) pure preparation of 2-oxo-2-(5-phenyl-1,3,4-oxadiazol-2-ylamino)ethyl 2-naphtho[2,1-b]furan-1-ylacetate (compound 1771) was obtained from Enamine (catalog no. T5526252).
Growth Inhibition.
Cultures of E. coli, S. aureus, Clostridium perfringens, E. faecalis, E. faecium, B. anthracis, and B. cereus were grown in the presence or absence of inhibitor in 96-well microplates at 37 °C for 18–22 h and monitored by measuring the optical density at 600 nm.
Inhibition of LTA Synthesis.
Bacteria grown in the presence or absence of compound 1771 were lysed in a bead beater, and cell extracts were subjected to Coomassie-stained SDS/PAGE or immunoblotting using a monoclonal antibody to detect LTA/polyglycerol-phosphate and polyclonal antibodies for LtaS and SrtA.
eLtaS Inhibition.
Size-exclusion HPLC was performed with a BioBasic SEC300 column equilibrated in a 50 mM Hepes-KOH buffer, pH 7.5, containing 10 µM MnCl2. The column was preequilibrated with 2 nmol 1-palmitoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-[phospho-rac-(1-glycerol)] (Avanti Polar Lipids). Purified eLtaS and SrtA (2 nmol) were injected, and chromatograms were recorded by measuring absorbance at 460 nm.
Electron Microscopy.
Samples were examined with a FEI Nova NanoSEM 200 scanning electron microscope operated with an acceleration voltage of 5 kV at a distance of 5 mm. Thin-sectioned samples were viewed with a Tecnai F30 (Philips/FEI) transmission electron microscope (field emission gun operating with a 300-kV accelerating voltage, using a magnification of 15,000–30,000×) and a high performance CCD camera with 4 k × 4 k resolution.
Animal Experiments.
Animal experiments were performed in accordance with the institutional guidelines following experimental protocol review and approval by the Institutional Biosafety Committee (IBC) and the Institutional Animal Care and Use Committee (IACUC) at the University of Chicago. S. aureus Newman (1 × 108 CFU suspended in 100 µL PBS) was injected into the periorbital venous plexus of BALB/c mice (n = 15), and animals were monitored for survival over 10 d. Animals received either two injections of inhibitor (32 mg/kg) or compound buffer (mock) separated by 12-h intervals before infection and an additional six doses after infection.
Expanded text describing all experimental procedures is provided in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Sue Chiang, members of the National Screening Laboratory for Regional Centers of Excellence in Biodefense and Emerging Infectious Disease, and Hannah Maier for technical assistance, as well as members of our laboratory for discussion. The authors acknowledge membership within and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217337110/-/DCSupplemental.
References
- 1.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 2.Neu HC. The crisis in antibiotic resistance. Science. 1992;257(5073):1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 3.Payne DJ, Holmes DJ, Rosenberg M. Delivering novel targets and antibiotics from genomics. Curr Opin Investig Drugs. 2001;2(8):1028–1034. [PubMed] [Google Scholar]
- 4.Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6(5):427–430. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CT. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406(6797):775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 6.Neuhaus FC, Baddiley J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(4):686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reichmann NT, Gründling A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett. 2011;319(2):97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer W. Bacterial phosphoglycolipids and lipoteichoic acids. In: Hanahan DJ, editor. Handbook of Lipid Research. Vol 6. New York: Plenum Press; 1990. pp. 123–234. [Google Scholar]
- 9.Duckworth M, Archibald AR, Baddiley J. Lipoteichoic acid and lipoteichoic acid carrier in Staphylococcus aureus H. FEBS Lett. 1975;53(2):176–179. doi: 10.1016/0014-5793(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 10.Gründling A, Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189(6):2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorasch P, Warnecke DC, Lindner B, Zähringer U, Heinz E. Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids and glycosylsterols. Eur J Biochem. 2000;267(12):3770–3783. doi: 10.1046/j.1432-1327.2000.01414.x. [DOI] [PubMed] [Google Scholar]
- 12.Kiriukhin MY, Debabov DV, Shinabarger DL, Neuhaus FC. Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: Role of YpfP, the diglucosyldiacylglycerol synthase. J Bacteriol. 2001;183(11):3506–3514. doi: 10.1128/JB.183.11.3506-3514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch HU, Haas R, Fischer W. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur J Biochem. 1984;138(2):357–363. doi: 10.1111/j.1432-1033.1984.tb07923.x. [DOI] [PubMed] [Google Scholar]
- 14.Gründling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(20):8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oku Y, et al. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol. 2009;191(1):141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb AJ, Karatsa-Dodgson M, Gründling A. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol. 2009;74(2):299–314. doi: 10.1111/j.1365-2958.2009.06829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schirner K, Marles-Wright J, Lewis RJ, Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28(7):830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garufi G, et al. Synthesis of lipoteichoic acids in Bacillus anthracis. J Bacteriol. 2012;194(16):4312–4321. doi: 10.1128/JB.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu D, et al. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc Natl Acad Sci USA. 2009;106(5):1584–1589. doi: 10.1073/pnas.0809020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96(22):12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karatsa-Dodgson M, Wörmann ME, Gründling A. In vitro analysis of the Staphylococcus aureus lipoteichoic acid synthase enzyme using fluorescently labeled lipids. J Bacteriol. 2010;192(20):5341–5349. doi: 10.1128/JB.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAdow M, et al. Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog. 2011;7(10):e1002307. doi: 10.1371/journal.ppat.1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand N, Davis BD. Damage by streptomycin to the cell membrane of Escherichia coli. Nature. 1960;185:22–23. doi: 10.1038/185022a0. [DOI] [PubMed] [Google Scholar]
- 25.Jones D, Metzger HJ, Schatz A, Waksman SA. Control of Gram-negative bacteria in experimental animals by streptomycin. Science. 1944;100(2588):103–105. doi: 10.1126/science.100.2588.103. [DOI] [PubMed] [Google Scholar]
- 26.Funatsu G, Wittmann HG. Ribosomal proteins. 33. Location of amino-acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J Mol Biol. 1972;68(3):547–550. doi: 10.1016/0022-2836(72)90108-8. [DOI] [PubMed] [Google Scholar]
- 27.McCormick MH, McGuire JM, Pittenger GE, Pittenger RC, Stark WM. Vancomycin, a new antibiotic. I. Chemical and biologic properties. Antibiot Annu. 1955-1956-1956;3:606–611. [PubMed] [Google Scholar]
- 28.Walsh CT. Vancomycin resistance: Decoding the molecular logic. Science. 1993;261(5119):308–309. doi: 10.1126/science.8392747. [DOI] [PubMed] [Google Scholar]
- 29.Yamakawa J, et al. Heterogeneously vancomycin-intermediate Staphylococcus aureus (hVISA) emerged before the clinical introduction of vancomycin in Japan: A retrospective study. J Infect Chemother. 2012;18(3):406–409. doi: 10.1007/s10156-011-0330-2. [DOI] [PubMed] [Google Scholar]
- 30.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375(9725):1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klevens RM, et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 32.Klevens RM, Edwards JR, Gaynes RP, System NNIS. National Nosocomial Infections Surveillance System The impact of antimicrobial-resistant, health care-associated infections on mortality in the United States. Clin Infect Dis. 2008;47(7):927–930. doi: 10.1086/591698. [DOI] [PubMed] [Google Scholar]
- 33.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302(5650):1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 34.Tenover FC, Biddle JW, Lancaster MV. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg Infect Dis. 2001;7(2):327–332. doi: 10.3201/eid0702.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, et al. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med. 2012;18(5):816–819. doi: 10.1038/nm.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arbeit RD, Maki D, Tally FP, Campanaro E, Eisenstein BI. Daptomycin 98-01 and 99-01 Investigators The safety and efficacy of daptomycin for the treatment of complicated skin and skin-structure infections. Clin Infect Dis. 2004;38(12):1673–1681. doi: 10.1086/420818. [DOI] [PubMed] [Google Scholar]
- 37.Stevens DL, et al. Linezolid versus vancomycin for the treatment of methicillin-resistant Staphylococcus aureus infections. Clin Infect Dis. 2002;34(11):1481–1490. doi: 10.1086/340353. [DOI] [PubMed] [Google Scholar]
- 38.van Hal SJ, Paterson DL. New Gram-positive antibiotics: Better than vancomycin? Curr Opin Infect Dis. 2011;24(6):515–520. doi: 10.1097/QCO.0b013e32834ab1de. [DOI] [PubMed] [Google Scholar]
- 39.Willems RJ, Hanage WP, Bessen DE, Feil EJ. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wörmann ME, Corrigan RM, Simpson PJ, Matthews SJ, Gründling A. Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol Microbiol. 2011;79(3):566–583. doi: 10.1111/j.1365-2958.2010.07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Projan SJ, Shlaes DM. Antibacterial drug discovery: Is it all downhill from here? Clin Microbiol Infect. 2004;10(Suppl 4):18–22. doi: 10.1111/j.1465-0691.2004.1006.x. [DOI] [PubMed] [Google Scholar]
- 42.Fischer W. Pneumococcal lipoteichoic and teichoic acid. Microb Drug Resist. 1997;3(4):309–325. doi: 10.1089/mdr.1997.3.309. [DOI] [PubMed] [Google Scholar]
- 43.Reid CW, et al. Structural characterization of surface glycans from Clostridium difficile. Carbohydr Res. 2012;354:65–73. doi: 10.1016/j.carres.2012.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.