Abstract
Antibiotic resistance genes (ARGs) are emerging contaminants posing a potential worldwide human health risk. Intensive animal husbandry is believed to be a major contributor to the increased environmental burden of ARGs. Despite the volume of antibiotics used in China, little information is available regarding the corresponding ARGs associated with animal farms. We assessed type and concentrations of ARGs at three stages of manure processing to land disposal at three large-scale (10,000 animals per year) commercial swine farms in China. In-feed or therapeutic antibiotics used on these farms include all major classes of antibiotics except vancomycins. High-capacity quantitative PCR arrays detected 149 unique resistance genes among all of the farm samples, the top 63 ARGs being enriched 192-fold (median) up to 28,000-fold (maximum) compared with their respective antibiotic-free manure or soil controls. Antibiotics and heavy metals used as feed supplements were elevated in the manures, suggesting the potential for coselection of resistance traits. The potential for horizontal transfer of ARGs because of transposon-specific ARGs is implicated by the enrichment of transposases—the top six alleles being enriched 189-fold (median) up to 90,000-fold in manure—as well as the high correlation (r2 = 0.96) between ARG and transposase abundance. In addition, abundance of ARGs correlated directly with antibiotic and metal concentrations, indicating their importance in selection of resistance genes. Diverse, abundant, and potentially mobile ARGs in farm samples suggest that unmonitored use of antibiotics and metals is causing the emergence and release of ARGs to the environment.
Keywords: concentrated animal feeding operations, horizontal gene transfer, growth-promoting antibiotics, tetracycline
The spread and aggregation of antibiotic-resistant genes into multidrug-resistant pathogens is challenging life-saving antibiotic therapies (1, 2). Indeed, the expansion of the antibiotic resistance gene reservoir in the environment has been caused by antibiotic use in humans and animals (3). Furthermore, a growing body of direct and indirect evidence from the past 35 y (4) establishes that farm antibiotic use correlates repeatedly with the rise and spread of associated resistance genes in human pathogens, as well as the direct transfer of antibiotic-resistant bacteria from animals to humans (5–8). Antibiotic use has increased the frequency of horizontal gene transfer and resistance gene fixation in genomes, leading to the development of diverse resistance genes in genomic islands (9). Acinetobacter baumannii is a case in point. In 30 y, it evolved from being completely antibiotic-susceptible to being multidrug-resistant by expanding a genomic island by 66 kb, including 45 resistance genes, which were horizontally transferred from various genera of bacteria, some of which likely originated from the environment (10). Antibiotic-resistant strains can then be distributed worldwide, aided by a number of human factors, but especially international travel for commerce, immigration, and recreation (11). Antibiotic resistance genes (ARGs) are becoming recognized as environmental pollutants, and action is being sought to preserve the efficacy of antibiotics. The World Organization for Animal Health, together with the US Food and Drug Administration and the World Health Organization, urge improved regulation of veterinary antibiotic use in over 100 developing countries (12).
China is the largest antibiotics producer and consumer in the world. In a 2007 survey, the estimated annual antibiotics production in China was 210 million kg, and 46.1% were used in livestock industries (13), at least four times the amount used in the US livestock industry in 1999 (14). In China, the use of antibiotics both for animal disease treatment and growth promotion is unmonitored, which often leads to high use, reflected by the high concentrations of antibiotic residues (hundreds of milligrams of tetracycline per kilogram) that are commonly detected in animal manures (15, 16). Manure is a major source of antibiotic pollution in the environment, and China produces an estimated 618 billion kg of swine manure annually (17). Most veterinary antibiotics are poorly absorbed by the animal and hence are excreted (18) and dispersed to soil when the manure is spread as fertilizer, the desired practice for recycling nutrients. Furthermore, the use of subtherapeutic levels of antibiotics in animal feeds causes an increase in antibiotic resistance traits in manure (19, 20), manure-amended soils (21), and downstream river waters and sediments (22). In addition, metals are added to swine feed for growth promotion and disease control and may provide a long-term coselective pressure for antibiotic resistance (23). The scale of the livestock industry in China and the volume of antibiotics use provide an opportunity to assess the impact of large-scale animal farm practices on antibiotic resistance genes in the environment. Previously, tetracycline resistance (tet) genes in soils adjacent to representative Chinese swine feedlots were positively correlated to concentrations of tetracycline residues (24), raising the question of whether the diversity and abundance of the antibiotic resistance reservoir extends beyond tetracycline resistance genes due to the use of additional antibiotics, possible coselection for other resistance genes, and/or recruitment of multidrug efflux pump genes.
Although antibiotic-resistant bacteria have been isolated and characterized from farm soils (21, 25), this method only samples microbes that are culturable and express their ARGs under those conditions. ARGs of noncultured populations, as well as “silent” or unexpressed ARGs (26), are sources of risk because they contribute to the resistance reservoir and could be horizontally transferred or expressed under other conditions. We used high-capacity quantitative PCR (qPCR) (19) with 313 validated primer sets, which target 244 ARGs (Table S1) from all major classes of ARGs, to extensively sample the antibiotic resistance reservoir. We sampled three large-scale commercial swine farms, each from a different region of China, at three stages of manure management: manure, manure compost, and soil receiving manure compost. Manure from pigs never fed antibiotics and soil from a pristine forest in Putian, China were used as experimental controls.
Results
Antibiotics and Metal Concentrations.
Antibiotics and their use as reported by the farmers are listed in Table S2. Total tetracycline concentrations in these manure and soil samples were as high as 15.2 mg⋅kg−1 and 0.78 mg⋅kg−1, respectively, as was determined previously (15). Of the sulfonamides analyzed in this study, sulfamethoxazole had the highest concentrations for all samples, ranging from 1.08 to 3.02 µg⋅kg−1 (Fig. S1). Sulfadiazine was also detected in all samples in a range of 0.50–4.81 µg⋅kg−1. Of the fluoroquinolones analyzed in this study, only ofloxacin and enrofloxacin were observed in most samples. The highest mean concentration of ofloxacin (335 µg⋅kg−1) and enrofloxacin (96.0 µg⋅kg−1) were observed in Putian compost and soil samples, respectively (Fig. S1). Zinc, copper, and arsenic, used as feed additives, were also elevated above background concentrations. The highest mean concentrations of copper, zinc, and arsenic were detected in Putian manure, Jiaxing compost, and Beijing manure, respectively, with copper up to 1,700 mg⋅kg−1 manure (Fig. S2). The concentration of copper, zinc, and arsenic were much higher in manure than in compost and soil samples, with the exception of the Jiaxing compost, in which copper and zinc had the highest concentrations of all of the samples.
Diversity of Antibiotic Resistance Genes.
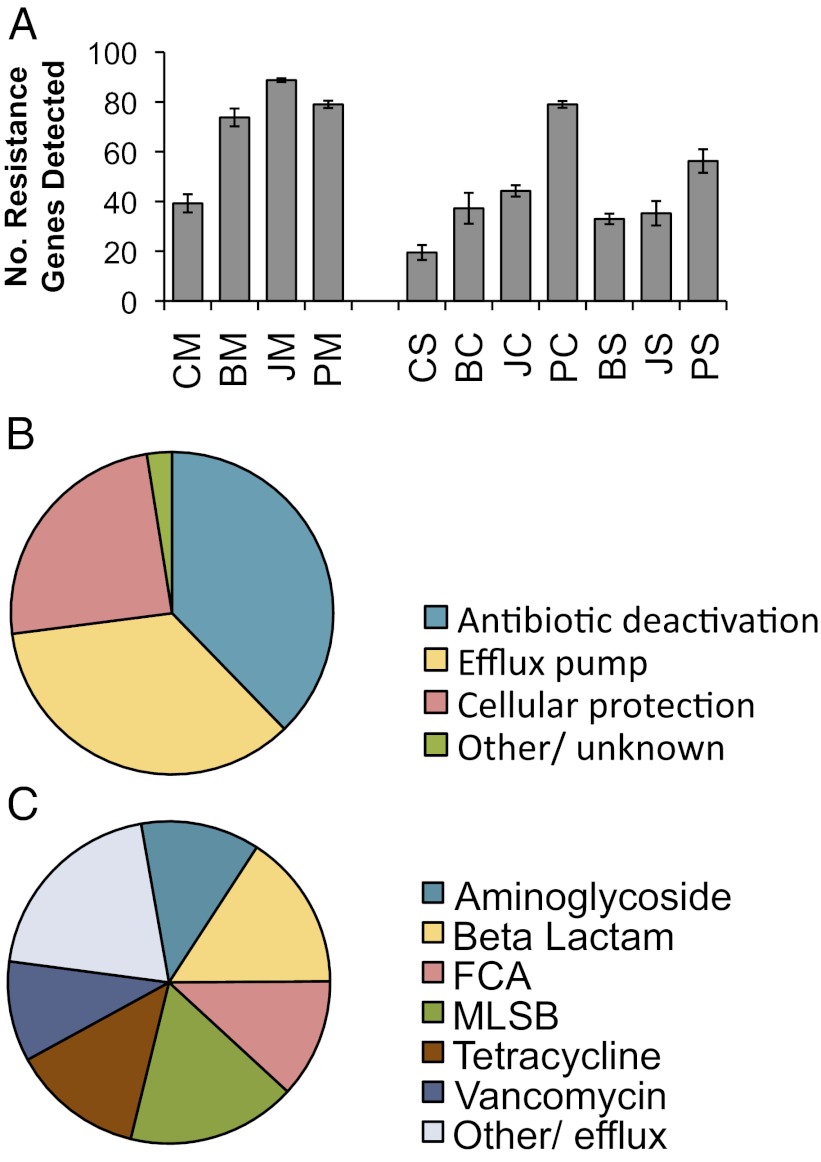
We detected 149 unique ARGs among all of the samples, which is three times more types of ARGs than were found in the control samples (Fig. 1A). The ARGs detected in these farms encompass the three major resistance mechanisms—efflux pumps, antibiotic deactivation, and cellular protection (Fig. 1B)—and potentially confer resistance to most major classes of antibiotics (Fig. 1C). Resistance gene profiles indicate the patterns and degrees of enrichment of ARGs for each site (Fig. 2) and that manure samples cluster separately from the other samples with the exception of the Putian compost. The compost and soil samples also cluster separately with the exception of one of the Beijing compost replicates, which grouped with the soil samples. Furthermore, Shannon diversity (indicating richness and abundance) of ARGs from farm samples was significantly higher than that of the control samples (Fig. S3).
Fig. 1.
Antibiotic resistance gene detection statistics. Sample names are abbreviated with two letters representing location and sample type: first C, B, J, and P (control, Beijing, Jiaxing, and Putian, respectively) and second M, C, and S [manure, compost, and soil (with compost amendment), respectively]. Because many resistance genes were targeted with multiple primers, if multiple primer sets detected the same gene, this was only counted as detection of a single unique resistance gene. (A) Average number of unique resistance genes detected in each sample. Error bars represent SEM of four field replicates. The resistance genes detected in all samples were classified based on (B) the mechanism of resistance, and (C) the antibiotic to which they confer resistance. FCA, fluoroquinolone, quinolone, florfenicol, chloramphenicol, and amphenicol resistance genes; MLSB, Macrolide-Lincosamide-Streptogramin B resistance.
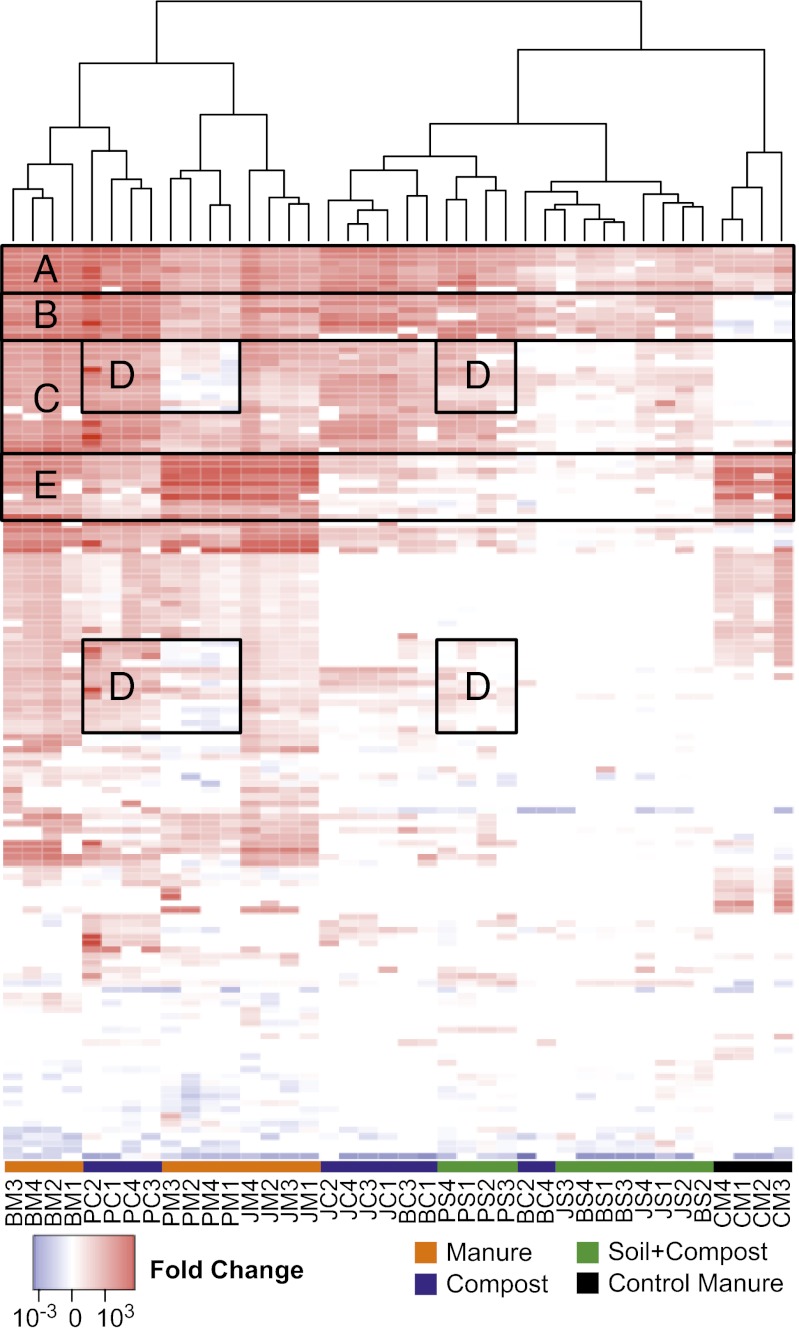
Fig. 2.
Resistance gene profile from the farm sites. Each column is labeled with the sample name (same abbreviation scheme as in Fig. 1, with numbers representing field replicates), and each row is the results from a single primer set. Values plotted are the ΔΔCT with the control soil being the reference sample for all samples. The legend denotes corresponding fold change values, which is a log scale. All primer sets (223) that showed amplification in at least one sample are shown. Columns were clustered based on Bray-Curtis diversity measures. Black boxes delineate resistance profiles: (A) enriched in all samples, including control manure (CM); (B) enriched in all farm samples, but not the CM; (C) widely enriched in most of the farm samples but not the CM; (D) genes that were enriched in the Putian compost but not the Putian manure; and (E) strongly enriched in CM and farm manures.
Abundance of Antibiotic Resistance Genes.
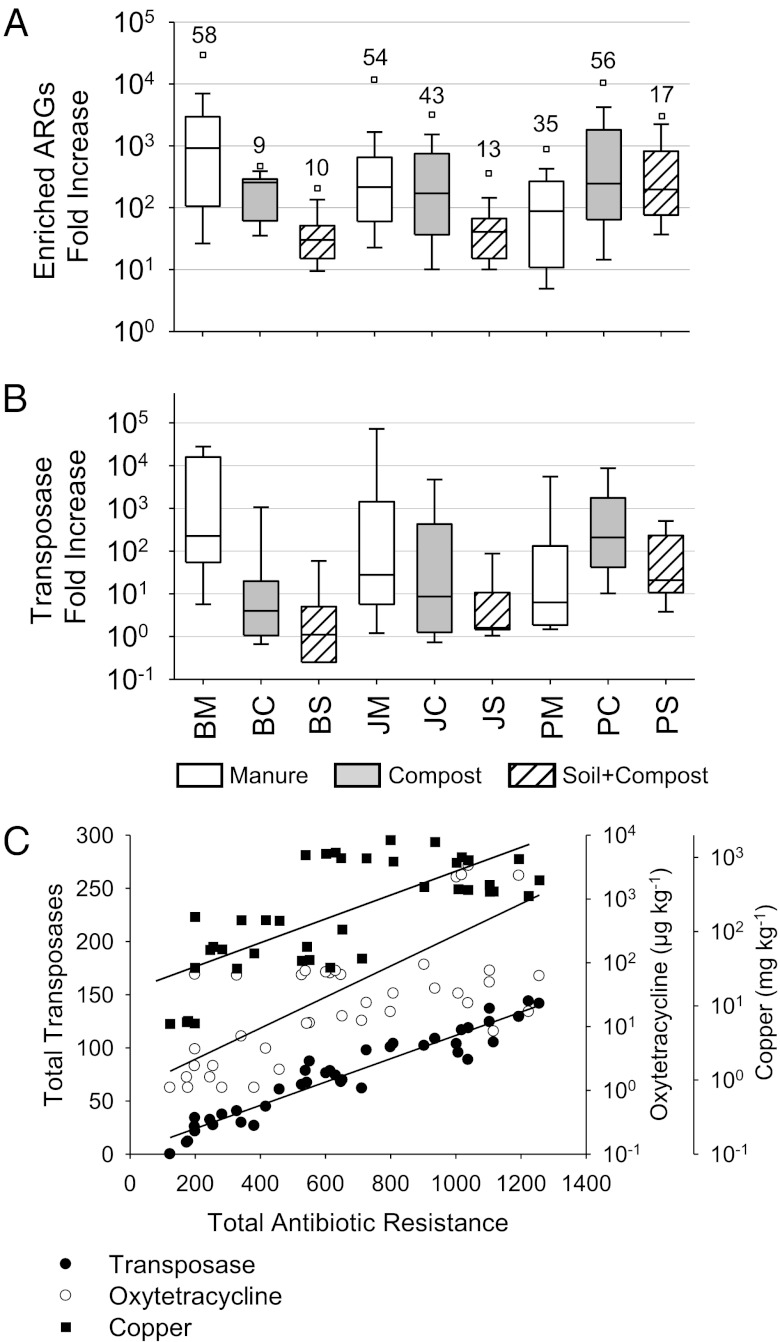
ARGs were highly enriched in the farm samples. We used the sum of the enrichment of all unique ARGs in a sample to approximate total enrichment in the farms. Maximum enrichment occurred in the manure samples at Beijing (121,000-fold) and Jiaxing (39,000-fold) farms, and in the compost at the Putian farm (57,000-fold enrichment), demonstrating the large expansion of the antibiotic resistance reservoir in these farms, including the enrichment of up to 19 unique tet genes in a single site (Table S3 gives enrichment details for all genes). A total of 63 unique ARGs were significantly enriched in at least one sample compared with controls at an overall median enrichment of 192-fold for all samples. The maximum enrichment of a single ARG was over 28,000-fold in the Beijing manure (Fig. 3A). In terms of absolute abundance, an aminoglycoside phosphorylation gene aphA3 is found 43% as frequently as the 16S rRNA gene in the manure samples, based on a 0.58 average value of the delta threshold cycle (∆CT) values (Table S4), meaning this single gene would be found in nearly one in every second bacterium, assuming a single copy of each gene in single genomes. In general, enrichment of individual ARGs decreases in soil samples but is still elevated, with average enrichment of nearly 100-fold, and some genes were enriched over 1,000-fold compared with the soil control. The Putian soil had more unique resistance genes enriched at a higher level than the other two farm soils. When combining the data from all farms, 56, 44, and 17 unique ARGs were statistically elevated in the manure, compost, and soil samples, respectively.
Fig. 3.
Abundance of resistance genes and transposases. In the box plots, the symbols indicate the following: box, 25th to 75th percentile; horizontal line, median; whiskers, 10th and 90th percentile; and square, maximum value. The y axis is a log scale of fold increase: farm manure compared with the control manure, and farm compost or soil compared with the control soil. (A) Only statistically enriched resistance genes are represented. The number above each site indicates the number of primer sets that yielded statistically significant results. (B) Summary of all nine primer sets used to target different transposase alleles (in B, top whisker represents the maximum value). (C) Correlation of total resistance and transposase abundances, oxytetracycline concentration, and copper concentration. Total antibiotic resistance and total transposases values are the sum of ∆∆CT values of all assays of that type in each sample. The sample identifiers below B apply to both A and B.
Transposase Enrichment.
Transposases, in parallel to ARGs, were highly enriched (Fig. 3B). Transposases were found in all samples (Fig. 2, subgroups A and B) and were enriched up to 90,000-fold in the manure samples and up to 1,000-fold in the soil samples. The abundance of ARGs is highly correlated to the levels of transposases in these farm samples (Fig. 3C) (e.g., as high as 0.970 for correlation between the abundance of tetracycline resistance genes and transposase genes) (Table S5).
Discussion
Feed Additive Use.
These swine farms use a complex mixture of growth-promoting chemicals, including antibiotics and metals. However, the individual dosage of each chemical, when considered alone, on these farms is not excessive compared with other farms globally. Total tetracyclines in manure and soil samples were as high as 15.2 mg⋅kg−1 and 0.78 mg⋅kg−1, respectively (15), which is within the range reported for some European manures between 2002 and 2005 (14). However, other farms in China use higher concentrations of antibiotics; for example, tetracycline and sulfonamide concentrations in manure reported previously (16) were as high as 764 mg⋅kg−1 and 20 mg⋅kg−1, respectively, whereas in this study their maximum concentrations were only 15 mg⋅kg−1 and 5 µg⋅kg−1 manure, respectively. However, the Jiaxing and Putian farms used 13 types of antibiotics, which is close to the estimate of the number of antibiotics used in fisheries along the entire Thai coastline (27). In addition to antibiotics, metals used as feed additives contributed to the complex mixture of selective pressures in these farms. The metal feed additives zinc, copper, and arsenic were elevated above background concentrations at levels typical in Chinese swine farms (28) and only slightly higher than concentrations reported in the United States and Europe [maximum values reported as 1,300–1,550 mg copper⋅kg−1 manure (29)]. Although no single antibiotic or metal concentration is excessive in these farms, it is the number of additives used that is striking. The effect of mixtures of resistance selecting agents is unknown but presumably increases the likelihood of coresistance in genetic elements (9).
Enlarged Diversity and Abundance of the Environmental Resistance Reservoir.
This study documents the breadth and extent of the antibiotic resistance reservoir in large-scale animal production facilities. Furthermore, we provide measures to estimate the field-scale response to composting and subsequent soil application representing typical manure management practices in China as a case study. The diverse set of resistance genes detected (Fig. 1) potentially confer resistance to all major classes of antibiotics, including antibiotics critically important for human medicine (30), such as macrolides (mphA and erm genes), cephalosporins (blaTEM and blaCTX-M), aminoglycosides (aph and aad genes), and tetracycline (tet genes). Although a number of vancomycin resistance genes were detected in these farm samples (Fig. 1C), we do not expect significant phenotypic resistance to vancomycin because detection levels were low and resistance is dependent on multigene van operons (1, 25), which we did not detect. However, our detection of individual van genes may be an indication that enrichment for van operons is possible under alternative conditions. In general, genes potentially conferring resistance to aminoglycosides, tetracyclines, sulfonamide, florfenicol, and quaternary ammonium compounds were enriched most broadly in all farm samples. Beta lactam and macrolide resistance genes were enriched primarily in manure samples, although they may still be present but at levels below detection in the downstream samples. A previous study using a similar qPCR method, sampling only a few individual pigs, detected 57 resistance genes, but only 8 were enriched (19). D’Costa et al. (25) found resistance to a broad range of antimicrobials but only considered cultured actinomycete strains. One specific type of resistance studied broadly is that for tetracyclines. In a survey of 14 tet genes among hundreds of tetracycline-resistant soil isolates, Ghosh and LaPara (21) found that the most common genes were tetL, tetA, tetM, and tetG (tetW was not included in their survey). We detected 22 of the 28 tetracycline resistance genes targeted on our array. The most abundant tet genes (based on ∆CT values, Table S4) in the manure were tetQ, tetW, tetX, tet(32), tetO, tetM, tetL, and tetG, whereas in the soil they were tetG, tetL, tetA, and tetW, the latter set being similar to those found in soil by Ghosh and LaPara (21). The increased number of resistance genes we detected compared with previous studies reflects our sampling at the herd and field levels and the use of a high-throughput qPCR method of detection.
The resistance genes found in our samples were not limited to the antibiotics administered. Aminoglycosides were not used in the Putian farm, but more than 10 aminoglycoside resistance genes were enriched in that farm up to more than 10,000-fold. Similarly, floR was enriched 500-fold in the Jiaxing compost but amphenicols were not known to be used at that farm. Coenrichment of these genes is most likely due to aggregation of resistance genes on mobile genetic elements (19, 31–34), as has been observed directly (35). In addition, the abundance of ARGs in these samples is correlated with the concentrations of antibiotics, as well as with copper, zinc, and arsenic (Fig. 3 and Table S5). The presence of heavy metals provides another coselective pressure for antibiotic resistance (23) and may aid in long-term persistence of ARGs during manure management and disposal (36). Only a few multidrug efflux pumps (qacE∆1 and dfrA1) were broadly enriched (Table S6). It seems that genes with specific mechanisms of resistance were preferably enriched in these high-selection-pressure environments. The diversity of resistance genes enriched and coenriched at the farm level is concerning because this broad set of ARGs or a subset thereof could be (co)enriched or transferred to pathogens under future selection conditions.
Swine farms are known hotspots for pervasive and abundant antibiotic resistance both in antibiotic-free animals (19, 37) and, especially, in antibiotic-treated animals (26, 38). The level of enrichment of individual resistance genes is on par with previous field-scale studies. Tetracycline resistance genes were enriched 102- to 104-fold in cattle waste lagoons (39), and the median enrichment of ARGs in this manure was about 104-fold. Of bacterial isolates from swine and chicken manure, 92% and >80%, respectively, were found resistant to at least one antibiotic (26, 31). We estimate about 43% of bacteria possess at least the aphA3 gene; hence, it is feasible that upward of 90% of the entire community would carry one of the other 148 resistance genes detected. Considering all antibiotic resistance genes combined in the manure or compost samples, we estimate a total of 50,000-fold enrichment (Table S3). Although enrichment of individual resistance genes is similar to that found in previous studies, we were able to capture a more complete picture of the total level of the antibiotic resistance reservoir.
Potential for Horizontal Gene Transfer of ARGs.
This study highlights that ARGs in swine farms are not only diverse but are also remarkably abundant, which together offers a higher statistical probability of dispersal, further selection, and/or horizontal transfer in the environment. The emergence and spread of ARGs are closely associated with mobile genetic elements such as plasmids, integrases, and transposases (20, 40, 41). The high degree of transposase enrichment and correlation with abundance of ARGs suggests that horizontal gene transfer may have aided the enrichment of ARGs. The transposases detected most frequently belong to the IS6 family of insertion sequences, which are typically found flanking an array of genes, often resistance genes (42). The most abundant member of the IS6 family in these samples, IS26, has been isolated, along with integrons in multidrug-resistant plasmids in enterobacteria (43). Integrons most commonly contain resistance cassettes encoding aadA genes (44), as well as qacE∆1 and sul2, which were among the most enriched genes in this study. The Putian farm ARGs that are more enriched in the compost than in the manure (Fig. 2, D boxes and Table S6) are predominately aadA and other aminoglycoside resistance genes and their enrichment may be due to their presence in integrons that also hold a resistance gene cassette relevant to the drugs used on the farm (34, 45). Additionally, the combination of antibiotics and metals may provide a stronger selection for realized horizontal gene transfer within the microbial community than either alone (9, 23, 34, 46). It appears that a number of factors in swine farms could contribute to elevated rates of horizontal gene transfer, including elevated concentrations of antibiotics, metals, ARGs, and mobile genetic elements, making subsequent dispersal, (co)enrichment, or horizontal transfer, including to human-associated bacteria, more probable.
Role of Manure Management in Controlling ARGs.
The long-term goal of manure management is to remove environmental contaminants while disposing of this high-volume waste product and capturing its value to improve soil fertility. The goal in the case of ARGs is to identify practices that decrease their concentration to a greater degree than by simple dilution (47). Manure composting decreased the abundance of ARGs at Beijing, but abundance remained nearly the same in the Jiaxing manure, while at Putian composting actually increased the abundance of ARGs. Composting concentrated sulfonamides (Fig. S1), sulfonamide resistance genes, and some metals (Fig. S2), consistent with the observation that sulfonamide resistance genes are more recalcitrant than tetracycline genes (22, 48, 49). The common practice of spreading compost on soil was not sufficient to reduce abundance of ARGs to background levels, and the Putian soil showed up to 3,000-fold enrichment. However, the practice decreased concentrations of ARGs substantially below compost levels. The relatively high enrichment of ARGs in Putian soil may be due to a higher manure/soil ratio and/or shorter time before sampling after amendment compared with other farms. These observations highlight the need to determine adequate composting time to reduce resistance levels before release to the more uncontrolled environment (50), as well as to define soil and landscape features that would minimize dispersal to the human food chain.
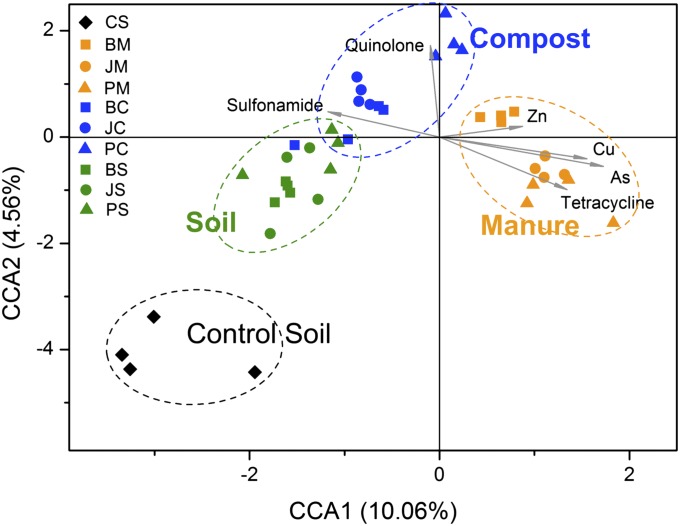
Resistance gene diversity and abundance patterns specific to each management type indicate the influence of the antibiotics as a selective pressure. These profiles show that generally samples of the same management type clustered together (Fig. 4). The relationships between the structure of detected ARGs and antibiotic and heavy metal concentrations were assessed with canonical correspondence analysis. Manure samples grouped separately by the first axis and were strongly affected by arsenic, copper, and tetracycline concentrations, which are likely among the dominant factors driving the changes in structures of ARGs on these farms (Fig. 4). Although only three farms are included in this study, regardless of their location (a separation of over 2,000 km), composting technique, or antibiotic dosage, the ARGs’ resistance profiles are similar, indicating that similar reservoirs of ARGs are likely common across China and in other countries where management practices are similar.
Fig. 4.
Canonical correspondence analysis (CCA) compares the abundance of detected resistance genes (symbols) and the concentration of heavy metals and antibiotics (arrows). The results showed that pig manure samples were positively correlated to the concentrations of copper, zinc, arsenic, and total tetracyclines. Environmental variables were chosen based on significance calculated from individual CCA results and variance inflation factors (VIFs) calculated during CCA. The percentage of variation explained by each axis is shown, and the relationship is significant (P = 0.005). CCA analyses were performed in R 2.13.0 with vegan package 1.17-9.
The diversity and abundance of ARGs reported in this study is alarming and clearly indicates that unmonitored use of antibiotics and metals on swine farms has expanded the diversity and abundance of the antibiotic resistance reservoir in the farm environment. The coenrichment of ARGs and transposases further exacerbates the risks of transfer of ARGs from livestock animals to human-associated bacteria, and then spread among human populations (4, 6). Policies and management tools to facilitate prudent use of antibiotics and heavy metals, including their combined use, in animal industries and animal waste management are needed. Decreased resistance levels have been observed in Europe after the disuse of agricultural antibiotics (51). Pig manure, with its abundant and diverse ARGs and sheer volume, is a major source of resistance genes and as such a public health hazard. Microbes from manure, compost, or soil containing the ARGs are subject to dispersal via runoff into rivers (22), leaching to subsurface waters, air dispersal via dust, human travel, and distribution of agricultural products, including compost for gardening, which could expand a local contamination to regional and even global scales (6, 11).
Materials and Methods
Sampling.
A total of 36 samples were collected in 2010 from three Chinese provinces including (from north to south) Beijing (Beijing farm), Zhejiang (Jiaxing farm), and Fujian (Putian farm). The manure and compost samples were obtained from representative swine farms with an animal intensity of 10,000 market hogs or more per year. Soil samples were collected from a nearby agronomic field to which manure-based compost had been applied. Four replicates were taken from each sample type and farm, and all of the samples were kept on dry ice during transportation and stored at −80 °C before DNA extraction and chemical analysis.
These are typical large-scale swine farms. Pigs are continuously housed on concrete. The manure was sampled within 1 d after excretion in all cases. In Beijing, compost was managed in outdoor windrows with aeration for 2 wk. In Jiaxing, pile composting was used with regular stirring (one or two times per day) for about 10 d. In Putian they used pile composting with limited aeration for 2–4 wk. In Jiaxing and Putian, compost products are packed and sold as commercial organic fertilizer for local farmers. For soil amendment, the composted manure spreading rate varies but is ∼10 tons/hectare, applied once per year. At the Beijing and Jiaxing farms, the soil had been receiving manure compost for more than 2 y, and the most recent application was 2 mo before sampling. At the Putian farm, the soil had been receiving manure compost for more than 3 y, and the most recent application was 1 wk before sampling.
Control samples received no known antibiotic input. The control soil is from a pristine forest in Putian, China. This soil has had no anthropogenic antibiotic input and has an abundance of ARGs and diversity profile similar to another temperate-region, antibiotic-free grassland soil we studied. The control pig manure samples were mixtures of DNA extracted from feces from pigs birthed from a mother with no antibiotic exposure and grown in facilities with no antibiotic exposure but fed a normal grower diet (ref. 19 gives further details). Sample CM1 was taken from six 84-d-old pigs not fed antibiotics. Samples CM2–4 were each taken from a single animal at three time points between 86 and 104 d of age. The control manure was used as a comparison against the farm manures, and the control soil was used as a comparison against both the farm compost and farm soil.
Antibiotic and Metal Quantitation.
Concentrations of sulfonamides and quinolones were analyzed in this study, including sulfadiazine, sulfamerazine, sulfamethoxydiazine, sulfamethazine, sulfamethoxazole, norfloxacin, ofloxacin, enrofloxacin, and ciprofloxacin. Previously, 5 target tetracyclines and 10 degradation products were analyzed (15).
Metals were analyzed in air-dried, milled samples after oxidative digestion in sealed tubes by inductively coupled plasma-mass spectrometry (7500cx; Agilent). Quantities were determined relative to reference standards. Sample extraction and analysis procedures for antibiotics and metals are described in SI Materials and Methods.
DNA Extraction.
High-molecular-weight community DNA was extracted by the freeze-grinding, SDS-based method (52) and was purified using a low-melting agarose gel followed by phenol extraction. DNA concentration and quality were determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc.).
Primer Design.
A majority of the primer sets (247) were designed, used, and validated in a previous study (19). For this study, 89 new primer sets were designed for categories of resistance genes not previously targeted as thoroughly. The same design parameters were used as before (19). Reference sequences were harvested from the Antibiotic Resistance Genes Database (http://ardb.cbcb.umd.edu/). Additional validation of the primer sets was performed and is described in SI Materials and Methods.
Quantitative PCR.
All quantitative PCR reactions were performed using the Applied Biosystems OpenArray platform, as described previously (19), except that a threshold cycle (CT) of 27 was used as the detection limit. Generally the technical triplicates were tested during separate testing occasions (plate and day of testing) as a method of quality control. The ΔΔCT method of comparison (53) was used to compare relative abundance between samples:
where CT is the threshold cycle, ARG is one of the 313 antibiotic resistance gene assays, 16S is the 16S rRNA gene assay, Target is the experimental sample, and Ref is the reference sample. The reference sample used as a comparison depended on the purpose of the analysis. When the purpose was to reveal changes among all farm types and the dynamics of ARGs because of manure management, the control soil was the reference sample for all farm samples, as was the case in Fig. 2. Average CT values were calculated by averaging the four field replicates. If there was no amplification in one of the four field replicates, it was considered a false negative and discarded. In calculation of the ∆CT of the reference sample, if there was no amplification, the detection limit CT (27.0) was used. Genes were considered statistically enriched if the range created by three SDs of the mean fold change was entirely >1.
Supplementary Material
Acknowledgments
We thank Benli Chai and Amanda Geaslin for technical assistance. Funding was provided by Ministry of Science and Technology of China (2011DFB91710) National Science Foundation of China (2121008) (to Y.-G.Z.), and at Michigan State University by the Pharmaceuticals in the Environment Initiative.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222743110/-/DCSupplemental.
References
- 1.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century—A clinical super-challenge. N Engl J Med. 2009;360(5):439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 2.Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally drug-resistant tuberculosis in India. Clin Infect Dis. 2012;54(4):579–581. doi: 10.1093/cid/cir889. [DOI] [PubMed] [Google Scholar]
- 3.Knapp CW, Dolfing J, Ehlert PAI, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol. 2010;44(2):580–587. doi: 10.1021/es901221x. [DOI] [PubMed] [Google Scholar]
- 4.Marshall BM, Levy SB. Food animals and antimicrobials: Impacts on human health. Clin Microbiol Rev. 2011;24(4):718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480(7376):241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 7.Levy SB. Emergence of antibiotic-resistant bacteria in the intestinal flora of farm inhabitants. J Infect Dis. 1978;137(5):689–690. [PubMed] [Google Scholar]
- 8.Price LB, et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3(1):00305–00311. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillings MR, Stokes HW. Are humans increasing bacterial evolvability? Trends Ecol Evol. 2012;27(6):346–352. doi: 10.1016/j.tree.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Fournier PE, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2(1):e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Church DL. Major factors affecting the emergence and re-emergence of infectious diseases. Clin Lab Med. 2004;24(3):559–586, v. doi: 10.1016/j.cll.2004.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert N. Rules tighten on use of antibiotics on farms. Nature. 2012;481(7380):125. doi: 10.1038/481125a. [DOI] [PubMed] [Google Scholar]
- 13.Hvistendahl M. Public health. China takes aim at rampant antibiotic resistance. Science. 2012;336(6083):795. doi: 10.1126/science.336.6083.795. [DOI] [PubMed] [Google Scholar]
- 14.Chee-Sanford JC, et al. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J Environ Qual. 2009;38(3):1086–1108. doi: 10.2134/jeq2008.0128. [DOI] [PubMed] [Google Scholar]
- 15.Qiao M, Chen W, Su J, Zhang B, Zhang C. Fate of tetracyclines in swine manure of three selected swine farms in China. J Environ Sci (China) 2012;24:1047–1052. doi: 10.1016/s1001-0742(11)60890-5. [DOI] [PubMed] [Google Scholar]
- 16.Pan X, Qiang Z, Ben W, Chen M. Residual veterinary antibiotics in swine manure from concentrated animal feeding operations in Shandong Province, China. Chemosphere. 2011;84(5):695–700. doi: 10.1016/j.chemosphere.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Wang FH, et al. The estimation of the production and amount of animal manure and its environmental effect in China. China Environ Sci. 2006;26:614–617. [Google Scholar]
- 18.Alcock RE, Sweetman A, Jones KC. Assessment of organic contaminant fate in waste water treatment plants. I: Selected compounds and physicochemical properties. Chemosphere. 1999;38(10):2247–2262. doi: 10.1016/s0045-6535(98)00444-5. [DOI] [PubMed] [Google Scholar]
- 19.Looft T, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA. 2012;109(5):1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binh CTT, Heuer H, Kaupenjohann M, Smalla K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol. 2008;66(1):25–37. doi: 10.1111/j.1574-6941.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh S, LaPara TM. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007;1(3):191–203. doi: 10.1038/ismej.2007.31. [DOI] [PubMed] [Google Scholar]
- 22.Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol. 2012;46(21):11541–11549. doi: 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 23.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14(4):176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Wu N, Qiao M, Zhang B, Cheng W-D, Zhu Y-G. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ Sci Technol. 2010;44(18):6933–6939. doi: 10.1021/es1007802. [DOI] [PubMed] [Google Scholar]
- 25.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311(5759):374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 26.Enne VI, Cassar C, Sprigings K, Woodward MJ, Bennett PM. A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol Lett. 2008;278(2):193–199. doi: 10.1111/j.1574-6968.2007.00991.x. [DOI] [PubMed] [Google Scholar]
- 27.Gräslund S, Holmström K, Wahlström A. A field survey of chemicals and biological products used in shrimp farming. Mar Pollut Bull. 2003;46(1):81–90. doi: 10.1016/s0025-326x(02)00320-x. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, et al. Potential risks of copper, zinc, and cadmium pollution due to pig manure application in a soil-rice system under intensive farming: A case study of Nanhu, China. J Environ Qual. 2011;40(6):1695–1704. doi: 10.2134/jeq2010.0316. [DOI] [PubMed] [Google Scholar]
- 29.Bolan NS, Khan MA, Donaldson J, Adriano DC, Matthew C. Distribution and bioavailability of copper in farm effluent. Sci Total Environ. 2003;309(1–3):225–236. doi: 10.1016/S0048-9697(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 30.Critically Important Antimicrobials for Human Medicine . Report of the WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance. 3rd Ed. Copenhagen: AGISAR; 2009. pp. 1–26. [Google Scholar]
- 31.Levy SB, FitzGerald GB, Macone AB. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N Engl J Med. 1976;295(11):583–588. doi: 10.1056/NEJM197609092951103. [DOI] [PubMed] [Google Scholar]
- 32.Varga C, et al. Associations between reported on-farm antimicrobial use practices and observed antimicrobial resistance in generic fecal Escherichia coli isolated from Alberta finishing swine farms. Prev Vet Med. 2009;88(3):185–192. doi: 10.1016/j.prevetmed.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol. 2009;532:397–411. doi: 10.1007/978-1-60327-853-9_23. [DOI] [PubMed] [Google Scholar]
- 34.Heuer H, Kopmann C, Binh CT, Top EM, Smalla K. Spreading antibiotic resistance through spread manure: Characteristics of a novel plasmid type with low %G+C content. Environ Microbiol. 2009;11(4):937–949. doi: 10.1111/j.1462-2920.2008.01819.x. [DOI] [PubMed] [Google Scholar]
- 35.Heuer H, et al. IncP-1ɛ plasmids are important vectors of antibiotic resistance genes in agricultural systems: Diversification driven by class 1 integron gene cassettes. Front Microbiol. 2012;3:2. doi: 10.3389/fmicb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berg J, et al. Cu exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ Sci Technol. 2010;44(22):8724–8728. doi: 10.1021/es101798r. [DOI] [PubMed] [Google Scholar]
- 37.Jackson CR, Fedorka-Cray PJ, Barrett JB, Ladely SR. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl Environ Microbiol. 2004;70(7):4205–4210. doi: 10.1128/AEM.70.7.4205-4210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol. 2011;14(3):236–243. doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Peak N, et al. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ Microbiol. 2007;9(1):143–151. doi: 10.1111/j.1462-2920.2006.01123.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang T, Zhang X-X, Ye L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE. 2011;6(10):e26041. doi: 10.1371/journal.pone.0026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heuer H, Smalla K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev. 2012;36(6):1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 42.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62(3):725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miriagou V, Carattoli A, Tzelepi E, Villa L, Tzouvelekis LS. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob Agents Chemother. 2005;49(8):3541–3543. doi: 10.1128/AAC.49.8.3541-3543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh R, et al. Identification of antimicrobial resistance and class 1 integrons in Shiga toxin-producing Escherichia coli recovered from humans and food animals. J Antimicrob Chemother. 2005;56(1):216–219. doi: 10.1093/jac/dki161. [DOI] [PubMed] [Google Scholar]
- 45.Binh CTT, Heuer H, Kaupenjohann M, Smalla K. Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res Microbiol. 2009;160(6):427–433. doi: 10.1016/j.resmic.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Petrova M, Gorlenko Z, Mindlin S. Tn5045, a novel integron-containing antibiotic and chromate resistance transposon isolated from a permafrost bacterium. Res Microbiol. 2011;162(3):337–345. doi: 10.1016/j.resmic.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Yu Z, Michel FC, Jr, Wittum T, Morrison M. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl Environ Microbiol. 2007;73(14):4407–4416. doi: 10.1128/AEM.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKinney CW, Loftin KA, Meyer MT, Davis JG, Pruden A. tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ Sci Technol. 2010;44(16):6102–6109. doi: 10.1021/es9038165. [DOI] [PubMed] [Google Scholar]
- 49.Dolliver H, Gupta S, Noll S. Antibiotic degradation during manure composting. J Environ Qual. 2008;37(3):1245–1253. doi: 10.2134/jeq2007.0399. [DOI] [PubMed] [Google Scholar]
- 50.Storteboom HN, et al. Response of antibiotics and resistance genes to high-intensity and low-intensity manure management. J Environ Qual. 2007;36(6):1695–1703. doi: 10.2134/jeq2007.0006. [DOI] [PubMed] [Google Scholar]
- 51.Aarestrup FM, et al. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob Agents Chemother. 2001;45(7):2054–2059. doi: 10.1128/AAC.45.7.2054-2059.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62(2):316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.