Abstract
Endoplasmic reticulum–associated degradation (ERAD) is a constitutive process that identifies misfolded proteins in the ER and shuttles them to the cytosol, where they can be degraded by the proteasome. The accumulation of misfolded proteins can be catastrophic at both the cellular and organismal level. Although the players involved and mechanistic details of ERAD are being characterized, much remains to be learned. Because of the complexity of the pathway, experimental studies generally require labor-intensive biochemical techniques. Here, we report the development of a system to analyze ERAD based on mutants of split or intact Venus fluorescent protein for which fluorescence depends on enzymatic deglycosylation. We have generated variants that only become fluorescent when they are first glycosylated in the ER and subsequently deglycosylated after retrotranslocation into the cytosol. The E3 ubiquitin ligase HMG-coA reductase degradation 1 homolog (Hrd1) and, consistent with the demonstrated glycosylation/deglycosylation requirement, the cytosolic deglycosylating enzyme peptide:N′glycanase are both required for fluorescence. Furthermore, although these deglycosylation-dependent fluorescent proteins are themselves ERAD substrates, they can also be fused to additional ERAD substrates to interrogate substrate-specific pathways. To validate the system we performed a genomewide siRNA screen that successfully identified known ERAD factors such as Hrd1; homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 (HERP); sel-1 suppressor of lin-12-like (SEL1L); and p97. These tools should greatly facilitate the identification of ERAD components and investigation of the mechanisms involved in this critical pathway.
It has been estimated that at least one-third of proteins expressed in cells are destined for the secretory pathway (1); and when they misfold, because of mutation, improper disulfide bond formation, or lack of coexpression of stabilizing interacting partners, they are targeted to the endoplasmic reticulum–associated degradation (ERAD) pathway. ERAD serves to translocate the misfolded proteins across the ER membrane into the cytosol where they are degraded by the proteasome (2, 3). The accumulation of misfolded proteins in the ER induces the unfolded protein response, which decreases translation, increases the levels of ERAD components, and prevents apoptosis (4). The aggregation of misfolded proteins and, by implication, problems with ERAD, is implicated in the development of many diseases, such as cystic fibrosis, Alzheimer’s, Parkinson’s, and potentially diabetes (5–7).
Terminally misfolded proteins or glycoproteins are recognized by ER chaperones and lectins, and targeted to the ERAD membrane complex (3, 8–11). This consists of chaperones, lectins, structural components, and other accessory factors, with a key component being an E3 ligase that ubiquitinates substrates. In yeast, the E3 ligases Hrd1 or Doa10 have been shown to be required for the degradation of all ERAD substrates studied to date (12). In mammalian cells the best studied are Hrd1 and gp78 (12, 13). Once ubiquitinated, substrates can be retrotranslocated across the ER membrane, potentially by the E3 ligase itself (14), by other translocon candidates, or via an unidentified channel. Most substrates are extracted from the retrotranslocation machinery into the cytosol by the AAA-ATPase p97, or vasolin containing protein (VCP), before being degraded by the proteasome (15–18).
Studies in yeast allow genetic screens to identify important ERAD components. However, screens using mammalian cells are much more difficult to perform. The fluorescent ERAD substrates generally available rely on the fusion of a fluorescent protein to a conventional ERAD substrate and they report the levels of fluorescent protein accumulating in the ER rather than the retrotranslocated cytosolic fraction. To circumvent this problem we have developed ERAD substrates that become fluorescent only when translocated to the cytosol. The system is based on glycosylation and deglycosylation of variants of either split or intact Venus protein, and ensures that essentially all of the fluorescence detected after inhibition of the proteasome derives from retrotranslocated protein. These tools should have a broad impact on the study of ERAD, and we demonstrate its utility in a high-throughput screen using mammalian cells.
Results
We initially used the split Venus system (19), where one-half of the protein was expressed in the cytosol and the other half was targeted to the ER through in-frame fusion with the H2-Kb signal sequence (Fig. 1 A and B and Fig. S1). In some constructs, a sequence corresponding to the invariant chain-derived class II-associated invariant chain peptide (CLIP) sequence was added. This generates an epitope reactive with the mAb CerCLIP.1 when the signal sequence is removed in the ER (Fig. S1). Because only half of Venus is expressed in the ER, it cannot achieve the proper folding state, and should be recognized by the ERAD machinery and translocated into the cytosol where heterodimerization of the split Venus components is driven by the fusion of both to a leucine zipper. FACS analysis demonstrated that this system does indeed lead to fluorescence, even in the absence of proteasome inhibition (Fig. S1). However, the signal sequence was incompletely cleaved from a large fraction of the ER-targeted Venus component (Fig. S1), suggesting that the majority of fluorescence was due to ER-targeted protein that aberrantly accumulated in the cytosol (supported by findings in Fig. 2). Although some experiments showed little fluorescence in the absence of proteasome inhibition (see SS-V1Z in Figs. 1D and 2 A and E), other experiments showed a population with bright fluorescence (Fig. S1), making it an unreliable ERAD reporter.
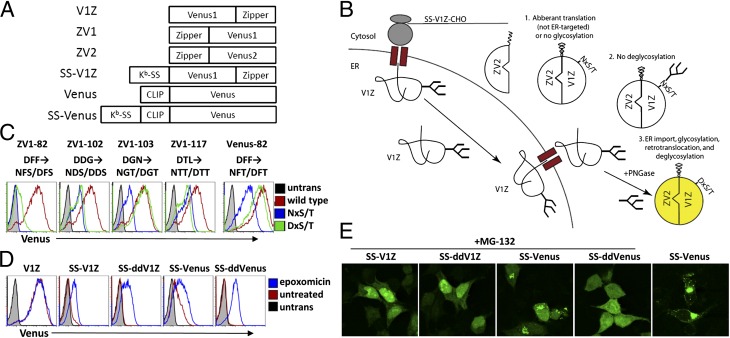
Fig. 1.
Identification of Venus mutants showing deglycosylation-dependent fluorescence. (A) Vectors used in this work. A leucine zipper (Z) was fused to either the N or C termini of the split Venus halves, with the N-terminal sequence of split Venus named Venus1 (V1) and the C-terminal sequence named Venus2 (V2). (B) Schematic depicting dd fluorescence. Fluorescence only occurs in the third scenario in which protein has reached the ER, been glycosylated, retrotranslocated, and deglycosylated in the cytosol. (C) HEK293T cells were cotransfected with wild type or the indicated mutant split Venus vector, the complementing Venus half, and MSCV-Thy1.1. Histograms show Thy1.1+ cells. (D and E) Cells were transfected with the indicated vectors (as well as ZV2 in the case of V1Z, SS-V1Z, and SS-ddV1Z) for 18–24 h, treated with 0.5 µM epoxomicin or 8 µM MG-132 for 6 h, and analyzed by FACS (D) or microscopy (E).
Fig. 2.
Fluorescence requires deglycosylation and Hrd1. (A and B) HEK293T cells were transfected with the indicated vectors as in Fig. 1, treated with 0.5 µM epoxomicin with or without 20 µM zVAD-fmk for 6 h, and analyzed by FACS. Histograms show Thy1.1+ cells. The geometric mean fluorescent intensity (GMFI) of untreated cells was subtracted from that of treated cells and these values used to determine the percent of epoxomicin-induced fluorescence reported in (B). (C and D) Cells transfected with ZV2 and SS-V1Z or SS-ddV1Z were solubilized in detergent and the lysates immunoprecipitated with the mAb 3E6 and protein A-Sepharose beads. Half of the beads were analyzed for fluorescence using a plate reader (C), and the other half were separated by SDS/PAGE, transferred to Immobolin-P, and blotted with anti-GFP-biotin (D). (E and F) Cells were transfected with the indicated vectors and DN Hrd1 or vector control. Thy1.1+ cells were analyzed by FACS after treatment for 6 h with 0.5 µM epoxomicin. Results in B and F are the mean+/−SEM of at least three independent experiments (***P < 0.001). Results in C and D are representative of at least three independent experiments. ND, not determined.
Identification of Deglycosylation-Dependent Venus Mutants.
To ensure that all of the fluorescence seen resulted from protein that had reached the ER and been retrotranslocated to the cytosol, we made use of the fact that removal of a glycan by the cytosolic enzyme Peptide:N′glycanase (PNGase) results in the deamidation of the glycosylated asparagine residue, converting it to an aspartic acid residue (Fig. 1B; refs. 20–22). First we screened for cytosolically coexpressed split Venus mutants, each equipped with a leucine zipper to induce dimerization, which showed reduced fluorescence when asparagine residues were substituted for aspartic acid residues. This was achieved by separately mutating every aspartic acid—or in two cases a glutamic acid—to an asparagine. In addition, we mutated the amino acid two residues downstream to a serine or threonine to create a glycosylation motif. Screening of 20 such mutants, expressed without a signal sequence in combination with the complementing Venus fragment in the cytosol, revealed 9 that had a drastic reduction in fluorescence (Fig. 1C and Fig. S2). To eliminate the possibility that the decrease in fluorescence was due to the introduction of serine or threonine, mutants were made that contain the wild-type aspartic acid residue and the corresponding serine/threonine mutation (Fig. 1C). Although some mutants showed partial fluorescence recovery, only that with aspartic acid at position 103 recovered to wild-type levels when converted to the potentially “deglycosylated” form; i.e., when position 105 was converted to a threonine. Overall, there was a 36.6+/−12.4-fold decrease in fluorescence between a construct with the leucine zipper N-terminal to the N-terminal half (ZV1) containing asparagine at position 103 versus that containing aspartic acid. When the same glycosylation mutation was introduced into single chain Venus, there was no effect on fluorescence. However, introduction of a glycosylation mutation at position 82 of single chain Venus led to an 8.7+/−2.5-fold difference between the construct containing an asparagine and that containing an aspartic acid (Fig. 1C). The reduced fluorescence was not due to decreased expression, as only minor differences were seen in protein levels (Fig. S2). These mutations (DGN→NGT at position 103–105 of ZV1 or V1Z and DFF→NFT at position 82–84 of Venus) will be referred to as deglycosylation-dependent, or dd mutants.
The dd mutations were then introduced into ER-targeted, signal sequence–containing, SS-ZV1, SS-V1Z, and SS-Venus vectors and transiently expressed in 293T cells, along with cytosolic ZV2 in the first two cases. Although there was slight basal fluorescence for the SS-ddZV1 vector, there was none in the SS-ddV1Z vector in the absence of proteasome inhibition (Fig. 1D). Addition of epoxomicin revealed fluorescence of SS-ddV1Z, with slightly less effect on SS-V1Z and little to no effect on the fluorescence of cytosolically expressed V1Z (Fig. 1D). Even though the ddVenus mutant showed fairly high fluorescence when expressed in the cytosol (Fig. 1C), very little was seen in the absence of proteasome inhibition when it was targeted to the ER (Fig. 1D). Treatment with epoxomicin-enhanced fluorescence of both SS-Venus and SS-ddVenus suggested that both wild-type and ddVenus are ERAD substrates. When analyzed by confocal microscopy, SS-ddV1Z fluorescence was mainly localized to the nucleus but also showed diffuse cytosolic labeling, and SS-ddVenus fluorescence was diffuse in the cytosol and nucleus (Fig. 1E). In untreated cells, SS-Venus was mainly localized to the Golgi apparatus, assessed by costaining with giantin, but was also present in punctate structures that may be secretory vesicles (Fig. 1E). Upon addition of epoxomicin, diffuse cytosolic and nuclear fluorescence were detectable along with the Golgi/vesicular labeling, suggesting a subpopulation of the protein was subject to ERAD. However, SS-ddV1Z and SS-ddVenus exhibited no vesicular staining, consistent with their being exclusively ERAD substrates.
dd Mutants Require Deglycosylation for Fluorescence.
Fluorescence should depend on glycosylation in the ER and deglycosylation in the cytosol. The requirement for deglycosylation was examined using the PNGase inhibitor zVAD-fmk (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone) (23). Similar to the data in Fig. 1D, addition of epoxomicin led to enhanced fluorescence in SS-V1Z–, SS-ddV1Z–, SS-Venus–, and SS-ddVenus–expressing cells (Fig. 2A). When cells were treated with zVAD-fmk plus epoxomicin, fluorescence of the dd mutants was decreased by at least 80%, and there was slight enhancement of fluorescence of the wild-type constructs by zVAD-fmk (Fig. 2 A and B). Furthermore, knockdown of PNGase using siRNA also led to a decrease in fluorescence of SS-ddV1Z. These data suggest that both glycosylation and deglycosylation, and, therefore, ER entry and exit, are required for fluorescence.
To provide a direct biochemical link between the glycosylation state of SS-ddV1Z and its fluorescence, cells coexpressing ZV2 and either SS-V1Z or SS-ddV1Z were treated with epoxomicin, zVAD-fmk, or a combination of both. Cells were lysed, and extracts immunoprecipitated (IP) with an anti-GFP antibody (3E6) that recognizes the N-terminal half of split Venus (Fig. S1). All of the fluorescence was associated with the IP material (Fig. 2C). When analyzed by Western blot, the epoxomicin-treated cells showed an enrichment of nonglycosylated SS-ddV1Z, and this population was reduced after cotreatment with zVAD-fmk (Fig. 2D). In contrast, the majority of SS-ddV1Z present in cell lysates was glycosylated. The 3E6 antibody only weakly reacts with glycosylated SS-ddV1Z for reasons that are not apparent as the antibody reacts with intact Venus containing the same glycosylation site (at position 103). Nevertheless, as all of the fluorescent material was associated with the IP beads, these results demonstrate that the fluorescent population after epoxomicin treatment is deglycosylated and indicate that both glycosylation and deglycosylation are required for fluorescence.
Hrd1 Is Required for SS-ddV1Z and SS-ddVenus Fluorescence.
The E3 ubiquitin ligase Hrd1 is required for the ubiquitination and degradation of many ERAD substrates (8, 24–28). To determine whether this was true for the dd mutants, cells were cotransfected with a dominant negative (DN) Hrd1 construct. Expression of DN Hrd1 inhibited epoxomicin-induced SS-ddV1Z and SS-ddVenus fluorescence by 86.2% and 72.3%, respectively (Fig. 2 E and F). DN Hrd1 slightly inhibited SS-V1Z fluorescence but did not affect V1Z fluorescence, demonstrating that the inhibition is specific for the retrotranslocated protein. Although a different E3 ligase could conceivably be required for retrotranslocation of SS-V1Z, it is likely that the limited effect is because very little fluorescence arises from protein that has reached the ER and been retrotranslocated, consistent with the data in Fig. S1. It is interesting to note that although DN Hrd1 slightly enhanced the fluorescence of SS-Venus, the epoxomicin-induced portion was eliminated by DN Hrd1 (Fig. 2E), consistent with the hypothesis that a population of wt Venus is constantly undergoing ERAD.
SS-ddVenus as a Tag for ERAD Substrates.
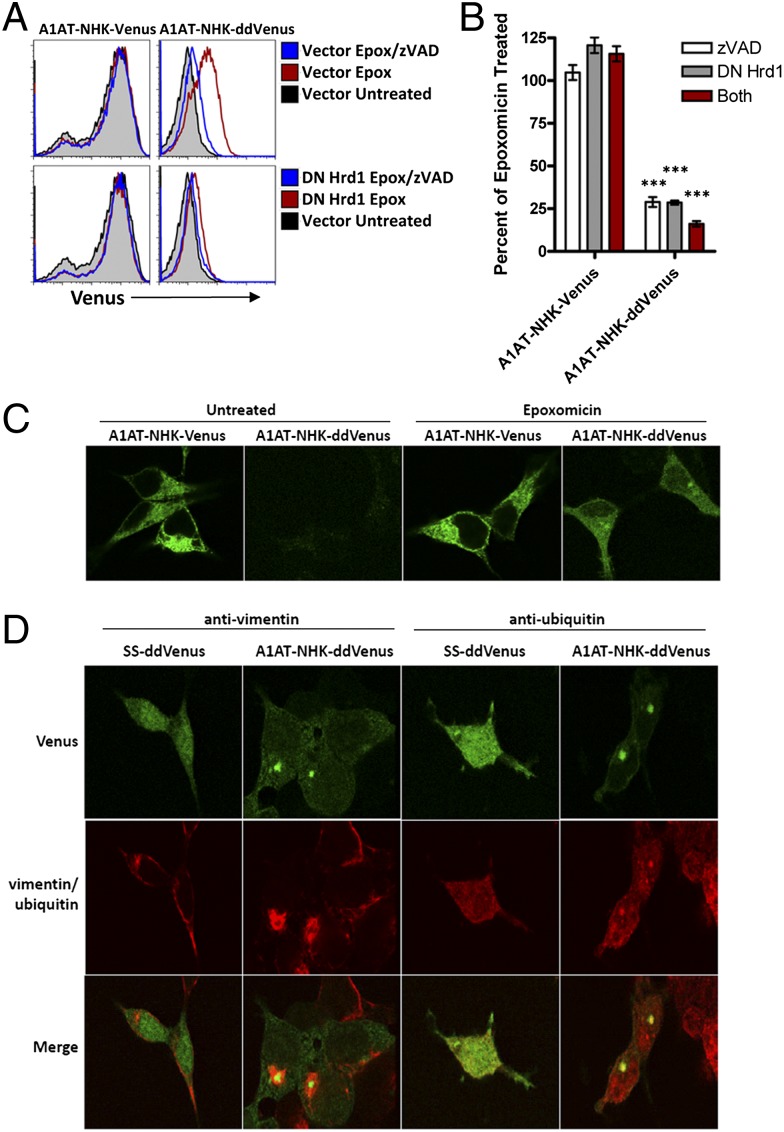
To determine whether the dd mutants could be useful in analyzing an established ERAD substrate, we fused wild-type Venus and ddVenus C-terminal to the unstable null Hong Kong variant of alpha-1–antitrypsin (A1AT-NHK; ref. 29). Single chain Venus was chosen instead of split Venus to obviate the need to cotransfect ZV2 as well as avoid the possibility that fusion with A1AT-NHK might affect split Venus dimerization. After treatment with epoxomicin, cells expressing A1AT-NHK-Venus showed a slight increase in fluorescence as assessed by FACS (Fig. 3A). A cytosolic population of A1AT-NHK-Venus was not distinguishable by microscopy after epoxomicin treatment, likely due to the presence of high levels of ER-localized A1AT-NHK-Venus (Fig. 3C) as assessed by costaining with calnexin. Cells expressing A1AT-NHK-ddVenus were completely nonfluorescent without the addition of epoxomicin, showing a four-to-sixfold increase in fluorescence with proteasome inhibition. This fluorescence was inhibited by over 70% when cells were treated with zVAD-fmk or transfected with DN Hrd1 (Fig. 3 A and B). Furthermore, when DN Hrd1 transfected cells were treated with epoxomicin and zVAD-fmk, fluorescence was decreased by more than 85%, demonstrating that essentially all of the fluorescence resulted from retrotranslocated protein. When analyzed by microscopy, a diffuse cytosolic pattern was seen, with a majority of cells also containing some punctate ddVenus fluorescence (Fig. 3C). The punctate structures were identified as aggresomes (30): many, but not all, were encapsulated by the intermediate filament protein vimentin and they colocalized with ubiquitin (Fig. 3D). In untransfected cells and cells expressing SS-ddVenus, the majority of vimentin was localized at the cell periphery, indicating that relocalization to aggresomes was associated with A1AT-NHK-ddVenus expression (Fig. 3D). Thus, ddVenus can be used to investigate the properties of other substrates, although additional properties could potentially arise in the fusion protein that are not evident in ddVenus alone nor in the linked substrate.
Fig. 3.
Deglycosylation-dependent Venus can be fused to an ERAD substrate. (A and B) HEK293T cells were transfected with the indicated vectors and Thy1.1+ cells were analyzed by FACS after treatment with 0.5 µM epoxomicin for 6 h. Results in B are the mean+/−SEM of at least three independent experiments (***P < 0.001 compared with A1AT-NHK-Venus controls). (C) Cells transfected with the indicated vectors were left untreated or treated with 0.5 µM epoxomicin and analyzed by microscopy. (D) Cells transfected with the indicated vectors were treated with 0.5 µM epoxomicin for 6 h and stained with antibodies to vimentin or ubiquitin (red).
Genomewide Screen Identifies Known ERAD Factors.
For screening purposes, we chose the split Venus system because it gives that best signal-to-noise ratio, and generated a stable cell line named 293T.FluERAD (for fluorescent ERAD) that coexpresses ZV2 and SS-ddV1Z. In the absence of proteasome inhibition, 293T.FluERAD cells were completely nonfluorescent (Fig. 4 A and C), demonstrating that all of the SS-ddV1Z is degraded at steady state. After addition of epoxomicin for 6 h, 293T.FluERAD cells showed a homogeneous peak of fluorescence (Fig. 4A). Furthermore, transfection of 293T.FluERAD cells with siRNAs targeting ERAD components demonstrated that: (i) PNGase and Hrd1, but not the E3 ubiquitin ligase gp78 were required for fluorescence, and (ii) these differences could be quantitated using an automated high-throughput microscope (Fig. 4 B and C). Similar to transiently transfected cells, the majority of fluorescence in 293T.FluERAD cells was localized to the nucleus with diffuse cytosolic labeling in some cells. Therefore, the nucleus was marked with Hoescht dye and nuclear intensity was used as a readout in a high-throughput siRNA screen.
Fig. 4.
High-throughput microscopic analysis of SS-ddV1Z fluorescence. (A) The 293T.FluERAD cells, stably expressing ZV2 and SS-ddV1Z, were transfected with the indicated siRNA for 66–72 h, treated with 0.5 µM epoxomicin for 6 h, and analyzed by FACS. (B and C) Stable cells were transfected with siRNA in 384-well plates, treated with epoxomicin and analyzed by microscopy using an Opera high-throughput microscope. The nucleus was marked with Hoescht dye and nuclear intensities determined. Nuclear intensities were normalized to cells transfected with RISC-free siRNA, a transfection control which does not get incorporated into the RISC. Results in B are the mean+/−SEM of at least three independent experiments (**P < 0.01, ***P < 0.001).
293T.FluERAD cells were reverse transfected in 384-well plates containing four gene-specific siRNAs per well. After 66–72 h of transfection, cells were treated with the proteasome inhibitor MG-132 for 6 h, fixed, and analyzed by high-throughput microscopy. The screen consisted of 58 plates accounting for 18,119 genes in total, and each plate was tested in triplicate. Using nuclear intensity as a read-out, robust Z (RZ) scores were determined as previously described using median intensities (31). The resulting RZ scores demonstrated that the assay was highly reproducible, yielding correlation coefficients of ∼0.9 for all subsets of plates run except one, which consisted of a single plate (Fig. S3). The screenwide Z′ (31), a composite measure of signal to noise and variation within plates, was 0.52 (with the max score being 1) demonstrating that the assay was robust. Based on the results of the screen, we set a threshold RZ score of −2 for genes that inhibited SS-ddV1Z fluorescence, which is at least two median absolute deviations from the median. Even though there was very little correlation between the cell numbers analyzed per well and the nuclear fluorescence (Fig. S3), any hit that had less than 15% of cells compared with the negative control siRNA was discounted.
Although the results of the entire screen are outside the scope of this paper, the results pertaining to known or suspected ERAD components are presented in Table 1. Similar to the data presented in Figs. 2 and 4, Hrd1 and PNGase were required for SS-ddV1Z fluorescence, being the 1st and 117th ranked hits in the screen, respectively. Also similar to the data presented in Fig. 4, gp78 siRNA had little effect. Other factors that have been previously described to play a role in ERAD were above the hit threshold, including SEL1L, HERP, FAS-associated factor 2/UBX domain containing 8 (FAF2)/UbxD8, and the p97 complex consisting of p97, ubiquitin fusion degradation 1 (UFD1), and nuclear protein localization 4 (NPL4). It was surprising that although there was a clear requirement for the p97 complex, knockdown of VCP-interacting membrane protein (VIMP), which can link p97 to the ERAD membrane complex (11, 32), actually enhanced Venus fluorescence. It is also interesting that the knockdown of sec61-γ and sec61-α1 inhibited fluorescence, but knockdown of sec61-α2 and sec61-β slightly enhanced fluorescence, suggesting that the Sec61 translocon is not absolutely required for retrotranslocation of this substrate. Knockdown of other genes linked to ERAD, such as: Derlin-1; Derlin-2; ER mannosidase I; Golgi mannosidase I; AUP1; Erdj3; Erdj5; ER degradation enhancer, mannosidase alpha-like (EDEM)2; and EDEM3 all showed some inhibition of fluorescence. Knockdown of EDEM1, Bip, and grp94 all enhanced fluorescence. Finally, two ER localized lectins, XTP3-B and osteosarcoma amplified 9 (OS-9), which are required for the recognition of many glycosylated ERAD substrates (8, 24, 33), only showed a marginal effect on fluorescence inhibition. This may be due to redundancy between the lectins.
Table 1.
Genomewide screen results for known or potential ERAD factors
| ERAD component | % cells* | Average RZ score | Average RZ score rank‡ |
| Hrd1 | 18.69 | −3.84 | 1 |
| HERP | 63.01 | −2.58 | 41 |
| SEL1L | 63.62 | −2.50 | 61 |
| UFD1 | 87.46 | −2.41 | 91 |
| PNGase | 60.45 | −2.36 | 117 |
| p97 | 33.68 | −2.31 | 143 |
| Sec61-γ | 51.48 | −2.21 | 215 |
| FAF2/UbxD8 | 113.39 | −2.08 | 336 |
| NPL4 | 56.73 | −2.04 | 409 |
| AUP1 | 60.13 | −1.74 | 1,012 |
| Sec61-α1 | 19.96 | −1.64 | 1,320 |
| Golgi mannosidase I | 39.55 | −1.53 | 1,686 |
| EDEM2 | 54.67 | −1.33 | 2,402 |
| Derlin-2 | 26.50 | −1.12 | 3,341 |
| HR23B | 30.47 | −1.07 | 3,582 |
| Erdj3 | 22.60 | −1.07 | 3,612 |
| EDEM3 | 55.67 | −0.88 | 4,507 |
| Erdj5 | 22.21 | −0.86 | 4,626 |
| ER mannosidase I | 15.60 | −0.84 | 4,741 |
| XTP3-B | 35.54 | −0.78 | 5,004 |
| OS-9 | 96.48 | −0.51 | 6,388 |
| Derlin-1 | 47.20 | −0.35 | 7,168 |
| VIMP | 68.70 | 2.48 | 1,627 |
| Hsp90 | 83.70 | 1.86 | 2,575 |
| Derlin-3 | 46.83 | 1.71 | 2,899 |
| EDEM1 | 63.33 | 1.70 | 2,909 |
| Gp78 | 78.26 | 1.23 | 4,156 |
| Sec61α2 | 32.26 | 0.91 | 5,237 |
| Bip/Grp78 | 52.20 | 0.75 | 5,868 |
| Grp94 | 34.03 | 0.58 | 6,561 |
| Sec61-β | 41.19 | 0.53 | 6,773 |
*Percentage of cells analyzed in knockdown wells versus the negative control siRNA wells.
‡A ranked list was sorted on inhibitors or enhancers.
To determine whether inhibition was due to off-target siRNA effects, ERAD components were knocked down using individual siRNAs rather than siRNA pools. siRNAs were scored positive if they inhibited fluorescence in 293T.FluERAD cells at least 50% as well as the positive control PNGase siRNA pool. Genes in which knockdown by only one siRNA decreased fluorescence were ruled off-target and those for which two or more decreased fluorescence were considered specific. Results from this analysis are presented in Fig. S3, and demonstrate the specificity of Hrd1, HERP, SEL1L, PNGase, UFD1, p97, sec61-γ, FAF2, NPL4, and AUP1 on SS-ddV1Z fluorescence. Similar to the results from the screen, none of the siRNAs against Derlin-1, gp78, VIMP, or EDEM1 inhibited fluorescence by more than 50%, suggesting that these proteins are not strictly required for SS-ddV1Z ERAD. However, all of the known ERAD components identified above the hit threshold were successfully validated, verifying the robustness of the screening system.
Discussion
The analytical approach to ERAD we developed depends on the substrate proteins undergoing two critical steps: ER entry resulting in glycosylation and retrotranslocation to the cytosol resulting in deglycosylation. We confirmed that both are necessary and demonstrated in multiple ways that Hrd1 is required for the acquisition of substrate fluorescence, suggesting that this E3 ligase is a key mediator of degradation. Both the split Venus and single chain Venus derivatives containing the described glycosylation motifs are genuine fluorescent ERAD substrates.
Our initial experiments used the split Venus system without the glycosylation motif, and we found that the majority of fluorescence arose because the ER-targeted component was aberrantly expressed in the cytosol. Overexpression may have exaggerated the problem because transfection of reduced amounts of DNA increased the fraction of protein with a properly cleaved signal sequence, although some fluorescence was still derived from uncleaved protein. While this manuscript was in preparation, a fluorescent ERAD system based on split GFP was reported (34). Here, one-half of GFP was fused to the ERAD substrates A1AT-NHK or CD3δ and the other half of GFP was expressed in the cytosol. These workers did not encounter the same problem with signal-sequence cleavage and, in fact, verified a requirement for Hrd1, SEL1L, and p97, confirming that fluorescence was due to ERAD. However, all of their experiments used cells stably expressing both halves of GFP, and stably expressing cells generally express less protein than transiently transfected cells. The deglycosylation-dependent system we describe works well with transient expression. Furthermore, fusion of split GFP to other ERAD substrates or expression in different vectors requires reverification of ER targeting in those settings. A major benefit of a deglycosylation-dependent substrate is that one can easily assess the proportion of fluorescence coming from deglycosylation by the addition of the PNGase inhibitor zVAD-fmk, eliminating the requirement for biochemical approaches to assess the efficiency on signal-sequence cleavage.
We verified the usefulness of split ddVenus by performing a genomewide siRNA screen. Two factors that we had already identified, namely Hrd1 and PNGase, were successfully confirmed in the screen, being the 1st and 117th ranked hits, respectively. Furthermore, the screen identified several other key mediators of ERAD with RZ scores above the hit threshold, including SEL1L, HERP, p97, NPL4, FAF2, and UFD1. Knockdown of other known or suspected ERAD components had variable effects in the screen. Of note, knockdown of different sec61 subunits had differing effects. Knockdown of sec61-β and sec61-α2 both led to a minor enhancement of fluorescent signal, but knockdown of sec61-α1 and -γ subunits decreased fluorescence. Although the sec61-γ and -α1 knockdown data might suggest a role for this translocon in retrotranslocation, fluorescence inhibition could also be due to decreased translocation into the ER as well as decreased glycosylation, as sec61 forms a ternary complex with oligosaccharyl transferase and ribosomes (35) to allow cotranslational glycosylation. Together, the sec61 data suggests that the sec61 translocon is not absolutely required for retrotranslocation of split ddVenus. Other factors, such as Derlin-1, EDEM1, EDEM2, EDEM3, ER mannosidase I, erdj3, erdj5, XTP3-B, and OS-9 had lower-than-expected scores in the screen, exhibiting only mild to moderate inhibition under siRNA knockdown. In a low-throughput assay these effects might be considered significant, but a high threshold must be set in high-throughput screens to define critical genes, a caveat for high-throughput screens in general. Overall, however, there is no doubt that the screen successfully identified many ERAD components.
The split ddVenus system may require factors not involved in ERAD to generate a fluorescent signal, e.g., chaperones that facilitate heterodimerization and folding. We therefore examined single chain Venus as a potential ERAD substrate. It was surprising to note that proteasome inhibition led to a two-to-threefold enhancement of wild-type ER-targeted Venus fluorescence, suggesting that a substantial portion of Venus is constantly undergoing ERAD. This was verified by the fact that epoxomicin-induced fluorescence was cytosolic and was inhibited by DN Hrd1. Although we cannot exclude the idea that the presence of the N-terminal CLIP sequence makes SS-Venus an ERAD substrate, the more likely explanation is aberrant disulfide bond formation. GFP and Venus contain two cysteine residues that when expressed in the ER can form interchain disulfide-linked aggregates accounting for up to 50% of the total protein (36, 37). We made glycosylation mutations involving the aspartic acid residues at positions 82 and 103 in Venus in attempts to generate a dd variant. Mutation at position 82 showed an 8.7-fold decrease in fluorescence that was restored to near wild-type levels when cytosolically expressed with an aspartic-acid residue rather than an asparagine residue, equivalent to the deglycosylated form. However, in contrast to the result with split Venus, the 103 mutation did not affect intact Venus fluorescence. Given that it is tolerated in single chain Venus, it may destabilize ZV1 or V1Z, not allowing proper tertiary structure when dimerized with ZV2. On the other hand, the aspartic acid at position 82 of GFP has been hypothesized to be part of the proton delivery pathway to the internal chromophore (38). Mutation of the negatively charged aspartic-acid residue to an uncharged asparagine may perturb proton delivery and subsequent fluorescence. However, the position 82 mutation clearly affects protein folding, causing a large reduction in secretion as well as increased interchain disulfide-linked aggregates. Whether these effects are due to the aspartic acid to asparagine mutation at 82 or phenylalanine to threonine mutation at 84 remains undetermined.
SS-ddVenus can be used as a tag to report the behavior of other ERAD substrates. When fused to A1AT-NHK the results observed upon proteasome inhibition were similar to those seen with SS-ddVenus alone, with two exceptions. First, aggresomes formed in the A1AT-NHK-ddVenus–transfected cells, and second, A1AT-NHK-ddVenus was mostly excluded from the nucleus while SS-ddVenus showed similar fluorescence in the nucleus and cytosol. Thus, ddVenus can be fused to an ERAD substrate in which the fluorescent protein component will enter the ER, and can potentially be used to report substrate-specific phenotypes that are independent of the characteristics of ddVenus itself. However, care must be ensure that observed phenotypes are consistent with the properties of the original, untagged substrate.
In summary, we have developed and characterized tools for the study of ERAD. Split or single chain Venus containing glycosylation mutations are ERAD substrates themselves and can be used for screening and potentially live cell imaging applications. Moreover, the substrates can be fused to traditional ERAD substrates to study substrate specific mechanisms of recognition, targeting, and retrotranslocation. In addition they may be valuable for the study of cytosolic factors that contribute to ERAD after retrotranslocation.
Materials and Methods
Details of experimental procedures, reagents, and constructs can be found in SI Materials and Methods.
Genome-wide siRNA Screen.
Stable 293T.FluERAD cells (1500/well) were added to wells of 384 well plates containing a final concentration of 20-nmol siRNA pools [5 nmol each siRNA (siGENOME, Dharmacon)] and Lipofectamine RNAiMAX (Invitrogen). After 66–72 h of transfection, MG-132 (8 μM) was added for 6 h. Cells were fixed and analyzed using a Perkin-Elmer Opera High Content Screening System confocal microscope.
Genome-wide siRNA screen data analysis.
Three fields were analyzed in each well. Calculations used to determine Z' and robust Z (RZ) scores were done according to ref. 31. For Z' determination, 16 replicate wells per plate containing positive control siRNA (PNGase) and the negative control siRNA (RNA-induced silencing complex [RISC]-free) were used. After normalization on a per plate basis, median values were used to determine the median absolute deviation of samples, with the hit threshold set at −2 for inhibitors, which is two median absolute deviations from the median intensity. The RZ scores from triplicate experiments were averaged and sorted by rank.
Supplementary Material
Acknowledgments
We thank Michael Wyler, Leena Kuruvilla, Ashima Bahn, and Marie-Aude Guié and the Yale Center for Molecular Discovery for their assistance with the genomewide siRNA screen, Susan Mitchell and James Cresswell for their technical assistance, and Nancy Dometios for help in manuscript preparation. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health Grant R01-AI097206, and a Cancer Research Institute postdoctoral fellowship (to J.E.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300328110/-/DCSupplemental.
References
- 1.Kanapin A, et al. RIKEN GER Group GSL Members Mouse proteome analysis. Genome Res. 2003;13(6B):1335–1344. doi: 10.1101/gr.978703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith MH, Ploegh HL, Weissman JS. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9(12):944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti A, Chen AW, Varner JD. A review of the mammalian unfolded protein response. Biotechnol Bioeng. 2011;108(12):2777–2793. doi: 10.1002/bit.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10(5):507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- 6.Schnell S. A model of the unfolded protein response: Pancreatic beta-cell as a case study. Cell Physiol Biochem. 2009;23(4-6):233–244. doi: 10.1159/000218170. [DOI] [PubMed] [Google Scholar]
- 7.Stefani M. Protein misfolding and aggregation: New examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta. 2004;1739(1):5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10(3):272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosokawa N, et al. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283(30):20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429(6994):834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 11.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429(6994):841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 12.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18(6):770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehnert M, Sommer T, Jarosch E. ERAD ubiquitin ligases: Multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays. 2010;32(10):905–913. doi: 10.1002/bies.201000046. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143(4):579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21(4):615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarosch E, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4(2):134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 17.Rabinovich E, Kerem A, Fröhlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22(2):626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414(6864):652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 19.Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch C, Blom D, Ploegh HL. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 2003;22(5):1036–1046. doi: 10.1093/emboj/cdg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. Identification of peptide:N-glycanase activity in mammalian-derived cultured cells. Biochem Biophys Res Commun. 1993;194(3):1124–1130. doi: 10.1006/bbrc.1993.1938. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi N. Demonstration of a new amidase acting on glycopeptides. Biochem Biophys Res Commun. 1977;76(4):1194–1201. doi: 10.1016/0006-291x(77)90982-2. [DOI] [PubMed] [Google Scholar]
- 23.Misaghi S, Pacold ME, Blom D, Ploegh HL, Korbel GA. Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem Biol. 2004;11(12):1677–1687. doi: 10.1016/j.chembiol.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188(2):223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burr ML, et al. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2011;108(5):2034–2039. doi: 10.1073/pnas.1016229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cattaneo M, et al. SEL1L and HRD1 are involved in the degradation of unassembled secretory Ig-mu chains. J Cell Physiol. 2008;215(3):794–802. doi: 10.1002/jcp.21364. [DOI] [PubMed] [Google Scholar]
- 27.Kikkert M, et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279(5):3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 28.Omura T, et al. A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J Neurochem. 2006;99(6):1456–1469. doi: 10.1111/j.1471-4159.2006.04155.x. [DOI] [PubMed] [Google Scholar]
- 29.Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263(15):7330–7335. [PubMed] [Google Scholar]
- 30.Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Y, et al. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102(40):14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284(25):17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong Y, Fang S. Live cell imaging of protein dislocation from the endoplasmic reticulum. J Biol Chem. 2012;287(33):28057–28066. doi: 10.1074/jbc.M112.381798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harada Y, Li H, Li H, Lennarz WJ. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc Natl Acad Sci USA. 2009;106(17):6945–6949. doi: 10.1073/pnas.0812489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aronson DE, Costantini LM, Snapp EL. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic. 2011;12(5):543–548. doi: 10.1111/j.1600-0854.2011.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain RK, Joyce PB, Molinete M, Halban PA, Gorr SU. Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells. Biochem J. 2001;360(Pt 3):645–649. doi: 10.1042/0264-6021:3600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agmon N. Proton pathways in green fluorescence protein. Biophys J. 2005;88(4):2452–2461. doi: 10.1529/biophysj.104.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.