The circadian clock is a time-keeping mechanism that allows organisms to organize their behavior and physiology around the 24-h day. In mammals, the generation of molecular rhythms in cells relies on interconnected transcriptional-translational feedback loops, which integrate posttranscriptional and posttranslational mechanisms within their “cogs” (1–3). Global analysis of these molecular oscillations have shown that approximately 10% of transcripts in a given tissue are under circadian regulation, whereas at the protein level the fraction has been estimated at around 20% in mouse liver (4). In PNAS, Masri et al. (5) reveal that the hepatic proteome exhibits circadian rhythms in reversible lysine acetylation, especially directed toward mitochondrial proteins.
Acetylation was initially recognized as a reversible post-translational modification in the 1960s and its function has mostly been discussed in the context of gene regulation, because histone tail acetylation is part of the “histone code,” the dynamic changes of which are believed to modulate gene expression epigenetically. Acetylation is also known to regulate the function of nonhistone proteins, including transcription factors and molecular chaperones (6). However, investigations aimed at assaying reversible acetylation of lysine residues in proteins in general have gained momentum, thanks to global proteomics studies suggesting that such acetylation is a key posttranslational modification, perhaps as prevalent as phosphorylation (7). Moreover, acetylation has emerged as an important regulator of metabolic function because the vast majority of enzymes involved in gluconeogenesis, fatty acid oxidation, and the tricarboxylic acid cycle can be acetylated in human liver tissues (8). Notably, the sirtuin SIRT3, a NAD+-dependent protein deacetylase located in mitochondria, has been identified as a regulator of fatty acid oxidation and an essential component of mitochondrial function (9).
In the context of circadian time-keeping, rhythms in the acetylation of histones were first reported at few specific clock-controlled loci activated by the transcriptional complex involving the BMAL1/CLOCK circadian transcription factors, and subsequently several reports highlighted genome-wide oscillations in histone acetylation across day and night (10–12). CLOCK itself has been reported to be a histone acetyltransferase (13), which not only catalyses the acetylation of histones, but also of its heterodimeric partner, BMAL1 (14). CLOCK acetyltransferase activity is thought to be counterbalanced by the NAD+-dependent deacetylase SIRT1, forming an autoregulatory loop contributing to energy homeostasis, because BMAL1/CLOCK modulate the levels of NAD+ in the cell through the transactivation of the rate-limiting enzyme in NAD+ biosynthesis (15). Moreover, SIRT1 promotes the deacetylation of the clock protein PERIOD2, and its subsequent degradation, reinforcing the likely role of reversible protein acetylation in the circadian clockwork.
Masri et al. (5) set out to measure the circadian acetylome (the set of acetylated proteins) in mouse liver using mass spectrometry-based proteomics. To this end, they collected liver samples at four time points during a 24-h cycle from wild-type and Clock-deficient mice kept in light-entrained conditions. Using label-free mass spectrometry, they measured the acetylation profiles of 179 proteins, the majority of them being acetylated at a single lysine residue. About 5% of the 306 acetylation sites were rhythmic in either the wild-type or Clock-deficient animals (19 and 15 sites, respectively), with two sites being shared between the two groups. Pathway analysis highlighted that the 30 proteins differentially acetylated in wild-type and mutant backgrounds were enriched for proteins participating in the tricarboxylic cycle and amino acid metabolism. The results thus substantiate the role of acetylation in the regulation of cellular metabolism, in particular mitochondrial function, and suggest that the liver clock drives rhythmic acetylation of several cytosolic and mitochondrial enzymes. However, the fact that a similar number of rhythmic acetylation sites were detected in the two genetic backgrounds indicates that systemic cues, such as food- and activity-related cues, might contribute to the daily acetylation patterns, given that Clock-deficient mice still exhibit robust circadian locomotor activity (16).
To gain further insight into the functionality of rhythmic acetylation, Masri et al. (5) integrated their acetylome data with published mouse liver transcriptome datasets (17) and with metabolomics data (18) that they generated from the same animals used in this study. Moreover, Masri et al. were also able to measure total protein abundance from their acetylome data, because unmodified peptides also copurify, albeit at low levels, during the enrichment of acetylated peptides. Whereas the distribution of metabolite/acetylation site pairs with high absolute correlation (that is, an absolute value of the Pearson correlation coefficient ≥ 0.9) displayed a similar number of highly correlated and anticorrelated pairs, both transcript/acetylation and protein/acetylation pairs had predominantly a positive correlation. Although there were examples, such as betaine-homocysteine methyltransferase, that displayed an antiphasic relationship between published protein accumulation (4) and acetylation profiles, this may be a result of acetylation-mediated degradation. Furthermore, a number of proteins were acetylated in a circadian fashion, yet their abundance remained approximately constant over time.
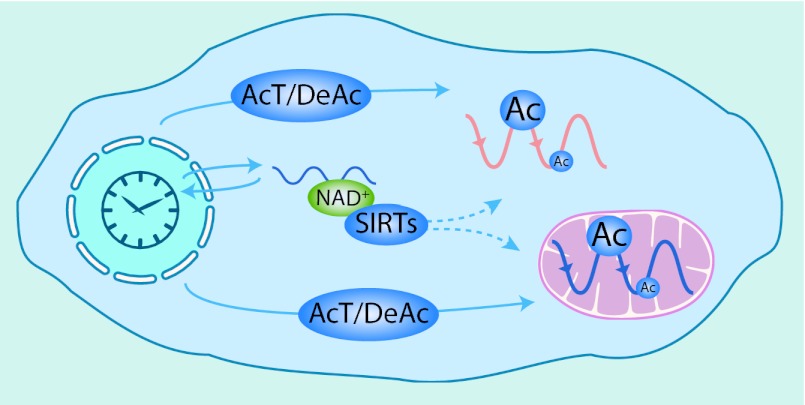
Taking these data together, Masri et al. (5) suggest an unanticipated role of the circadian clock in modulating mitochondrial function through reversible acetylation (Fig. 1). These results support the growing importance of posttranslational and nontranscriptional mechanisms for circadian time-keeping (4, 19, 20). The integration of the acetylome data with other datasets suggests that acetylation might have various functions in circadian physiology, including rhythmic activation of constitutively expressed proteins and fine-tuning protein function. Such mechanisms may act to reshape metabolic networks to fulfill dynamic demands on a daily timescale, without paying the cost of expressing genes, thus licensing certain biosynthetic pathways at specific times of the day. This is indeed a key function of circadian clocks: temporal sequestration of cellular tasks that are immiscible. Mechanistically, CLOCK’s histone acetyltransferase activity might contribute to the generation of acetylation rhythms. However, it is very likely that other acetylases/deacetylases play an important role, because a significant number of acetylation sites are still rhythmic in Clock-deficient animals. For example, the mitochondrial deacetylase SIRT3 might contribute to rhythmic acetylation because its cofactor NAD+ undergoes circadian oscillations in the cytosol (15), and perhaps also in the mitochondria of liver cells. Further studies will surely help to identify the coupling between transcription/translation loops within the clockwork and acetylome oscillations, and thus shed light on how circadian acetylation modulates cellular metabolism.
Fig. 1.
Circadian acetylation in the cell. Scheme representing plausible mechanisms that the circadian clock might use to generate cytosolic and mitochondrial acetylation rhythms. The activity of acetyltransferases (AcT), including CLOCK, together with deacetylases (DeAc), might be controlled through transcriptional and posttranscriptional mechanisms. In addition, the circadian clock may also use the NAD+-dependant deacetylases SIRTs to modulate protein acetylation, because NAD+ levels are rhythmic in the cytosol.
Footnotes
The authors declare no conflict of interest.
See companion article on page 3339.
References
- 1.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(Pt 3):311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34(10):483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 5.Masri S, et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci USA. 2013;110:3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 8.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschey MD, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44(2):177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47(2):158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16(6):833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 15.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50(3):465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 17.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rey G, Reddy AB. Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.01.003. 10.1016/j.tcb.2013.01.003. [DOI] [PubMed] [Google Scholar]