Abstract
Syncytins are envelope genes of retroviral origin that have been co-opted for a role in placentation and likely contribute to the remarkable diversity of placental structures. Independent capture events have been identified in primates, rodents, lagomorphs, and carnivores, where they are involved in the formation of a syncytium layer at the fetomaternal interface via trophoblast cell–cell fusion. We searched for similar genes within the suborder Ruminantia where the placenta lacks an extended syncytium layer but displays a heterologous cell-fusion process unique among eutherian mammals. An in silico search for intact envelope genes within the Bos taurus genome identified 18 candidates belonging to five endogenous retrovirus families, with one gene displaying both placenta-specific expression, as assessed by quantitative RT-PCR analyses of a large panel of tissues, and conservation in the Ovis aries genome. Both the bovine and ovine orthologs displayed fusogenic activity by conferring infectivity on retroviral pseudotypes and triggering cell–cell fusion. In situ hybridization of placenta sections revealed specific expression in the trophoblast binucleate cells, consistent with a role in the formation—by heterologous cell fusion with uterine cells—of the trinucleate cells of the cow and the syncytial plaques of the ewe. Finally, we show that this gene, which we named “Syncytin-Rum1,” is conserved among 16 representatives of higher ruminants, with evidence for purifying selection and conservation of its fusogenic properties, over 30 millions years of evolution. These data argue for syncytins being a major driving force in the emergence and diversity of the placenta.
Keywords: synepitheliochorial, placental evolution, phylogeny, placentome, ERV
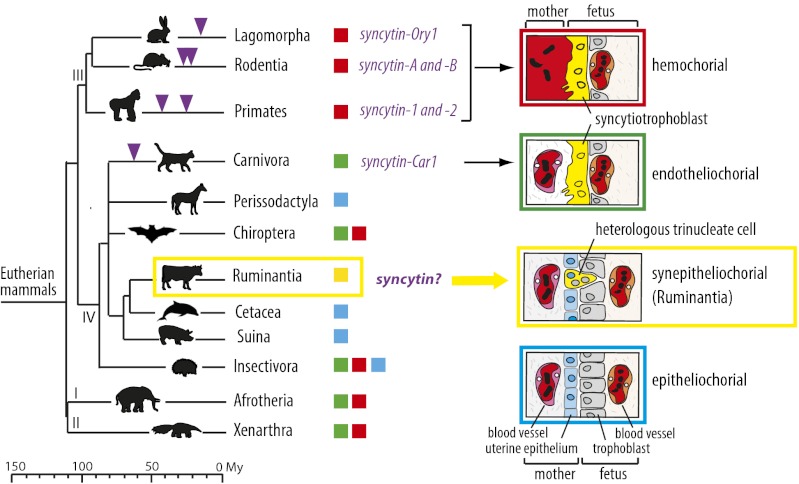
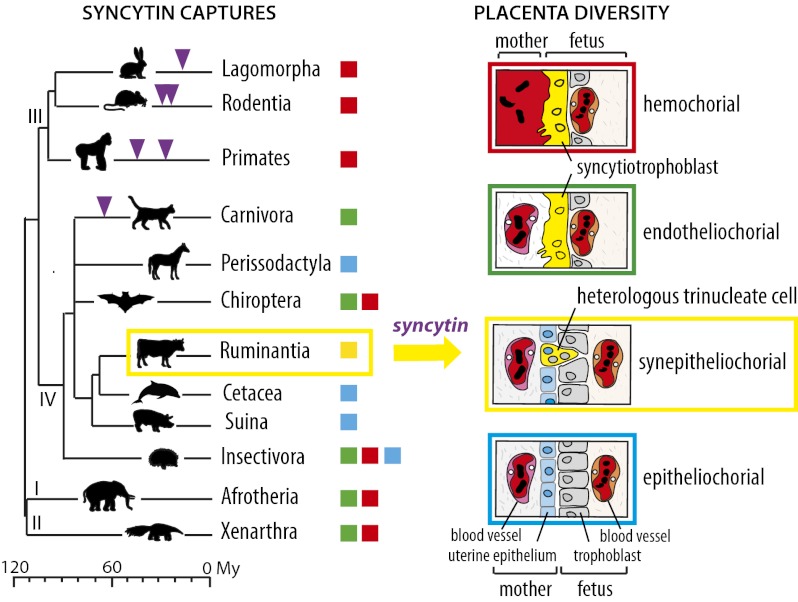
Infection by exogenous retroviruses throughout vertebrate evolution has led to a significant portion of the genome (e.g., 8% in human and 10% in the mouse) now being composed of retroviral sequences. Over time, these sequences were altered via genetic mutations, insertions, and recombinations, either passively or in response to host-defense mechanisms. Consequently, the majority of integrated retroviral sequences are nonfunctional remnants (1, 2). However, among this large pool of highly reiterated endogenous retrovirus (ERV) sequences, a few examples exist of single-copy retroviral genes that have been preserved since their integration and can be considered as bona fide cellular genes contributing to the host physiology. Among them are the syncytin genes encoding the envelope (Env) protein of ERVs, initially required for intracellular entry of exogenous retroviruses that have been co-opted independently in eutherian mammals for a key role in placenta formation (3). Syncytin-1 and syncytin-2 integrated into the genome of primates 25 and >40 Mya, respectively, and retained their coding capacity during speciation (4–6). They are expressed specifically in the placenta, are fusogenic in ex vivo assays, and one of them, syncytin-2, displays immunosuppressive activity (7). Remarkably, homologous counterparts have been identified in two other orders of Euarchontoglires, namely within Rodentia (syncytin-A and -B) (8) and Lagomorpha (syncytin-Ory1) (9), as well as in the order Carnivora (syncytin-Car1) (10) which belongs to the Laurasiatheria clade that diverged from Euarchontoglires around 100 Mya (Fig. 1). All identified syncytins share closely related functional properties, although they have a completely distinct origin, are integrated at separate genomic locations, and differ in their sequence. Recently, we have demonstrated unambiguously that the mouse syncytin-A and -B genes are essential for placentation, with syncytin-KO mice displaying a lack of cell–cell fusion at the level of the syncytiotrophoblast layers that separate maternal and fetal blood spaces, resulting in impaired embryonic survival (11, 12). Remarkably, exogenous retroviral envelope (env) genes therefore have been integrated independently and adapted for similar essential placental functions via a convergent evolution process. One challenging hypothesis is that this stochastic acquisition of genes of exogenous origin has been instrumental in establishing the remarkable structural and functional diversity of the mammalian placenta. Actually, one of the major divergent traits among modern eutherian mammal placentae is the number and nature of the layers at the fetomaternal interface, leading to four major placental types (Fig. 1) (reviewed in refs. 13–15). In the simplest type, the fetal trophoblast cells simply are apposed to the intact uterine epithelium (noninvasive epitheliochorial placentation), whereas in endotheliochorial and hemochorial placentation, these trophoblast cells fuse together to form an invasive syncytiotrophoblast layer which is apposed to the endothelial wall of the maternal blood vessel (endotheliochorial placenta of carnivores) or in direct contact with maternal blood (hemochorial placenta of simians, rodents, and lagomorphs). At present, all the characterized syncytins have been found to be associated with the latter two invasive placentation types, where they are involved in syncytiotrophoblast formation. Interestingly, a fourth unusual type, the synepitheliochorial placenta, displays a unique process of trophoblast fusion: Some specific trophoblast cells (the binucleate cells, BNCs) fuse with a single uterine epithelial cell, giving rise to trinucleate cells (TNCs) or even multinucleate structures of mixed fetal and maternal origin that release proteins into the maternal circulation (16–18). This placentation type is observed in all studied families of higher ruminants [the Pecora, including the majority of ruminants (e.g., the Bovidae, Cervidae, Giraffidae, and Antilocapridae)], with the few species belonging to the Tragulina being considered as a primitive extant group with slightly different placental structures (diffuse or noncotyledonary placenta) (15, 19–22). The suborders the closest to the ruminants (e.g., Cetacea, Suina, and Tylopoda) display a noninvasive epitheliochorial placentation suggesting that the synepitheliochorial placentation emerged only once—or at least rarely—in the course of eutherian mammal evolution. To address the role of retroviral gene capture in this process, we asked whether mammals possessing a synepitheliochorial placenta have “captured” a common retroviral env gene with syncytin-like properties for the generation of this specific placental structure. Among the ruminants, we selected the cow (Bos taurus) because its genome has been sequenced, it can be reared and investigated easily at different stages of gestation, and its placental physiology has been described extensively.
Fig. 1.
Multiple Syncytin captures and diversity of placental structures in eutherian mammals. (Left) The phylogeny of eutherians that can be grouped into four major clades: (I) Afrotheria, (II) Xenarthra, (III) Euarchontoglires, and (IV) Laurasiatheria (adapted from ref. 46). The different types of placentation are indicated by colored squares (the color code is given in the scheme to the right). The time of insertion of the different syncytin genes is shown. Branch length is proportional to time (in million years, My). (Right) Schematic color-coded representation of the fetomaternal interface in the four main types of placental structures. Placental types are classified from top to bottom in the order of decreasing invasive properties and extent of syncytialization. The synepitheliochorial placentation of Ruminantia is unique among eutherians and is characterized by a heterologous cell-fusion process between cells of fetal and maternal origin and a very limited extent of syncytialization.
Results
In Silico Search for Retroviral env Genes Within the Cow (Bos taurus) Genome.
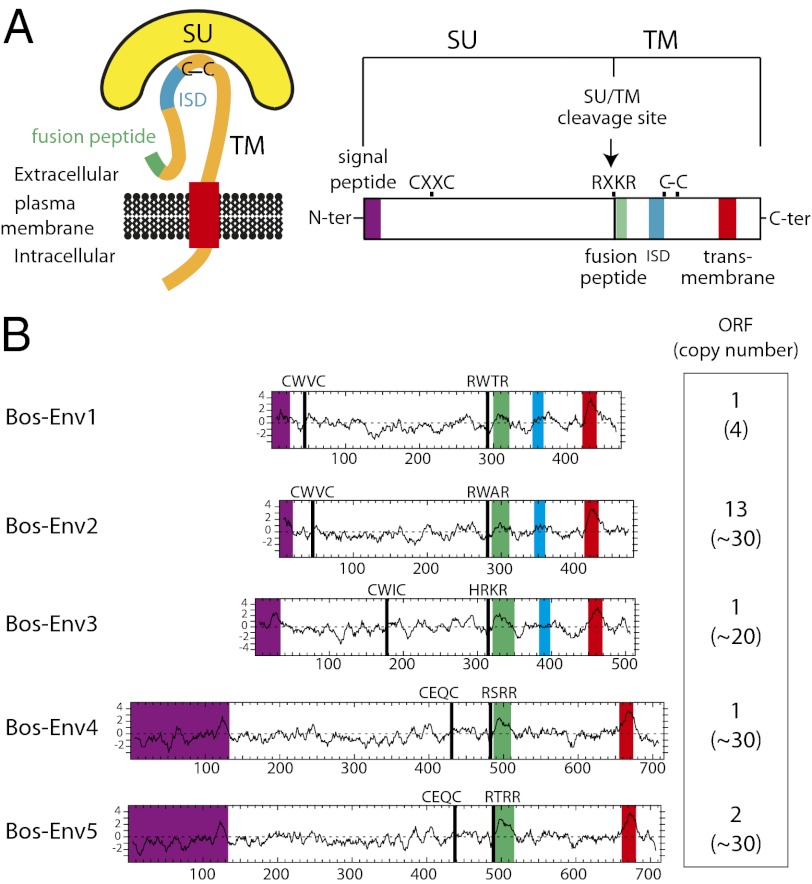
To identify putative env-derived syncytin genes, we made use of the available cow genome sequence [9.5× coverage assembly of the Bos taurus genome, University of California Santa Cruz (UCSC) UMD3.1/BosTau6, Nov. 2009]. A BLAST search for ORFs (from the Met start codon to the stop codon) longer than 400 aa was performed using a selected series of Env sequences, including all presently identified syncytins (Methods). It resulted in a series of sequences that were selected further for the presence of an hydrophobic domain >20 aa located 3′ to a C-X5,6,7-C motif, corresponding to a highly conserved motif of retroviral envelopes [C-C and transmembrane domain; see scheme in Fig. 2]. It yielded 18 sequences incorporated into the phylogenetic Env tree shown in Fig. 3. Some of the sequences can be grouped into single families, resulting finally in five families that we named “Bos-Env1” to “Bos-Env5” (Fig. 3). Interestingly, two of the families, Bos-Env4 and Bos-Env5, correspond to previously identified sequences (23, 24); the other three families had not been described previously. Analysis of the overall structure of the five identified Env families (Figs. 2 and 3) strongly suggests that they indeed correspond to bona fide retroviral Env proteins, with all their characteristic features (25, 26), including the presence of a predicted signal peptide sequence at the N terminus, a putative Furin cleavage site delineating a surface (SU) and a transmembrane (TM) subunit, and a CXXC motif in the SU subunit corresponding to a binding domain between the two subunits. Hydrophobicity plots identify the hydrophobic transmembrane domain within the TM subunits required for anchoring the Env protein within the plasma membrane and a putative hydrophobic fusion peptide at the N terminus of the TM subunit. Some of the TM subunits contain a canonical immunosuppressive domain (ISD) (7). Finally a BLAST search reveals that only the bos-env1 gene family is present at a low copy number (four copies, with only one containing a full-length ORF); the four other bos-env gene families display a much higher copy number (>20) (Fig. 2).
Fig. 2.
Structure of a canonical retroviral Env protein and characterization of the identified bovine candidates. (A) Schematic representation of a retroviral Env protein, delineating the SU and TM subunits. The furin cleavage site (consensus: R/K-X-R/K-R) between the two subunits, the C-X-X-C motif involved in SU–TM interaction, the hydrophobic signal peptide (purple), the fusion peptide (green), the transmembrane domain (red), the putative ISD (blue), and the conserved C-X5/6/7-C motif are indicated. (B) Characterization of the candidate bovine Env proteins. (Left) The hydrophobicity profile for each candidate is shown with the position of the canonical structural features highlighted in A shown when present. The color code is as in A. (Right) number of full-length env gene ORFs within each family of elements. The total number of genomic copies is shown in parentheses.
Fig. 3.
Retroviral Env protein-based phylogenetic tree with the identified Bos-Env protein candidates. The maximum-likelihood tree inferred with the RAxML software was constructed using Env protein amino acid sequences from murine and human ERVs and from a series of infectious retroviruses. The horizontal branch length is proportional to the percentage of amino acid substitutions from the node (the scale bar is shown on the left), and the percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. The clusters of 13 and 2 Env proteins that were grouped into single families of elements (Bos-Env2 and Bos-Env5, respectively) are distinguished by indication of their chromosomal position (Table S2). Bos-Env4 and Bos-Env5 (chr24) correspond to the Env of the previously identified bovine ERVs BERV-K1 and -K2 (23, 24), respectively. BFV, bovine foamy virus; BLV, bovine leukemia virus; ENTV, enzootic tumorigenic virus; FeLV, feline leukemia virus; JSRV, Jaagsiekte retrovirus; HERV, human ERV; HFV, human foamy virus; HIV, human immunodeficiency virus; MoMLV, Moloney murine leukemia virus; mIAPE, Mus musculus intracisternal A-type particle with an envelope gene; MMTV, murine mammary tumor virus; MPMV, Mason–Pfizer monkey virus; OMVV, ovine Maedi-Visna virus; PERV, porcine ERV.
Transcription Profiles for the Identification of Placenta-Specific env Genes.
We then examined the transcript levels of the five candidate env gene families in bovine placenta and in a panel of other bovine tissues. Quantitative RT-PCR (qRT-PCR) analyses were performed using primers designed to be specific for the ORF-containing sequences within each family of elements (Table S1). Placenta samples were analyzed at day 62 (d62) of gestation (delivery is at d280). As illustrated in Fig. 4, genes encoding Bos-Env1 and Bos-Env4 have the characteristic profile of bona fide syncytin genes because they are expressed in the placenta, with only very limited expression in the uterine endometrium (<5% of the placental level) and no expression in the oviduct and other tissues. Bos-env2 displays only limited expression (at least 20-fold lower) in the placenta and is expressed in other somatic tissues (e.g., spleen, muscle, and skin), whereas bos-env3 and bos-env5 have no or very low expression in all tested organs (Fig. 4). Taken together, in silico analyses combined with qRT-PCR assays for the bovine retroviral env genes clearly identify bos-env1 and bos-env4 as putative syncytin genes; bos-env2, -env3, and -env-5 were not considered further in the present study.
Fig. 4.
qRT-PCR analysis of the candidate env gene transcripts from the cow. Transcript levels are expressed as the ratio of the expression level of each env gene to that of the SDHA control gene (Methods). Note the enlarged vertical scale for the two placenta-specific bos-env1 and bos-env4 genes. Fetal villous tissue at d62 was used as the bovine placental sample, and the corresponding interplacentomal uterine endometrium from the mother was analyzed in parallel. The results obtained for the five env gene candidates in the same series of tissues are shown (tissues are displayed in the same order in all panels; tissue names are abbreviated in the upper right and three lower panels). Values for the placenta are the means of at least three samples; error bars show the mean ± SEM.
Characterization of the bos-env1 and bos-env4 Candidate Genes.
Bos-env1.
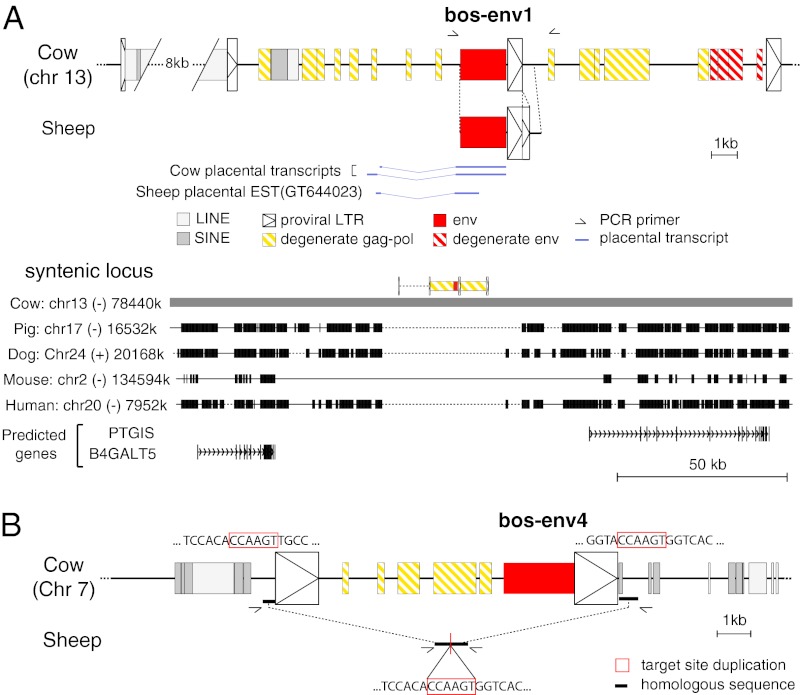
Examination of the Bos taurus genomic sequence revealed that bos-env1 is part of a proviral structure, positioned on chromosome 13, with degenerate but identifiable LTRs and gag-pol gene sequences (Fig. 5A). Its 5′ LTR is truncated by the insertion of a long interspersed nucleotide element (LINE), and its 3′ LTR corresponds, rather unexpectedly, to the 5′ LTR of a second tandem provirus displaying 92% identity with the bos-env1–carrying provirus but with a noncoding env gene (Fig. 5A). As commonly observed for retroviruses, a primer binding site sequence that is used for priming reverse transcription (26) can be identified downstream of the 5′ LTRs using the Genomic tRNA Database (http://lowelab.ucsc.edu/GtRNAdb/) and in the present case is found to be complementary to a bovine glycine tRNA (Fig. 5A). As classically observed for retroviruses, a putative acceptor splice site for the generation of a subgenomic env transcript can be identified just upstream of the Env initiation codon using the splice site prediction program at www.cbs.dtu.dk/services/NetGene2. Its position and functionality were ascertained by RT-PCR analysis of Bos-Env1–encoding transcripts in the placenta, using appropriate primers (Fig. 5A and Table S1).
Fig. 5.
Characterization of the bos-env1–, ovis-env1–, and bos-env4–containing ERVs and of their genomic location in the cow and sheep. (A) Characterization of the bos-env1–containing ERV. (Upper) Structure of the bos-env1–containing ERV and evidence for orthology between the cow and sheep env1 sequences. Homologous regions common to both sequences are aligned. Repeated mobile elements (gray) as identified by the RepeatMasker web program are positioned. Of note, bos-env1 is in a provirus directly followed by a tandem repeat of a homologous provirus sharing a common LTR. The proviral LTRs, the degenerate gag-pol and env genes, and the env1 gene ORF sequence are indicated (the key for symbols used is shown below the panel). PCR primers used to identify the bos-env1 orthologous copy in the sheep (black half arrows) and splice sites for the env subgenomic transcripts as determined by RT-PCR of cow placental RNA or by alignment with an EST sequence for the sheep are indicated. The nonhomologous sequence in the sheep LTR positioned 3′ to the env1 sequence compared with the cow corresponds to a 200-bp direct tandem duplication of part of the LTR 3′ end in the sheep. (Lower) Absence of the bos-env1–containing ERV in the genomes of distant mammalian lineages. The bos-env1–containing provirus (in yellow, with its LTRs schematized by boxed triangles and the env gene in red) was used as a reference, and synteny between the cow, pig, dog, mouse, and human genomes was determined with the Comparative Genomic tool of the UCSC Genome Browser (http://genome.ucsc.edu/). The positions of exons (vertical lines) of the resident B4GALT5 and PTGIS gene and the sense of transcription (arrows) are indicated. Homologous regions are shown as black boxes, nonhomologous regions as thin lines (not to scale), and gaps as dotted lines. (B) Structure of the bos-env4–containing ERV and evidence of the absence of proviral integration in the sheep syntenic locus. The symbol code is as in A. Homologous sequences flanking the provirus in the cow and colinear in the sheep are indicated by bold lines. Comparison of the flanking sequences of the cow provirus with those of the empty locus in the sheep (obtained by PCR with the indicated primers) provides evidence for target-site duplication in the cow (red boxes), a characteristic feature of retroviral integration.
Cow genomic DNA PCR with a forward primer located within the provirus at the 5′ end of the env gene and a reverse primer positioned ∼500 bp downstream of the 3′ LTR (Fig. 5A), resulted in amplification of a product of the expected size whose sequence matched that from the Bos taurus genomic database. Interestingly, PCR carried out with the same primers on sheep (Ovis aries) DNA resulted in the amplification of a homologous sequence (Fig. 5A) with a full-length env gene ORF followed by a 3′ LTR (highly related to that in the Bos taurus genome with the exception of a 200-bp duplication of its 3′ end) further followed by a 500-bp flanking sequence with 89% nucleotide identity to the corresponding bovine sequence. Comparison of the cow and sheep env sequences discloses 86% nucleotide identity, with all the canonical sites and domains characteristic for a retroviral Env protein showing high conservation (Fig. S1). These results show that bos-env1 has an ortholog in the Ovis aries genome, which we will now refer to as “ovis-env1.” Of note, a qRT-PCR analysis of ovis-env1 transcripts in a panel of tissues including the placenta at d55 of gestation (delivery is at d150), carried out as in Fig. 4 for the cow genes, also demonstrates placenta-specific expression in this other ruminant species, as with the orthologous bos-env1 gene, with notably no detectable expression in tissues of the genital tract (endometrium, oviduct, and cervix) (Fig. S2).
As further shown in Fig. 5A, the bos-env1 provirus is integrated close to (but not in an intron of) the putative PTGIS and B4GALT5 genes, in the sense orientation. By performing a search with the Comparative Genomic tool of the UCSC Genome Browser (http://genome.ucsc.edu), we identified the syntenic genomic locus in the pig, dog, mouse, and human genomes. Both the bos-env1 orthologous copy and the associated proviruses are missing in all these species (Fig. 5A).
Bos-env4.
Examination of the Bos taurus genomic sequence indicates that bos-env4 is part of a proviral structure positioned on chromosome 7, with identifiable LTRs and degenerate gag-pol gene sequences (Fig. 5B). It corresponds to the previously identified BERV-K1 sequence (23) and, as clearly shown in Fig. 3, its envelope is more closely related to that of beta- and delta-retroviruses [e.g., murine mammary tumor virus (MMTV) and Jaagsiekte retrovirus (JSRV)] than to that of gamma-retroviruses, at variance with what has been observed for all previously identified syncytins. The bos-env4 gene could not be retrieved by in silico screening of the available Ovine genome database (3× coverage assembly of the Ovis aries genome; UCSC International Sheep Genomics Consortium (ISGC)/oviAri1 Dec. 2010). Because the latter corresponds to only a low-coverage release, we directly investigated the presence of an orthologous bos-env4 gene in the sheep genome via PCR of sheep DNA with primers bracketing the bos-env4 locus (see position of the primers in Fig. 5B). A single band corresponding to the “empty” locus (1.4 kb) was obtained and, as illustrated in Fig. 5B, its sequencing allowed us to infer the position of the exact site of insertion of the bovine ERV, accompanied by a 6-bp “target site duplication,” as classically observed for retroviruses. The sequencing also excludes the possibility that the gene might have been present in the sheep and then removed by homologous recombination, in which case a solo LTR should have been found at the integration site. This result clearly indicates that bos-env4 has no ortholog in the sheep, consistent with a probable recent endogenization of the bovine retrovirus. Thus, bos-env4 was not considered further as a candidate ancestral gene for ruminant placentation.
Assay for the Fusogenic Activity of bos-env1 and ovis-env1.
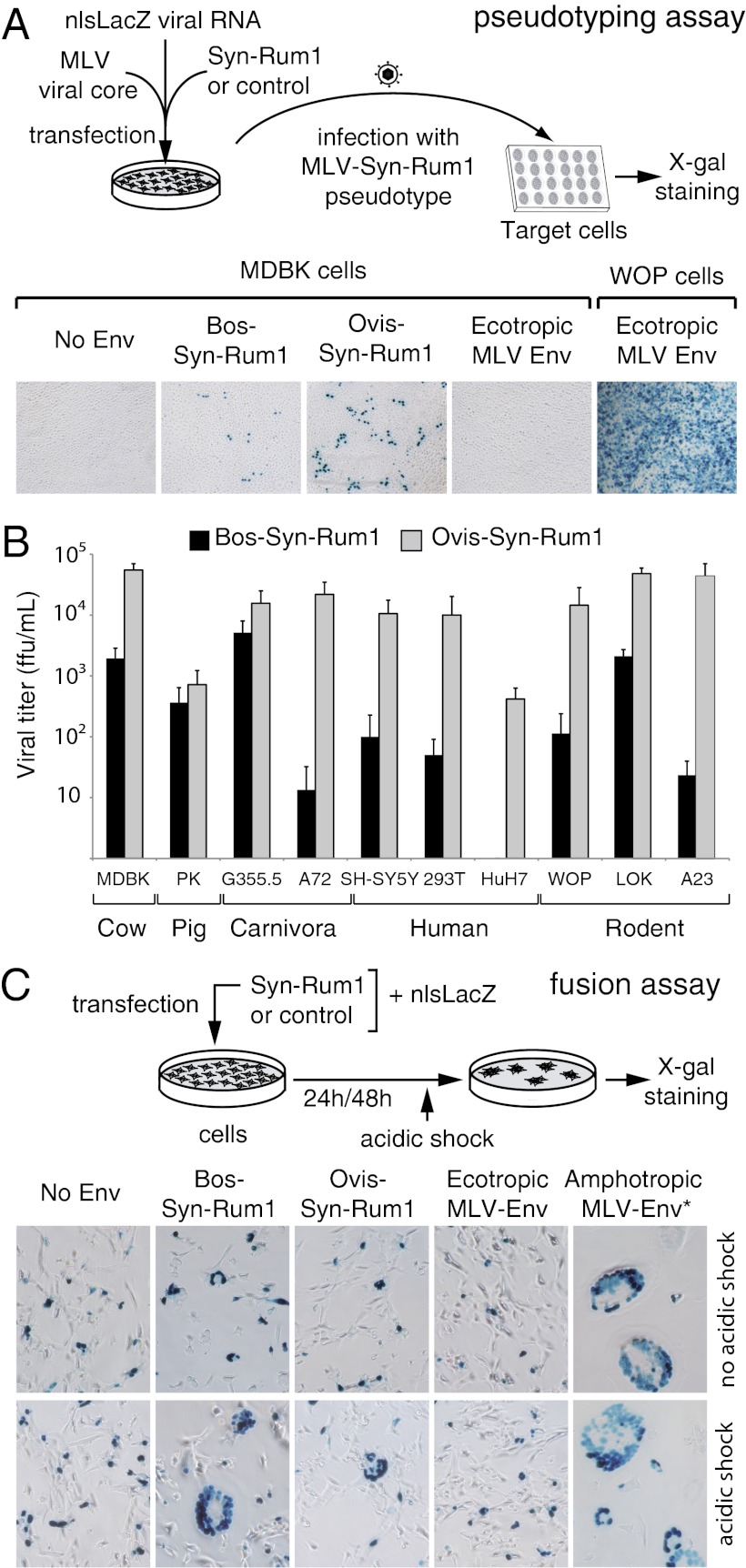
The functionality of the protein encoded by the bos-env1 and ovis-env1 orthologs as an ancestral, retrovirally derived, fusogenic Env protein was assayed ex vivo as previously described for other syncytins (9, 10, 27). Basically, we tested whether these Env proteins added in trans could render a recombinant retrovirus, deprived of its own native env gene, able to infect test target cells (Fig. 6A; scheme). To do so, the amplification products of bos-env1 and ovis-env1 (ORF followed by 1.2 kb of untranslated 3′ sequences containing the 3′ LTR) were cloned into an expression vector containing the CMV promoter (Methods), and plasmids with a full-length env gene ORF 100% identical to the genomic sequences were assayed. As expected for an Env protein of retroviral origin, both Bos-Env1 and Ovis-Env1 can produce infectious particles. Indeed, as illustrated in Fig. 6A, pseudotypes generated in human 293T cells with a murine leukemia virus (MLV) core are able to infect the bovine MDBK cell line. Pseudotypes without an envelope or with the ecotropic MLV envelope (otherwise infectious for WOP murine cells) were used as negative controls. The infectious titers obtained are significant, namely 1.9 × 103 focus-forming units (ffu)/mL for Bos-Env1, with even higher values, 5.5 × 104 ffu/mL, for Ovis-Env1. As illustrated in Fig. 6B, a large series of cell types from different species (e.g., cow, pig, carnivores, primates, and rodents) could be infected, thus suggesting that the as-yet unidentified receptor for the Env protein is a widespread and conserved protein. The infection rate is variable, even among cells from the same animal species (see the human SH-SY5Y, 293T, and HuH7 cells), a result most probably associated with variations in the levels of expression of the cognate receptor. Fig. 6B also shows that the difference between the cow and sheep Env pseudotype titers is observed systematically for all target cells tested, although to a variable extent depending on the cells (see for instance the very low relative Bos-Env1 values in the dog A72 or hamster A23 cells compared with the cat G355.5 cells). These differences most probably are caused by intrinsic differences between the two envelopes, and their variability might result from subtle divergences in the parallel coevolution of the env and receptor genes. The fusogenic properties of Bos- and Ovis-Env1 were tested further in a cell–cell fusion assay involving transfection of cells with expression vectors for each env gene and detection of syncytium formation 24–48 h posttransfection. The cells used were selected from those with the highest susceptibility to pseudotype infection (Fig. 6B) and included G355.5, LOK, and MDBK cells. In the former two cell lines (MDBK could not be efficiently transfected) only very limited fusion activity could be detected, but, interestingly and as previously observed for a series of retroviral envelope proteins (28, 29), fusion activity could be detected unambiguously after a brief (5-min) acid treatment of the cells (Fig. 6C). Both Ovis- and Bos-Env1 induced large syncytia in G355.5 cells with a pH 5 shock, an effect not observed in the absence of Env or with the ecotropic (mouse only) MLV Env as negative controls. The extent of fusion was similar to that of a C-terminally truncated amphotropic MLV Env (30) (Fig. 6C, Lower Right) (30) used as a positive control. Fusion activity also was detected with the LOK cells, at least with Bos-Env1. Although the observed enhancement in cell–cell fusion activity under low pH conditions is not an unexpected result given previous observations for some retroviral Envs (28, 29), including that of the infectious ovine JSRV retrovirus, it is not common among syncytins. In the present case, the enhancement could suggest that specific local pH conditions prevail at the fetomaternal interface between the BNCs and maternal uterine epithelial cells. Taken together, the results of these assays show that the bos- and ovis-env1 genes have conserved their fusogenic properties and accordingly can be considered bona fide syncytin genes; therefore the ruminant family of orthologous genes was named “syncytin-Rum1.” Of note, the bos-env4 gene was found to be negative in the above fusogenicity assays, i.e., in both the pseudotyping experiment and the cell–cell fusion assay, even under low-pH conditions.
Fig. 6.
Bos- and Ovis-Env1 are fusogenic retroviral Env proteins. (A) Bos-Env1 and Ovis-Env1 are fusogenic in pseudotyping assays. (Upper) Schematic representation of the assay for cell infection with Bos-Env1– or Ovis-Env1–pseudotyped virus particles. Pseudotypes are produced by cotransfection of human 293T cells with expression vectors for the MLV core, the Bos-Env1 or Ovis-Env1 proteins (or either ecotropic-MLV Env or an empty vector as controls), and a plasmid expressing a nuclear β-galactosidase encoded by a nlsLacZ-containing retroviral transcript. Pseudotyped virus particles in cell supernatants then are assayed for infection of the indicated target cells, which are stained with X-Gal (3-d postexposure). (Lower Left) X-Gal–stained target cells exposed to particles pseudotyped with Bos-Env1, Ovis-Env1, or, as a negative control, Env from an ecotropic MLV (infecting only murine cells) on cow MDBK target cells. (Lower Right) As a positive control, the viral titer of particles pseudotyped with ecotropic MLV-Env was tested on murine WOP cells. (B) Viral titers for particles pseudotyped with Bos-Env1 or Ovis-Env1 assayed on a panel of target cells from cow (MDBK), pig (PK), carnivores [cat (G355.5) or dog (A72)], human (HuH7, 293T, and SH-SY5Y), or rodents [mouse (WOP and LOK) or hamster (A23)]. Titers, expressed as focus-forming units (ffu) per milliliter ± SD, are corrected for the background values of control particles without an Env protein and are the means from at least three independent experiments. (C) Bos-Env1 and Ovis-Env1 are fusogenic in a cell–cell fusion assay. (Upper) Schematic representation of the cell–cell fusion assay with cells cotransfected with a plasmid expressing Bos-Env1 or Ovis-Env1 (or an ecotropic-MLV Env, a C-terminal–truncated amphotropic-MLV Env, or an empty vector as controls) and a plasmid expressing a nuclear β-galactosidase (nlsLacZ). Twenty-four hours after transfection, cells were treated transiently (5 min) with neutral or acidic PBS buffer (pH 7 or pH 5) and were stained with X-Gal 4–6 h posttreatment. (Lower) G355.5 cat cells were transfected with Bos-Env1, Ovis-Env1, or, as a negative control, Env from an ecotropic MLV or an empty vector, and neutral (Upper Row) or acidic (Lower Row) pH treated. As a positive control, cells were transfected with the C-terminal–truncated (R-less) amphotropic-MLV Env (Amphotropic-MLV Env*) (Right) (30).
Localization and Quantitative Analysis of the Bovine and Ovine syncytin-Rum1 Transcripts.
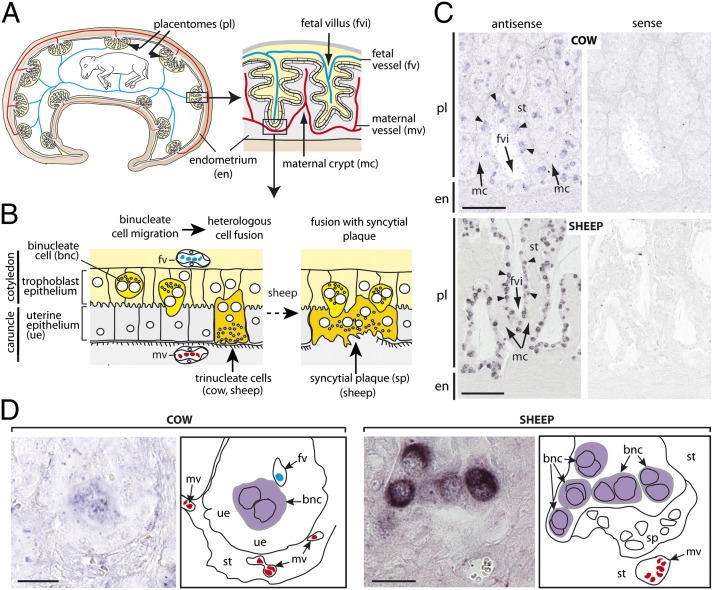
As schematized in Fig. 7A for the cow, the typical ruminant placenta contains specialized zones of fetomaternal contact, called “placentomes,” consisting of branched fetal (or cotyledonary) villi interdigitating with maternal (or caruncular) crypts. Fetal and maternal epithelia are in close contact, allowing efficient exchanges between the fetal and maternal vascular systems. In ruminants, 15–20% of the fetal trophoblast epithelium consists of BNCs that originate from an endoreplication process without any fusion. BNCs have two large polyploid nuclei and produce granules with large amounts of hormones and effectors (Fig. 7B) (31). Remarkably, throughout gestation, mature BNCs migrate and fuse with a single uterine epithelial cell to form fetomaternal hybrid TNCs, which release BNC granules by exocytosis into the maternal circulation (Fig. 7B). Interestingly, the extent of fusion differs among species: In the cow, TNCs form only transiently and degenerate rapidly after granule release, whereas in the sheep, the fusion process involving the BNCs proceeds further and generates multinucleate syncytial plaques of limited size (5–25 nuclei), which replace part of the uterine epithelium (Fig. 7B) (16, 17).
Fig. 7.
Structure of the synepitheliochorial placenta of higher ruminants and in situ hybridization for syncytin-Rum1 expression on cow and sheep placental sections. (A) (Left) Schematic drawing of the bovine fetus and placenta in utero. The yellow and gray areas represent the fetus and mother membranes, respectively; the localized areas of formation of the fetomaternal villous units, the placentomes (pl), are indicated. The maternal (red) and fetal (blue) vessels are schematized. (Right) Detailed scheme of a bovine placentome. The placental fetal villi are intimately enmeshed with preformed maternal endometrial crypts (both covered by their respective epithelium) for maximal exchanges between the fetal and maternal blood circulations. Of note, the placentome organization in the cow and sheep is identical, except that on the fetal side it is convex in the cow and concave in the sheep. (B) Diagram of the synepitheliochorial fetomaternal barrier of ruminants. Binucleate cells (BNCs) residing in the trophoblast epithelium and possessing characteristic granules migrate into the uterine epithelium and fuse with the apex of a single uterine epithelial cell forming a fetomaternal hybrid trinucleate cell. BNC granules then are released on the maternal side of the placenta. In the sheep, the fusion process involving the BNCs proceeds further and generates multinucleate syncytial plaques which partially replace the uterine epithelium. (C and D) ISH on serial sections of placenta from cow and sheep, respectively, at the first trimester, observed at different magnifications, using digoxigenin-labeled antisense or sense riboprobes revealed with an alkaline phosphatase-conjugated anti-digoxigenin antibody. (C) Partial view of a placentome (pl) and endometrium (en) zona, with the fetal villi (fvi), the maternal crypts (mc), the uterine stroma (st), and the labeled BNCs (arrowheads) indicated. (D) Higher magnification of a fetomaternal interhemal area (see scheme in B); the maternal crypts and fetal villi, the labeled BNCs, the fetal and maternal vessels, the uterine stroma, and the position of the uterine epithelium in the cow or of the syncytial plaque in the sheep are schematized to the right of each panel. Note that the syncytial plaque, the uterine epithelium, and the stroma are not labeled. (Scale bars: 200 µm in C and 25 µm in D.)
To assess the physiological relevance of syncytin-Rum1 expression, we performed in situ hybridization (ISH) experiments on paraffin sections of cow and sheep placentomes (gestation d62 for the cow and d55 for the sheep). Specific digoxigenin-labeled antisense probes were synthesized for detection of bovine and ovine syncytin-Rum1 transcripts. The corresponding sense probes were used as negative controls. As shown in Fig. 7C, labeling was observed only with the antisense probe. In the cow and sheep, syncytin-Rum1 expression was restricted to dispersed trophoblastic cells lining the fetal villi (Fig. 7C) that displayed two nuclei (see higher magnification in Fig. 7D) and therefore were identified as BNCs; this identification also was confirmed by IHC using pregnancy-associated glycoprotein as a specific marker of BNCs. Fig. 7D shows, for the sheep, syncytin-Rum1 expression in a series of BNCs that are located at the tip of a fetal villus and apposed to a basal syncytial plaque, which itself is not labeled, thus indicating tightly regulated expression of syncytin-Rum1 in BNCs, which stops as fusion proceeds. In the cow, the equivalent fused TNCs could not be identified unambiguously enough for such a conclusion to be drawn. Of note, no labeling was detected in the maternal uterine tissues (uterine epithelium, stroma, and endometrium) interdigitating with the fetal villi in the placentome (Fig. 7 C and D). Similarly, the uterine maternal tissues (uterine epithelium, stroma, and glandular epithelium) of the interplacentomal areas, assayed in the same conditions, also were negative (Fig. S3). Conclusively, the syncytin-Rum1 expression profile is consistent with a role in triggering the fusion of BNCs with uterine epithelial cells to form the fetomaternal TNCs (cow) or syncytial plaques (sheep).
It is notable that the ISH experiments reveal a higher number of positive BNCs with a higher intensity of labeling for the syncytin-Rum1 transcripts of Ovis aries than for those of Bos taurus although both tissue samples were processed using similar experimental conditions (Fig. 5C). To quantify further the relative levels of the bovine and ovine syncytin-Rum1 transcripts throughout gestation, we performed a qRT-PCR analysis of RNA extracted from sheep and cow placentae at several comparable gestational stages using a pair of primers designed in regions that were 100% identical in the two orthologs (Fig. 8). The levels of transcripts were found to be higher in Ovis aries than in Bos taurus throughout gestation, although to a variable extent depending on the gestational stage. Interestingly, at early placentation stages, ovine and bovine env transcript levels differ only weakly and show a parallel increase along with initiation of BNC heterologous fusion [which takes place at d16–17 in the sheep and at d20 in the cow (16, 17)]. Then, a strong up-regulation is observed for the ovine (but not the bovine) syncytin-Rum1 gene, which is expressed at a 15- to 20-fold higher level than the bovine gene throughout the remaining pregnancy. This up-regulation correlates with the remarkable increase in the size of the syncytial plaques (mean nuclei number rising from 5 to 25 nuclei) observed at the onset of placentome formation in the sheep (d25) (17). Both observations suggest a direct correlation between the level of syncytin-Rum1 expression and the extent of the fusion process.
Fig. 8.
Relative expression level of the syncytin-Rum1 gene in the bovine and ovine placenta during gestation. The mRNA levels were quantified by qRT-PCR using a unique pair of primers designed from regions that are identical in the bovine and ovine syncytin-Rum1. Transcript levels are expressed relative to the lowest value taken as unity and were normalized relative to the amount of the gene encoding succinate dehydrogenase, subunit A (SDHA) (logarithmic scale). Placenta tissues were obtained at the indicated days of gestation (comparable gestation stages are grouped). Vertical arrows indicate two major quantitative changes in the BNC fusion processes occurring during pregnancy (16, 17). Note the significant 3.5- to 10-fold increase in both the bovine and ovine syncytin-Rum1 transcript levels observed at the initiation of the BNC fusion processes and the strong six- to sevenfold increase in the ovine (but not bovine) syncytin-Rum1 transcript level concomitant with the increase in size of the syncytial plaques in sheep placenta. The higher level of syncytin-Rum1 expression could not be explained by a higher proportion of BNCs in the sheep than in the cow, because the percentage of BNCs in the trophectoderm (around 20%) is almost identical in both species throughout the gestation period (31). dpc, days post coitum.
Search for syncytin-Rum1 in Ruminantia.
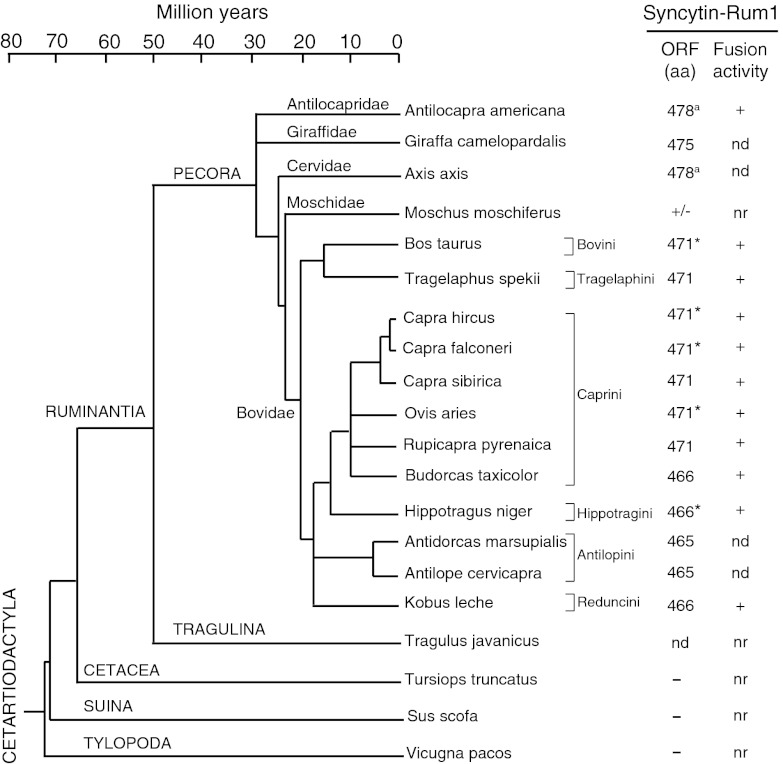
To characterize syncytin-Rum1 further and to determine its status and evolution in the suborder Ruminantia, we searched for the orthologous gene in several species of the family Bovidae and in representative species of each of the five other families of Ruminantia (Fig. 9). Genomic DNA from these different species was PCR-amplified using a pair of primers bracketing the bovine syncytin ORF and designed to amplify the ORF-containing sequences specifically (Table S1). A single amplification product with the expected size was obtained for all species (with the exception of Tragulus and Moschus). In all Bovidae species, sequencing the PCR products revealed the presence of a unique, full-length Syncytin-Rum1–encoding ORF (465–471 aa long), most probably corresponding to the orthologous syncytin-Rum1 copy, as was further demonstrated unambiguously for four species—Capra hircus, Capra falconeri, Ovis aries, and Hippotragus niger—using a reverse primer downstream of the 3′ LTR (Fig. 5). In species belonging to the more divergent Cervidae, Giraffidae, and Antilocapridae families, sequencing of the PCR products demonstrated the presence of more than one sequence. Cloning of the PCR fragments into the pGEM-T vector and sequencing of individual clones disclosed, for each species, noncoding sequences as well as a few sequences harboring full-length syncytin-Rum1 ORFs (one in Giraffa, five in Antilocapra, four in Axis axis). For the latter two species, the full-length syncytin-Rum1 ORFs shared high nucleotide identity (>96%) and possibly correspond to PCR-induced variants of a unique coding sequence in each species. Finally, a pair of primers internal to the ORF and designed in the regions most highly conserved among all available syncytin-Rum1 orthologs (Table S1) resulted in the amplification of a PCR product of the expected size (450 bp) in Moschus moschiferus, disclosing high sequence identity (84%) with the bovine gene, and therefore most probably corresponding to amplification of a syncytin-Rum1 family gene. With the same set of internal primers, no amplification product could be obtained from the distant species Tragulus javanicus. Although this result suggests the absence of syncytin-Rum1 sequences in this species, it is possible that such sequences could not be amplified because of a too great sequence divergence in the primitive Tragulina (divergence >50 Mya) (Fig. 9). In conclusion, syncytin-Rum1 sequences are unambiguously present in Bovidae, Moschidae, Cervidae, Giraffidae, and Antilocapridae, i.e., in the Pecora corresponding to the so-called “higher ruminants,” indicating conservation for at least 30 million years of evolution.
Fig. 9.
Status of syncytin-Rum1 during the radiation of the Ruminantia. The Ruminantia phylogenetic tree is shown with the two infraorders, Pecora (higher ruminants; most of the ruminant families) and Tragulina (only one extant family), as well as the outgroups Cetacea, Suina, and Tylopoda (adapted from refs. 47 and 48). The phylogeny of Pecora is not fully resolved (44, 45). Branches whose interrelationship is still under question are represented as paraphyletic. The names of the 17 Ruminantia species tested for the presence of the syncytin-Rum1 gene are given, together with the length (in amino acids) of the Syncytin-Rum1 proteins that were identified for each species (all sequences were deposited in GenBank, accession numbers JX412964–JX412985); the fusogenic activity for each cloned gene, as determined by the pseudotyping assay in Fig. 6 C and D, is provided. Asterisks indicate syncytin-Rum1 genes PCR-amplified with their 3′ flanking sequence; a indicates PCR-amplification of more than one syncytin-Rum1 full-length ORF. For Antilocapra americana, one gene (GenBank accession number JX412981) among the five tested was found to be fusogenic. n.d., not demonstrated; n.r., not relevant; ±, only partial syncytin-Rum1 sequence was obtained.
The presence of a syncytin-Rum1 gene also was investigated in other species of the order Cetartiodactyla, i.e., among Cetacea (Tursiops truncatus), Suina (Sus scrofa), and Tylopoda (Vicugna pacos) (Fig. 9), as well as in different mammal orders i.e., Perissodactyla (Equus caballus), Primates (Homo sapiens), and Rodentia (Mus musculus), by an in silico search of the corresponding Ensembl databases using the BLAST program. No sequence with a homology >50% could be found, suggesting that syncytin-Rum1 capture took place only once in the course of eutherian mammal evolution.
Purifying Selection and Functional Conservation of syncytin-Rum1.
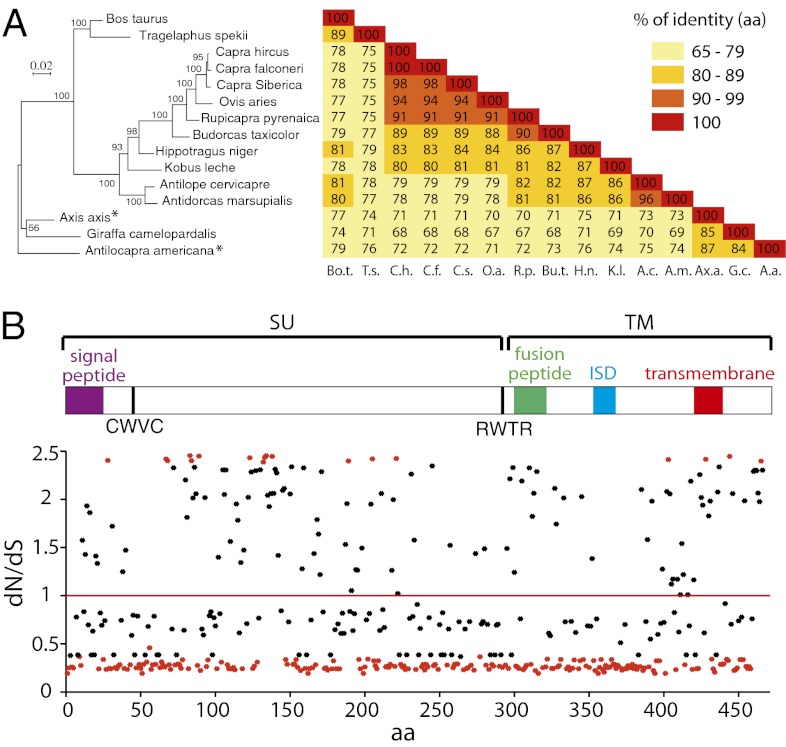
Sequence analysis of the syncytin-Rum1 genes identified here demonstrates high similarities, 67–98% (Fig. 10A), as expected for a bona fide cellular gene. Interestingly, the phylogenetic tree generated from an alignment of these sequences (Fig. 10A) is congruent with the Ruminantia phylogenetic tree in Fig. 9, with minor differences for some poorly resolved nodes.
Fig. 10.
Evidence for sequence conservation of and selection pressure on the syncytin-Rum1 gene. (A, Left) Syncytin-Rum1–based maximum-likelihood phylogenetic tree determined using amino acid alignment of the Syncytin-Rum1 proteins identified in Fig. 9, inferred with the RAxML program. The horizontal branch length and scale indicate the percentage of amino acid substitutions. Percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. An asterisk indicates that the sequence used for Axis axis is a consensus of the four full-length gene ORFs (otherwise branching together) and, for Antilocapra americana, the one shown to be fusogenic in the pseudotyping assay (Fig. 9). (A, Right) Double-entry table for the pairwise percentage of amino acid sequence identity between the syncytin-Rum1 genes belonging to the indicated species. (B) Site-specific analysis of selection on syncytin-Rum1 gene codons, using the PAML (M8 model) package. The relevant selection indexes are provided for each codon. A schematic representation of the Syncytin-Rum1 protein domains is given also; conventions are as in Fig. 2. Significant values (P ≥ 0.95) are represented as red dots.
To characterize further the conservation/evolution of the syncytin-Rum1 gene, we performed a refined analysis of the sequences using methods based on the rate of nonsynonymous versus synonymous substitutions (dN/dS) within the complete set of sequences and allowing differences in selection pressure between different domains of the proteins to be revealed [site-specific selection (32, 33)]. Such an analysis, using the PAML package (34), provided support for a model [model M8 versus model M7: χ2 = 40.8, degree of freedom (df) = 2, P value = 1.38e−9] in which most of the codons are under purifying selection (dN/dS ≤1, 67% of the codons) and some are under positive selection (dN/dS >1, 33% of the codons). This analysis is illustrated in Fig. 10B; the dN/dS values are indicated for each amino acid position, and values significantly lower or higher than unity are shown in red. Although 43.7% of all codons are under strong purifying selection, a few (3.8%) can be identified with a dN/dS value significantly higher than unity, essentially in a definite domain of the SU subunit and not found in the ISD, suggesting positive selection for some env function. Analyses using the HyPhy package (33) with slightly different site-specific models [random-effect likelihood (REL) and fixed-effect likelihood (FEL)] led to similar conclusions, although those analyses are less sensitive in detecting which specific codons have a dN/dS value significantly higher than unity. Conclusively, the syncytin-Rum1 genes are mainly under strong purifying selection, with some evidence for site-restricted positive selection. In the case of HIV, several domains of the env gene have been demonstrated to be subject to positive selection [e.g., the variable regions of the SU subunit (35)], with mutations in these domains favoring virus escape from the host immune response. For syncytin-Rum1 the observed limited positive selection might correspond instead to the necessary conformational adaptation of the Env receptor-binding domain to its as-yet unidentified cellular receptor.
To determine whether the strong selective pressure exerted on the syncytin-Rum1 gene does correlate with the conservation of its functional properties, a pseudotyping ex vivo assay, as illustrated in Fig. 7 for the cow and sheep representatives, was tentatively performed. The PCR-amplified syncytin-Rum1 genes from the species listed in Fig. 8 were cloned into the same eukaryotic expression vector, and pseudotypes were assayed similarly using either bovine (MDBK) or cat (G355.5) cells as a target. As indicated in Fig. 9, 11 of these genes, including the distant Antilocapra gene, were found to be positive in this assay. However, we could not provide evidence for fusogenic activity in the case of four of the sequences (but for two of them evidence for their identity to the native genomic sequences is lacking), possibly because of PCR-introduced mutations in the cloned genes or impaired/reduced interaction between these Envs and the receptor present in the cell lines tested.
Taken together, the data suggest that syncytin-Rum1 is a bona fide cellular gene co-opted for a physiological role in placentation.
Discussion
Here we have identified syncytin-Rum1, the env gene from an ERV that integrated into the genome of ruminants more than 30 Mya. This gene has been maintained as a functional retroviral env gene since that time, being conserved in the 16 species tested that were selected from the major Ruminantia families. This gene displays all the canonical characteristics of a syncytin gene: (i) it exhibits fusogenic activity, because it can functionally replace a present-day retroviral env gene within a recombinant infectious retrovirus and mediate cell–cell fusion; (ii) it has been subject to selective pressure in the course of evolution, displaying essentially low rates of dN/dS and conservation of its fusogenic property; and (iii) it is specifically expressed in the placenta, as evidenced by both RT-PCR analyses and ISH of cow and sheep placental tissue sections. Syncytin-Rum1 can be added to the previously identified syncytins, namely the two primate syncytin-1 and syncytin-2 genes (4–6), the two syncytin-A and syncytin-B genes in Muroidea (8), the syncytin-Ory1 gene in Leporidae (9), and the syncytin-Car1 in Carnivora (10). Importantly, all syncytins are unrelated and correspond to independent captures in separate mammalian lineages of genes of retroviral origin. Of note, all six previously identified syncytins belong to mammals displaying a placental architecture with an extended syncytium layer (syncytiotrophoblast) formed at the fetomaternal interface and associated with the hemochorial or endotheliochorial placental types. Syncytin-Rum1 is associated with a third placental type, i.e., the synepitheliochorial placenta, which is unique among eutherian mammals. In the case of the ruminant synepitheliochorial placenta, limited fusion activity indeed is detected in the form of TNCs in the cow, or multinucleate cells in the sheep, resulting from the heterologous fusion of specific trophoblastic cells (the BNCs) with cells of maternal origin (15). The resulting structures are indeed intermediates between the epitheliochorial structure, with no trace of syncytium, and the hemochorial and endotheliochorial structures, both of which possess an extended syncytiotrophoblast layer and (in all cases in which it has been investigated) a bona fide syncytin. Therefore it is tempting to propose that the emergence of the specific ruminant placental architecture among mammals results precisely from the acquisition of syncytin-Rum1. The present results support this hypothesis. First, in the genome of all of the ruminants tested (with the exception of the ancestral extant Tragulus), syncytin-Rum1 is conserved as a functional env gene; it is not found in other Cetartiodactyla groups, including Cetacea, Suina, and Tylopoda, or in more distant eutherian mammals, such as primates and rodents. Second, the pattern of expression of syncytin-Rum1 as revealed by ISH of placenta sections, namely in the BNCs that undergo heterologous fusion, is precisely the pattern expected for the generation of the TNCs and multinucleate cell structures observed at the bovine and sheep fetomaternal interface, respectively. Like syncytin-2 in the human placenta, which is expressed only at the level of the mononucleate trophoblast cells and not in the syncytiotrophoblasts with which they fuse (36), Syncytin-Rum1 expression is restricted to the fetal trophoblast BNCs with no expression in the TNCs or multinucleate cells, thus ensuring that fusion will not extend indefinitely to all the cells of the maternal epithelium. Third, a refined analysis of the expression of the syncytin-Rum1 orthologs demonstrates a >10-fold higher level of expression in the sheep than in the cow, as revealed by qRT-PCR using primers common to both genes (also see the more intense labeling in the sheep in ISH experiments). This result is consistent with the higher extent of fusion in the sheep (multinucleation in the sheep versus trinucleation for the cow). Finally, although this argument is a negative one, in our in silico search within the cow genome we found no other env gene of retroviral origin that fulfilled the requested criteria for being an ancestral ruminant syncytin: Among the five candidate genes, only two, syncytin-Rum1 and bos-env4, were expressed specifically in the placenta, and bos-env4 was absent in the sheep and, furthermore, was nonfusogenic.
Altogether, the data strongly suggest that syncytin gene capture has been a widespread process that took place in several widely separated lineages in the course of eutherian evolution and that the remarkable variability in placental structures might result simply from the diversity of the syncytin genes that have been stochastically captured in the course of mammalian evolution. In this respect, parameters such as the intrinsic level of fusogenicity of the Env proteins, the specific levels of syncytin expression in the appropriate tissues, the presence of the receptor required for Env-mediated fusion at the surface of the right neighboring cell, or, in the present case, the local pH at the fusion site would control and finely tune the placentation process. For instance, the cloning of syncytin-Rum1 in a significant number of species provides molecular clues to account for the subtle differences in placental structures observed in these species and opens interesting research perspectives. Identification of the cognate receptor for Syncytin-Rum1 and analysis of its species-specific interactions with the proteins encoded by the corresponding syncytin orthologs probably will be necessary for a complete understanding of the evolution of this syncytin. Actually, as illustrated in Fig. 10B, there are limited but significant domains in which positive selection can be identified, in addition to the overall purifying selection acting on the full-length protein. Part of the SU subunit is one of these domains, and it will be interesting to determine whether this region(s) indeed corresponds to the region involved in the binding to the receptor (once the latter is identified) and whether this positive selection actually corresponds to a convergent coevolution of the Env protein and its cognate receptor. Conversely, it also was observed (Fig. 10B) that the ISD domain clearly is not subject to positive selection and that the amino acids critical for immunosuppressive activity are fully conserved among all the characterized orthologs (Fig. S1), suggesting a role for the associated immunosuppressive function. This characteristic feature of retroviral envelopes, which has been demonstrated to be essential for immune escape in the case of infectious retroviruses [e.g., MLV (37)] also is carried by previously identified syncytins (7) and may contribute to the establishment of fetomaternal tolerance. Another unanswered question that should be addressed concerns the enhanced level of expression of syncytin-Rum1 in the sheep versus cow. It will be of interest to determine whether there are functional binding sites in the syncytin-Rum1–regulatory regions for transcription factors such as Glial cell missing-1 (Gcm-1), which are responsible for fetal villus initiation/formation in rodents (38, 39) and that have been shown to regulate the expression of several syncytin genes [e.g., primate syncytin-1 (40) and muroid syncytin-B (41)]. Clearly, specific acquisition of Gcm-1–responsive element(s) by ovis-env1 in the course of Ruminantia evolution could account for its high level of expression.
Finally, a pending question concerns bos-env4: This previously identified (23, 24) env gene has a placenta-specific expression but is not conserved in ruminants, being absent in the sheep (our present data), and therefore cannot have been the driving force involved in the emergence of the synepitheliochorial placenta in the course of evolution. However, as illustrated in Fig. 3, bos-env4 is close to the envelope gene of beta/delta retroviruses and in particular to the envelope gene of the ovine JSRV retrovirus. The latter is an unusual element, because it exists both as an oncogenic infectious retrovirus responsible for pulmonary carcinoma in sheep and as recently endogenized elements integrated during the last 5–7 million years in the sheep/goat lineage (reviewed in ref. 42). These endogenous JSRV elements (enJSRVs) are expressed in the placenta but are not placenta-specific, being expressed also in the genital tract (e.g., uterus, oviduct, and cervix) (43). However, in vivo inhibition of their expression via injection of antisense morpholino oligonucleotides into the pregnant ewe interrupted placentation (44), and therefore it has been suggested that enJSRVs contribute to some early steps of placenta formation at the peri-implantation stage. One interesting hypothesis is that the recently captured enJSRV in the sheep and bos-env4 in the cow (although there still is no conclusive evidence for their activities in cell–cell fusion) might be “nascent” syncytins that possibly today cooperate with and even, in the course of evolution, will replace the bona fide syncytins that have been and still are the active fusogenic genes involved in ruminant placenta formation. This hypothesis would be consistent with the even bolder scenario that we proposed in which the emergence of eutherian mammals was made possible by the capture of a founding retroviral env gene, subsequently replaced in the diverse mammalian lineages upon successive and independent germline infections by new retroviruses and de novo co-optation of their env gene, assuming that each new gene provides its host with a positive selective advantage. This scenario would account for both the diversity of the captured syncytins that can be found at the present time, concomitant with the diversity of the observed placental architectures, and the directional evolution of the reproductive modes from oviparity to placental viviparity.
Methods
Biological Samples, Quantitative RT-PCR, ISH, Search for syncytin-Rum1 in Other Species, Syncytin-Rum1 Expression Vector, and Fusion Assays.
See SI Methods.
Database Screening and Sequence Analyses.
Retroviral endogenous env gene sequences were searched by BLAST on the cow genome (9.5× coverage assembly of the Bos taurus genome, UCSC UMD 3.1/BosTau6, Nov. 2009). The sequences containing an ORF longer than 400 aa (from start to stop codons) were extracted from the BosTau6 genomic database using the getorf program of the EMBOSS package (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) and translated into amino acid sequences. These sequences were BLASTed against the TM subunit amino acid sequences of 35 retroviral envelope glycoproteins (from representative ERVs, among which are known syncytins, and infectious retroviruses), using the blastp program of the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/BLAST). Putative envelopes then were selected based on the presence of a hydrophobic (TM) domain (located 3′ to a highly conserved C-X5,6,7-C motif (Fig. 2). The identified Env-encoding sequences are given in Dataset S1, and their coordinates are listed in Table S2. Signal peptides were predicted using the SignalP 4.0 Server (www.cbs.dtu.dk/services/SignalP).
The cow and sheep genomes (3× coverage assembly of the Ovis aries genome, UCSC ISGC/oviAri1 Dec. 2010) were screened further with the identified Env glycoprotein sequences using the BLAST program from the NCBI. Multiple alignments of amino acid sequences were carried out by using the Seaview program and the ClustalW protocol (www.ebi.ac.uk). Maximum-likelihood phylogenetic trees were constructed with RaxML 7.3.2 (45), with bootstrap percentages computed after 1,000 replicates using the GAMMA + GTR model for the rapid bootstrapping algorithm. PAML4 (34) was used to run site-specific selection tests and obtain dN/dS ratios for all syncytin-Rum1 sequences. PAML models analyzed assumed no molecular clock (clock = 0) and a single dN/dS for all tree branches (model = 0). We used likelihood ratio tests to compare the improvement in likelihood for a model that allows for positive selection (M8) compared with a model (NS site = 7–8) that does not (M7). Each analysis ran until convergence (Small_Diff = 0.5e-6). The control file is available upon request. HyPhy (33) was used on the datamonkey web server (www.datamonkey.org) to run site-specific REL and FEL tests.
The genome assemblies of the dolphin (Tursiops truncatus; Ensembl, turTru1, 2008), alpaga (Vicugna pacos; Ensembl, vicPac1, 2008), pig [Sus scrofa; UCSC Swine Genome Sequencing Consortium (SGSC) Sscrofa 9.2], horse (Equus caballus; UCSC Broad/equCab2, 2007), dog (Canis lupus familiaris; UCSC Broad CanFam3.1, 2011), human (Homo sapiens; UCSC GRCh37/hg19, 2009), and mouse (Mus musculus; UCSC NCBI37/mm9, 2007) were screened for the presence of the identified syncytin-Rum1 sequence either by locating syntenic genomic regions using the UCSC genome browser (http://genome.ucsc.edu/) or by identifying homologous sequences using the Blast program.
Supplementary Material
Acknowledgments
We thank E. Campion for contributions to the ISH experiments; S. Camous, P. Chavatte-Palmer, Y. Heyman, N. Mansouri-Attia, and O. Sandra for providing bovine and ovine tissues; P. Bolifraud, K. Al Gubory, and B. Mandon-Pepin for help in collecting ovine tissues; the staff of the experimental farms of Bressonvilliers and Brouessy for animal management; Dr. I. Arroyo Lopez, J.-L. Berthier, G. Dousseau, B. Guffond, F. Hergueta-Claro, L. E. Olson, Dr. R. Potier, C. Rejaud, J. Rigoulet, and C. Voisin for tissue or blood samples; S. Blaise, J. Richardson, and S. Fichelson for the gift of A72, PK, and LOK cell lines; O. Bawa for her contribution to the histological analyses; and P. Dessen for assistance in the computing work. This work was supported by the Centre National de la Recherche Scientifique and the Institut National de la Recherche Agronomique (INRA) and by grants from the Ligue Nationale contre Le Cancer (Equipe Labellisée) (T.H.), the INRA (Agenae-2), the European Union (European Network of Excellence on Embryo Implantation Control), and the Agence Nationale de la Recherche MECABPA. As a post-doctoral fellow, S.A.D. was supported by the PremUp Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX412964–JX412985).
See Author Summary on page 3227 (volume 110, number 9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215787110/-/DCSupplemental.
References
- 1.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26(3):291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 2.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 3.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta. 2012;33(9):663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74(7):3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 6.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100(22):13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangeney M, et al. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104(51):20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102(3):725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci USA. 2012;109(7):E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106(29):12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupressoir A, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA. 2011;108(46):1164–1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leiser R, Kaufmann P. Placental structure: In a comparative aspect. Exp Clin Endocrinol. 1994;102(3):122–134. doi: 10.1055/s-0029-1211275. [DOI] [PubMed] [Google Scholar]
- 14.Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod Biol Endocrinol. 2004;2:46. doi: 10.1186/1477-7827-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wooding P, Burton GJ. Comparative Placentation: Structures, Functions and Evolution. Berlin: Springer; 2008. [Google Scholar]
- 16.Wathes DC, Wooding FB. An electron microscopic study of implantation in the cow. Am J Anat. 1980;159(3):285–306. doi: 10.1002/aja.1001590305. [DOI] [PubMed] [Google Scholar]
- 17.Wooding FB. Role of binucleate cells in fetomaternal cell fusion at implantation in the sheep. Am J Anat. 1984;170(2):233–250. doi: 10.1002/aja.1001700208. [DOI] [PubMed] [Google Scholar]
- 18.Wooding FB. Current topic: The synepitheliochorial placenta of ruminants: Binucleate cell fusions and hormone production. Placenta. 1992;13(2):101–113. doi: 10.1016/0143-4004(92)90025-o. [DOI] [PubMed] [Google Scholar]
- 19.Hradecký P, Mossman HW, Stott GG. Comparative histology of antelope placentomes. Theriogenology. 1988;29(3):693–714. doi: 10.1016/s0093-691x(88)80015-3. [DOI] [PubMed] [Google Scholar]
- 20.Wooding FB, Morgan G, Adam CL. Structure and function in the ruminant synepitheliochorial placenta: Central role of the trophoblast binucleate cell in deer. Microsc Res Tech. 1997;38(1-2):88–99. doi: 10.1002/(SICI)1097-0029(19970701/15)38:1/2<88::AID-JEMT10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho AF, Klisch K, Miglino MA, Pereira FTV, Bevilacqua E. Binucleate trophoblast giant cells in the water buffalo (Bubalus bubalis) placenta. J Morphol. 2006;267(1):50–56. doi: 10.1002/jmor.10387. [DOI] [PubMed] [Google Scholar]
- 22.Wooding FB, Kimura J, Fukuta K, Forhead AJ. A light and electron microscopical study of the Tragulid (mouse deer) placenta. Placenta. 2007;28(10):1039–1048. doi: 10.1016/j.placenta.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Baba K, et al. Identification of novel endogenous betaretroviruses which are transcribed in the bovine placenta. J Virol. 2011;85(3):1237–1245. doi: 10.1128/JVI.01234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshi K, et al. Bovine trophoblastic cell differentiation and binucleation involves enhanced endogenous retrovirus element expression. Reprod Biol Endocrinol. 2012;10:41. doi: 10.1186/1477-7827-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanstrom R, Wills JW. Synthesis, assembly and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 26.Vogt VM. 1997. Retroviral virions and genome. Retroviruses, ed Coffin JM, Hughes SH, Varmus, HE (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), pp 27–69.
- 27.Blaise S, Ruggieri A, Dewannieux M, Cosset F-L, Heidmann T. Identification of an envelope protein from the FRD family of Human Endogenous Retroviruses (HERV-FRD) conferring infectivity on retroviral particles and functional conservation among simians. J Virol. 2004;78(2):1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redmond S, Peters G, Dickson C. Mouse mammary tumor virus can mediate cell fusion at reduced pH. Virology. 1984;133(2):393–402. doi: 10.1016/0042-6822(84)90405-7. [DOI] [PubMed] [Google Scholar]
- 29.Côté M, Zheng Y-M, Liu S-L. Receptor binding and low pH coactivate oncogenic retrovirus envelope-mediated fusion. J Virol. 2009;83(22):11447–11455. doi: 10.1128/JVI.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rein A, Mirro J, Haynes JG, Ernst SM, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68(3):1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wooding FB. Frequency and localization of binucleate cells in the placentomes of ruminants. Placenta. 1983;4(Spec No):527–539. [PubMed] [Google Scholar]
- 32.Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155(1):431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosakovsky Pond SL, Frost SD. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida I, et al. Change of positive selection pressure on HIV-1 envelope gene inferred by early and recent samples. PLoS ONE. 2011;6(4):e18630. doi: 10.1371/journal.pone.0018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malassiné A, et al. Expression of the fusogenic HERV-FRD Env glycoprotein (syncytin 2) in human placenta is restricted to villous cytotrophoblastic cells. Placenta. 2007;28(2-3):185–191. doi: 10.1016/j.placenta.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Schlecht-Louf G, et al. Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc Natl Acad Sci USA. 2010;107(8):3782–3787. doi: 10.1073/pnas.0913122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anson-Cartwright L, et al. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet. 2000;25(3):311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber J, et al. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol Cell Biol. 2000;20(7):2466–2474. doi: 10.1128/mcb.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu CY, et al. GCMa regulates the syncytin-mediated trophoblastic fusion. J Biol Chem. 2002;277(51):50062–50068. doi: 10.1074/jbc.M209316200. [DOI] [PubMed] [Google Scholar]
- 41.Simmons DG, et al. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development. 2008;135(12):2083–2091. doi: 10.1242/dev.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spencer TE, Palmarini M. Endogenous retroviruses of sheep: A model system for understanding physiological adaptation to an evolving ruminant genome. J Reprod Dev. 2012;58(1):33–37. doi: 10.1262/jrd.2011-026. [DOI] [PubMed] [Google Scholar]
- 43.Palmarini M, et al. Expression of endogenous betaretroviruses in the ovine uterus: Effects of neonatal age, estrous cycle, pregnancy, and progesterone. J Virol. 2001;75(23):11319–11327. doi: 10.1128/JVI.75.23.11319-11327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunlap KA, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103(39):14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 46.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 47.Decker JE, et al. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci USA. 2009;106(44):18644–18649. doi: 10.1073/pnas.0904691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hassanin A, et al. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol. 2012;335(1):32–50. doi: 10.1016/j.crvi.2011.11.002. [DOI] [PubMed] [Google Scholar]