Abstract
Allopolyploidization has been a driving force in plant evolution. Formation of common wheat (Triticum aestivum L.) represents a classic example of successful speciation via allopolyploidy. Nevertheless, the immediate chromosomal consequences of allopolyploidization in wheat remain largely unexplored. We report here an in-depth investigation on transgenerational chromosomal variation in resynthesized allohexaploid wheats that are identical in genome constitution to common wheat. We deployed sequential FISH, genomic in situ hybridization (GISH), and homeolog-specific pyrosequencing, which enabled unequivocal identification of each of the 21 homologous chromosome pairs in each of >1,000 individual plants from 16 independent lines. We report that whole-chromosome aneuploidy occurred ubiquitously in early generations (from selfed generation S1 to >S20) of wheat allohexaploidy although at highly variable frequencies (20–100%). In contrast, other types of gross structural variations were scant. Aneuploidy included an unexpected hidden type, which had a euploid chromosome number of 2n = 42 but with simultaneous loss and gain of nonhomeologous chromosomes. Of the three constituent subgenomes, B showed the most lability for aneuploidy, followed by A, but the recently added D subgenome was largely stable in most of the studied lines. Chromosome loss and gain were also unequal across the 21 homologous chromosome pairs. Pedigree analysis showed no evidence for progressive karyotype stabilization even with multigenerational selection for euploidy. Profiling of two traits directly related to reproductive fitness showed that although pollen viability was generally reduced by aneuploidy, the adverse effect of aneuploidy on seed-set is dependent on both aneuploidy type and synthetic line.
Keywords: chromosome dynamics, hidden aneuploidy, synthetic wheat, wheat evolution
Hexaploid common wheat (Triticum aestivum L.) is a major food crop with international significance, the evolution of which is characterized by two sequential allopolyploidization events: one leading to formation of allotetraploid wheat (T. turgidum L.) and the other to allohexaploid wheat (T. aestivum) (1, 2). Despite decades of research, the mechanisms by which the initial allopolyploid individuals became stabilized, established, and accumulate to successful speciation remains largely unknown in this important crop.
In theory, chromosome-level perturbation should be among the first manifestations of nascent allopolyploidization. Indeed, two recent molecular cytogenetic studies, in resynthesized allotetraploid Brassica napus lines (3) and young natural allotetraploid Tragopogon miscellus populations (4), respectively, have provided unique insights into the chromosomal dynamics associated with nascent allotetraploidy. Being at the resolution of individual chromosomes, these studies have documented a surprisingly high incidence of both structural and numerical changes in nascent allotetraploid plants (3, 4). It was found that early generations of resynthesized allotetraploid Brassica plants already involved extensive aneuploidy, which further accrued in successive generations and reached up to 95% by the 10th selfed generation (3). Even more surprising, all six naturally established allotetraploid Tragopogon populations investigated, which were approximately 40 generations old, still contained 69% aneuploidy (4). Several common features were associated with chromosomal variations between these two phylogenetically divergent plant taxa, including concomitant numerical and structural changes and near exclusive dependency on compensating homeologous relationships for chromosome loss and gain, implicating gene dosage balance as a major constraining mechanism for aneuploid retention (3–5).
Because common wheat is an allohexaploid, it evolved a unique genetic control of chromosome pairing, the Pairing homeologs 1 (Ph1) gene that restricts meiotic pairing exclusively to intragenomic homologous chromosomes (6). Thus, one might expect fundamental differences in chromosomal stability during the early generations of allohexaploid wheat formation from that of allotetraploid plants, such as Brassica and Tragopogon, which probably also lack a strong genetic system for strict homologous pairing control (3, 4; but see also ref. 7). Unexpectedly, however, a previous study showed that the initial generations (S0 to S2) of newly synthesized allohexaploid wheat containing the active Ph1 gene was also associated with meiotic irregularity and aneuploidy that occurred in a parent-dependent manner (8). In particular, this study reported what they called “apparent aneuploidy,” referring to the plants that had an euploid chromosome number (2n = 42) but with loss and gain of chromosomes (8). Nonetheless, being based on genomic in situ hybridization (GISH) analysis that only enables discrimination at the subgenome level (A, B, and D), this study was unable to monitor loss, retention, or gain of individual chromosomes, nor was it able to monitor relationships between the aneuploid chromosomes. Furthermore, duration of the detected aneuploidy was not systemically investigated, which would entail pedigree analysis across multiple generations.
To learn more about the immediate chromosomal consequences following allohexaploidization in wheat, we conducted a detailed investigation on chromosomal composition of a set of 16 independently synthesized allohexaploid wheat lines and on transgenerational chromosome dynamics in two representative lines; all these lines had the same genome constitution (2n = 6x = 42, BBAADD) as natural common wheat, and hence, are likely recapitulating the initial chromosomal trajectories of speciation in T. aestivum. We performed sequential FISH and GISH analyses followed by homeolog-specific pyrosequencing, which enabled identification of each individual homologous chromosome pair. Using these analyses, genome and chromosome bias for aneuploidy was determined. Furthermore, by pedigree analysis and selection for euploidy across multiple successive generations, we studied propensities of euploidy vs. aneuploidy. We also measured fitness consequences of the various types of aneuploidy relative to euploidy in two representative lines. We discuss our findings in the context of common as well as unique features of nascent aneuploidy in allohexaploid wheat compared with other plant taxa, and possible mechanisms causing the extensive aneuploidy.
Results
Whole-Chromosome Aneuploidy Was Associated with All Studied Synthetic Allohexaploid Wheat Lines.
We established a combination of FISH and GISH procedure that allows us to routinely identify all 21 wheat chromosome pairs (Fig. 1 and Fig. S1). Using this methodology, we karyotyped a set of 16 independent synthetic allohexaploid wheat lines. These lines were independently developed using diverse accessions of tetraploid wheat T. turgidum ssp. durum or ssp. carthlicum (2n = 4x = 28, BBAA) and Aegilops tauschii (2n = 2x = 14, DD). For each line we karyotyped from 30 to 75 arbitrarily selected individual plants from the latest generation available, which ranged from S8 to >S20 among different lines (Dataset S1).
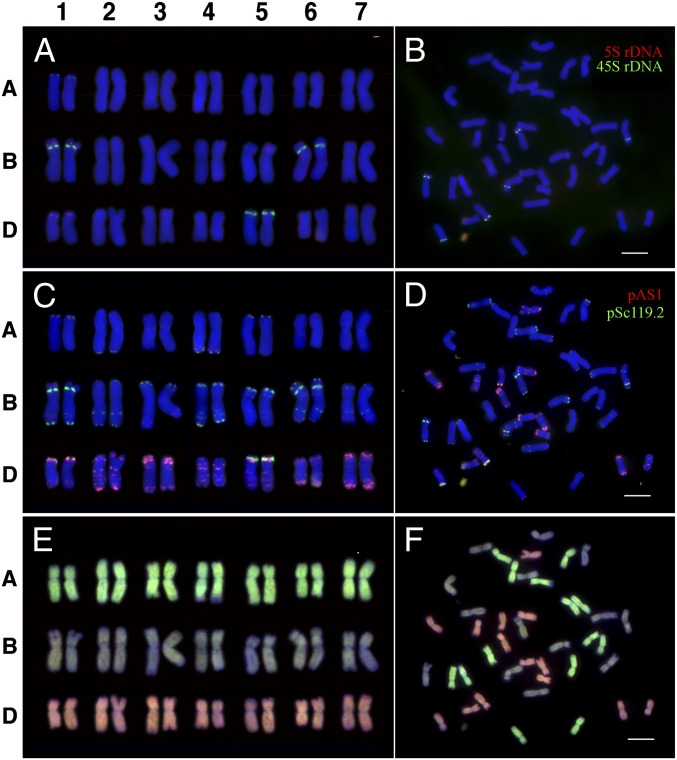
Fig. 1.
An example of karyotyping of a single plant from the nascent allohexaploid wheat line 960. (A) Karyotype based on FISH using 5S and 45S rDNA probes. Chromosomes 1A, 1B, 1D, 5A, 5B, 5D, and 6B can be identified. (B) The original FISH image used for the karyotype in A. (C) Karyotype based on FISH using two repetitive DNA probes, pSc119.2 and pAS1. All 21 wheat chromosomes can be identified based on these two repeats and the two rDNA probes. (D) The original FISH image used for the karyotype in C. (E) Karyotype based on GISH. The A (green), B (blue), and D (pink) subgenome chromosomes can be distinguished. (F) The original FISH image used for the karyotype in E. (Scale bars, 10 µm.)
We analyzed the chromosome composition of 580 plants and found that whole-chromosome aneuploidy occurred in all 16 lines (Dataset S1). The frequencies of aneuploidy varied from approximately 20–100% among the plants from each line, with chromosome numbers ranging from 39 to 45 (Dataset S1). All aneuploidy occurred in a per plant basis; that is, no plant contained two or more types of aneuploid cells, which is in contrast with synthesized Arabidopsis allohexaploids (9). Surprisingly, we did not identify any unambiguous chromosomal translocation/inversion or duplication/deletion in any of the 580 plants, although we cannot exclude the presence of submicroscopic chromosomal rearrangements. These results showed that whole-chromosome aneuploidy is generally associated with nascent allohexaploidization in wheat, but other gross structural changes, if they exist, are minimal.
Transgenerational Chromosome Number Dynamics and Hidden Aneuploidy.
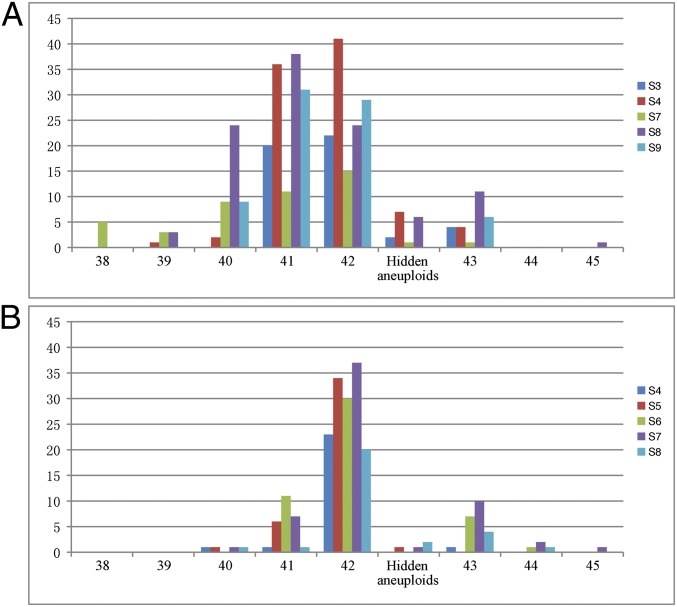
We chose two lines (960 and AT5) to investigate transgenerational chromosome number dynamics, because seeds from different generations were available for both of these lines. We analyzed five different generations each of 960 (S3, S4, and S7–S9) and AT5 (S4–S8). For each generation, from 26 to 107 randomly selected individuals were karyotyped, giving a total of 571 individual plants analyzed from both lines, of which 366 belong to 960 and 205 belong to AT5 (Fig. 2). Both 960 and AT5 showed variable chromosome numbers in each generation; the chromosome numbers across all five generations ranged from 38 to 45 for 960, and 40–45 for AT5 (Fig. 2). However, 960 showed much higher frequencies of aneuploidy than AT5 in each generation. The loss or gain of a single chromosome [i.e., monosomy (2n = 41) or trisomy (2n = 43)] was the most frequent aneuploidy types if both lines were considered together, but the types with 2n = 40 chromosomes were also prominent in 960 (Fig. 2).
Fig. 2.
Chromosome number distribution in five different selfed generations of (A) 960 (S3, S4, and S7–S9) and (B) AT5 (S4–S8). Chromosome numbers were determined based on karyotyping of individual plants. Hidden aneuploids refer to those with a euploid chromosome number (2n = 42) but simultaneous loss and gain of different chromosomes. For each generation, from 26 to 107 randomly selected individuals were karyotyped (SI Materials and Methods). x axis = number of chromosomes and y axis = number of individuals.
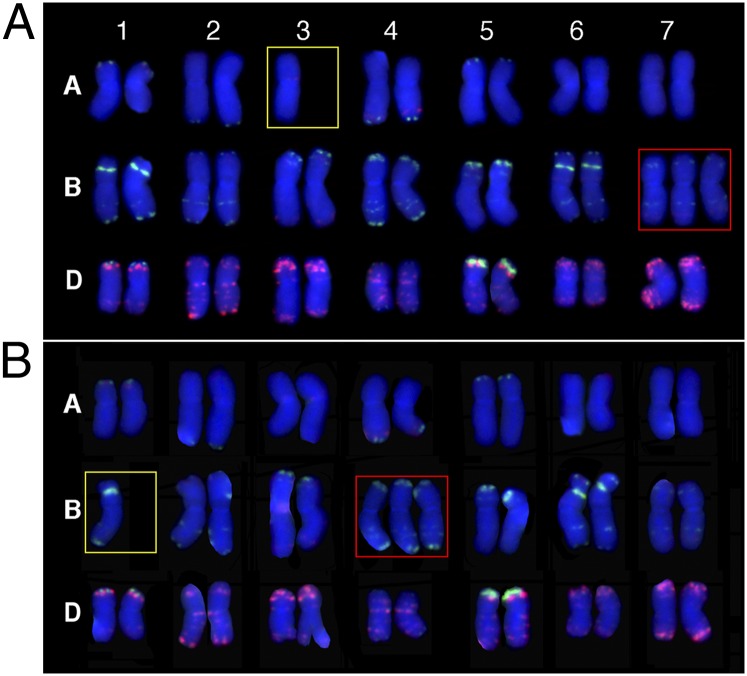
We discovered a unique class of aneuploidy in both lines, which contained a chromosome number of 2n = 42 but included a combination of loss of one chromosome and gain of another chromosome. We hereby define this category of aneuploidy as “hidden aneuploidy,” as it would have been identified as euploidy by simple chromosome counting. The hidden aneuploidy could be further divided into two types: type I with the aneuploid chromosomes from different genomes, and type II with the aneuploid chromosomes from a single genome, which would be undetected by GISH analysis alone (Fig. 3). Surprisingly, of a total of 20 hidden aneuploid individuals identified in the two lines, none was involved in chromosomes associated with the same homeologous group.
Fig. 3.
Examples of the two types (I and II) of “hidden aneuploidy” observed in 960. (A) Karyotype of a type I hidden aneuploid with a 2n chromosome number of 42, but a genomic constitution of 38 + 1 (3A) + 3 (7B). (B) Karyotype of a type II hidden aneuploid with a 2n chromosome number of 42, but a genomic constitution of 38 + 1(1B) + 3(4B). Yellow and red boxes respectively mark the lost and gained chromosomes.
Chromosome Loss or Gain Showed Genome and Chromosome Biases.
Although the 16 lines differed substantially in aneuploid frequencies, gain or loss of individual chromosomes in all lines as a whole was associated with recognizable genome biases (Fig. S2). Specifically, the B subgenome showed the highest frequency of chromosome loss (P < 2.2e-16) (Fig. S2), but A was also significantly higher than D (P < 2.2e-16) (Fig. S2); all three genomes were significantly different from each other in chromosome gain (P = 8.383e-06), with B showing the highest frequency, followed by D and A (Fig. S2). Taking both loss and gain together across the 16 lines, the B genome showed the most lability and the D genome was most stable in these nascent allohexaploid wheats (Fig. S2).
In the two lines, 960 and AT5, for which we obtained data for multiple consecutive generations, individual chromosome biases for loss or gain were analyzed both as a constituent genome and as an individual chromosome. For genome biases, the B subgenome in 960 showed a significantly higher frequency of chromosome loss than did the A and D subgenomes (P < 2.2e-16), but no significant difference was observed between A and D (P = 0.09538) (Fig. S3A). No significant difference was found among the three subgenomes for chromosome gain (P = 0.1274) in 960 (Fig. S3A). In AT5, significant difference was detected for chromosome loss among the three genomes (P value = 0.01787), with D showing the highest frequency of loss (Fig. S3B), but A and B did not show significant difference (P = 1). Genomes B and A in AT5 showed a similar frequency of chromosome gain (P = 1). A significant difference was detected for chromosome gain between each two of the three genomes in AT5 (P = 0.002821 for A vs. B, P = 1.327e-06 for B vs. D, and P = 0.02219 for A vs. D). For individual chromosome biases, in 960 chromosome 1B showed the highest frequency of aneuploidy (Fig. S3C). Gain or loss of 1B was found in 25% of the plants. Aneuploidy of chromosomes 4B and 6B were associated with 14% and 10% of the plants. Among the A genome chromosomes, 6A was most frequently associated aneuploidy (9% of the plants) (Fig. S3C). Only one chromosome (2D) showed complete euploidy in the total of 366 plants analyzed in 960 (Fig. 4C). In AT5, chromosome 5B showed the highest frequency (41%) of aneuploidy, followed by chromosome 1A (20%) (Fig.S3D). Eight chromosomes (2A, 2D, 3A, 3B, 3D, 5D, 7A, and 7D) did not show aneuploidy in the total of 205 plants analyzed in AT5.
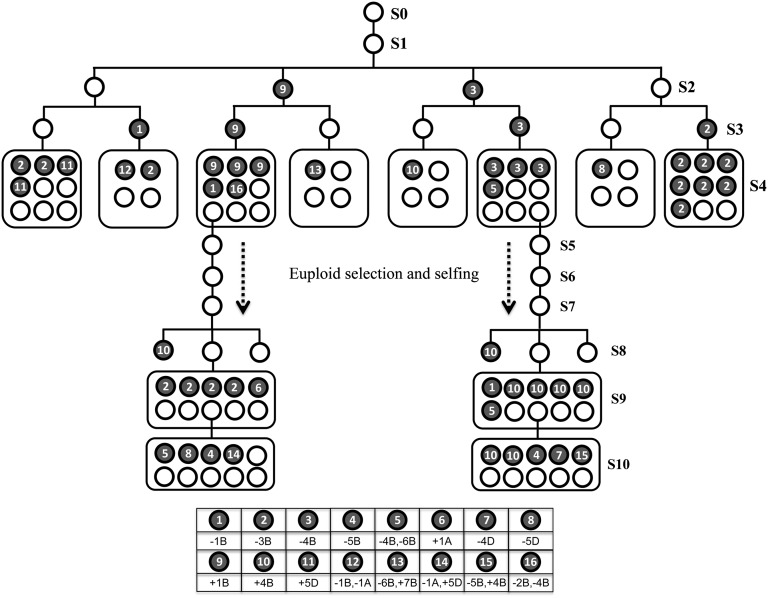
Fig. 4.
Transgenerational chromosome number variation based on karyotyping of a total of 111 individual plants across 11 consecutive selfed-generations (S0–S10) of 960. A single euploid S1 plant was selected from a euploid S0 plant. Then, two euploid S2 plants and the first two S2 aneuploid plants (identified as a trisomic 1B plant and a monosomic 4B plant) were selected to produce S3. In S3, one euploid plant and the first S3 aneuploid plant from each S2 plant were selected (the exact aneuploid karyotypes were shown). From four to nine S4 plants derived from each of the eight S3 plants were randomly selected and karyotyped (the exact aneuploid karyotypes were shown). Two independent euploid S4 plants of different lineages were then used to generate euploidy S5, S6, S7, and S8 plants sequentially (transgenerational persistent selection for euploidy). Ten randomly selected S9 plants from each of the two euploid S8 plants were karyotyped (the exact aneuploid karyotypes were shown). Empty and filled circles denote for euploid and aneuploid plants, respectively, and the exact euploid karyotypes were designated at the bottom.
Karyotype Stabilization Was Not Accomplished by Transgenerational Selection.
We next explored whether karyotype stabilization can be accomplished by transgenerational selection for euploidy. We started with a single euploid S0 plant from 960 and conducted pedigree analysis for propensities of euploid vs. aneuploid siblings (Fig. 4). We identified a single euploid S1 plant, and selected four S2 plants, including one trisomic 1B plant, one monosomic 4B plant, and two euploid plants. We then selected one euploid and one aneuploid S3 plants from each S2 plant. Several S4 plants derived from each of the eight S3 plants were karyotyped. Both euploidy and aneuploidy plants were found in each of the eight S4 populations. Two independent euploid S4 plants were then used to generate euploid S5, S6, S7, and S8 plants sequentially (Fig. 4). Ten randomly selected S9 plants from each of the two euploid S8 plants were karyotyped. Nearly 50% of aneuploid plants were found in the two S9 populations, and in the two similarly developed S10 populations (Fig. 4). The same analysis was performed in AT5 and similar results were obtained (Fig. S4). Thus, the karyotypes of 960 and AT5 were not stabilized after 9–10 consecutive generations of selection for euploidy.
Molecular Cytogenetic Karyotyping Was Validated by Homeolog-Specific Pyrosequencing.
A set of 15 pyrosequencing primers (Dataset S2) was designed based on the homeolog-specific SNPs identified from the 15 unique genes representing seven groups of homeologous chromosome in the allohexaploid wheat (SI Materials and Methods). The locus-specific gene primers amplify products from the three homeologous copies of a gene, which later serve as temple for pyrosequencing to enable unequivocal diagnosis of both loss and gain of any individual chromosome from the wheat chromosome complement (Fig. S5).
We performed pyrosequencing on a subset of 211 plants from 960, which were individually karyotyped and represent eight (S2–S9) of the 11 selfed generations (S0–S10). The sequencing data perfectly matched with the karyotyping results (Fig. S6). However, in a few cases the pyrosequencing data indicated loss or gain of additional chromosomes that were incongruent with their karyotypes. However, independent cytological examination of these plants confirmed their chromosome numbers being consistent with the original karyotyping results, suggesting that the occasional incongruent pyrosequencing data may have been caused by localized deletions, duplications, or rearrangements of the targeted sequences (10). Pyrosequencing data were also obtained for a subset of 128 plants of AT5, which were individually karyotyped and represent five (S4–S8) of the nine selfed generations (S0–S8). We observed similar high levels of consistency between the two analytical methods for this line (Fig. S7).
Impact of Aneuploidy on Pollen Viability and Seed-Setting.
We assessed the impact of aneuploidy on two traits related to reproductive fitness, pollen viability and seed-setting, in both 960 and AT5 under glasshouse conditions. The pollen viability of all aneuploid plants (2n = 40, 41, 43, and hidden aneuploidy) as a whole was significantly reduced compared with that of euploid plants (P = 2.04E-04) in 960 (Fig. S8A). Aneuploidy as a whole was significantly associated with reduced seed-set (P = 0.003319), further analysis indicated that only two types of aneuploid plants: for example, those missing two chromosomes (2n = 40), either nullisomy (missing a pair of chromosomes) or double monosomy (missing two nonhomologous chromosomes), and hidden aneuploidy, produced significantly less seeds than that of euploid plants (P = 1.50E-06 for 2n = 40 vs. euploidy, and P = 0.00024 for hidden aneuploidy vs.. euploidy) in this line (Fig. S8B).
We also examined the impact of aneuploidy associated with each of the 21 chromosomes on seed-setting in 960. Aneuploidy of three chromosomes (3A, 3B, and 5A) produced significantly lower seed-set than euploidy (P = 0.00351, 0.00499, and 0.00032, respectively), but aneuploidy of the remaining 18 chromosomes did not cause a significant reduction in seed-set compared with euploid plants (Fig. S8C).
We conducted similar analysis for AT5. The pollen viability of all four aneuploid types (2n = 40, 41, 43, and hidden aneuploidy) as a whole was also significantly reduced compared with that of euploidy (P = 1.08E-05) (Fig. S9). Similar to 960, significant reduction in seed-set was observed between all types of aneuploidy as a whole and euploid plants (P = 4.77E-07). Different from 960, however, further analysis showed that all four types of aneuploidy (2n = 40, 41, 43, and hidden) showed significantly lower seed-set than euploidy (P values ranging from 0.00026 to 0.0032) (Fig. S9).
Discussion
Persistence of Whole-Chromosome Aneuploidy in the Early Generations Postallohexaploidization in Wheat and Its Possible Cause.
Previous studies have reported that allopolyploidization in wheat induces extensive genomic changes at the molecular level, including rapid elimination of coding and noncoding DNA sequences, epigenetic modifications and altered patterns of gene expression (11–14). Conceivably, these rapid genetic and epigenetic alterations have played important roles in overcoming the unforeseeable genomic conflicts encountered in nascent allopolyploidy (12, 15–21). Nonetheless, chromosome-level variations subsequent to the allopolyploidization have not been fully investigated in wheat (8). One possible reason for this situation likely lies in the generally accepted assumption that, because of the presence of the Ph1 gene in polyploid wheats, which suppresses meiotic pairing between homeologous chromosomes and ensures strict homologous pairing (6, 11), chromosome stability is automatically achieved upon allohexaploidization. Thus, the demonstration that chromosomal aneuploidy widely occurred at the initial generations of newly synthesized allohexaploid wheat lines came as a surprise (8). However, because of limitations of resolution (GISH) and number of generations (S0 to S2) studied in Mestiri et al. (8), the issues of generality and persistency of aneuploidy at the individual-chromosome level associated with nascent wheat allohexaploidization remained unknown.
We addressed these issues by using a set of 16 independently synthesized allohexaploid wheat lines, and by karyotyping >1,000 individual plants at different selfing-generations after allohexaploidization. This large-scale karyotyping at the resolution level of individual chromosomes has established that whole-chromosome aneuploidy occurred ubiquitously in nascent allohexaploid wheat, albeit at highly variable frequencies across the diverse parental combinations. More importantly, we demonstrate that aneuploidy was persistent, and karyotypic stabilization was not achieved even by consecutive selection for euploidy over multiple generations. To our knowledge, no previous investigation has been conducted to purposely select for euploidy over multiple generations in any nascent allopolyploid taxon, thus leaving the general impression that karyotype stabilization for euploidy could be accomplished by selection. Our results indicate that this might not be the case, and suggests that additional mechanisms are likely required for karyotype stabilization postallopolyploidization. Our findings, however, raise a conundrum, as all natural common wheat varieties are known to be chromosomally stable, producing only 1–3% aneuploid individuals under normal conditions (22). Thus, the mechanisms by which karyotype stabilization has been achieved in natural common wheat remain an open question. Although we cannot exclude the possibility that the actual parental genotypes involved in the formation of the allohexaploid individuals leading to common wheat speciation did not generate the persistent aneuploidy as we observed in this study, this seems unlikely given the diverse parental combinations we analyzed.
With respect to the generality of aneuploidy associated with nascent allopolyploidization, our results are in full agreement with recent studies in resynthesized Brassica allotetraploid lines (3) and the newly formed (approximately 40 generations old) natural Tragopogon allotetraploid populations (4). Both studies showed unexpected high frequencies of chromosomal aneuploidy, and in the case of Brassica also transgenerational accumulation of aneuploidy, which were not detected in previous studies with lower resolution cytogenetic tools (3, 4). Notably, in the Brassica study, all 50 lines analyzed were derived from a single parental combination, albeit each line being independent starting from F1 (3). Although for Tragopogon, it is not possible to know the exact generation or to follow transgenerational trend as they are natural populations (4). Thus, our data in wheat, being based on multiple independent lines, pedigree analysis, as well as purposeful selection for euploidy across multiple generations, have added a new layer of complexity to the immediate chromosomal consequences of allopolyploidization; that is, aneuploidy not only generally occurs but also is transgenerationally persistent, and moreover, karyotype stabilization cannot be rapidly accomplished by selection for euploidy.
The occurrence of organismal aneuploidy on a per plant basis (i.e., lack of somatic mosaics) is apparently because of compromised fidelity of the meiotic machinery, but mitosis remains unaffected. For example, both multivalent formation because of nonexclusive homologous pairing and homologous pairing but with relaxed bivalent formation may cause chromosome mis-segregation and, hence, lead to production of unbalanced gametes. If these unbalanced gametes are still functional and competitive, then zygotic, and by extension organismal aneuploidy is almost ensured. Indeed, lack of strict control on exclusive homologous pairing and multivalent formation have been suggested as a major cause for aneuploidy, as well as other structural changes, in most newly formed plant allopolyploids (3, 4, 23). In wheat, however, because of the presence of the Ph1 gene, exclusive homologous pairing and bivalent formation at metaphase I (MI) are secured, which seemed also true in newly synthesized allohexaploid wheats (8). However, bivalent formation at MI does not imply absence of nonspecific interactions of homeologs or even nonhomologs at earlier stages of meiosis. In fact, it has been clearly demonstrated by electron micrographs of serial sections from nuclei that extensive multivalent formation and interlocking between nonhomologous chromosomes generally occur during zygotene in the standard cultivar (Chinese Spring) of common wheat. These multivalents are, however, subsequently corrected through turnover of the central region of the synaptominal complex, to yield only bivalents at pachytene and MI (24) (see, for example, figure 11 in ref. 24). Thus, crossing over occuring after (not before) completion of the pairing correction is essential to ensure exclusive bivalent formation in natural common wheat (24). It has been shown in yeast nascent polyploids that essential components of the cellular machinery may not scale proportionally with a sudden change of ploidy level (25), which can result in geometric constraint and cause mis-segregation of chromosomes during meiosis (25). Thus, it is conceivable that if similar geometric constraint is also associated with newly formed wheat allopolyploids, then correcting of the zygotene multivalents might be undermined in respect to efficacy and timing, hence, resulting in chromosome mis-segregation.
Genome and Chromosome Biases of Aneuploidy in Nascent Allohexaploid Wheat.
We found that chromosomal aneuploidy (loss or gain) showed clear biases both between the two parental genomes and across the individual chromosomes in the resynthesized Brassica allotetraploid lines (3). In contrast, aneuploidy did not show a consistent genome bias in Tragopogon across the six studied natural populations (4). We found in wheat that of the three constituent subgenomes, B showed the highest frequencies of chromosome loss or gain, followed by A, but the recently added D genome showed the most stability in the majority of the lines analyzed. This result is surprising given that genomes B and A, constituting the natural allotetraploid wheat species (T. turgidum), have coexisted for about 0.5 million y since allotetraploidization-mediated speciation (26), and therefore, one might think that the newly introduced D genome from A. tauschii would be more unstable. Nonetheless, our unexpected observation is reminiscent of the classic work that showed that whenever the D genome of A. tauschii was a constituent genome of a naturally occurring allopolyploid species in the Aegilops-Triticum complex (including T. aestivum), it functions as the so-called “pivotal genome” that underwent little alteration in terms of its encoded phenotypic traits over evolutionary time, but those of its coexisting genomes were often significantly modified (27). This finding is also in line with the recently proposed hypothesis that whenever whole genome duplication occurs (irrespective of auto- or allopolyploidy), physical diploidization occurs preferentially in only one of the two constituent genomes, the so-called under-dominant genome (28). Thus, in our case it is likely that the D genome of A. tauschii possessed the unique property of being self-stable (pivotal) but causing instability of its coinhabiting genomes once being merged with other genomes by allopolyploidization.
Features of Chromosomal Variation Uniquely Found in Nascent Allohexaploid Wheat.
We uncovered in the nascent allohexaploid wheat lines two features of chromosomal variation that were not shared by previous studies of other plant taxa. In allotetraploid Brassica (3) and Tragopogon (4), chromosome variations were found to include equally prominent numerical changes (aneuploidy) and other gross structural alterations (e.g., intra- and intergenomic rearrangements, chromosome breakage/fusion, and loss of rDNA and other repeats). In contrast, gross structural changes in the form of intra- or intergenomic rearrangements were not identified among the >1,000 plants we studied. Moreover, discernible alteration targeted by the four DNA repeats (including 45S rDNA, 5S rDNA, pAS1, and pSc119.2) that were used as FISH probes were not observed; instead, loss or gain of signals for these repeats was always consistent with loss or gain of the chromosomes bearing the signals. Although we cannot exclude the occurrence of small-scale structure alterations, especially those confined to a given genome or chromosome (e.g., small inversions), it appears safe to conclude that chromosome-level variation associated with nascent allohexaploidization in wheat is predominantly numerical (i.e., whole-chromosome aneuploidy). This finding is consistent with the established genome constitution of natural common wheat (T. aestivum), in which intergenomic translocations are uncommon (29, 30). In contrast, there are extensive intergenomic translocations associated with many other established allopolyploid crops. For example, at least nine intergenomic translocations were detected in Nicotiana tabacum (31), 18 in Avena sativa (32), and numerous in B. napus (7). A possible explanation for the near-absence of structural chromosomal changes in nascent allohexaploid wheat is function of the Ph1 gene inherited from the tetraploid wheat. By restricting meiotic pairing to homologous chromosomes only, and hence minimizing pairing between homeologs, intergenomic rearrangements are prevented, which indeed were found as a major cause of structural changes in the Brassica synthetic allotetraploids (3, 7, 20, 23).
Another salient feature of aneuploidy found in wheat is the lack of dependency on homeologous compensation of the variable chromosomes. This feature is especially striking in the case of hidden aneuploidy: of a total of 20 such plants identified (2n = 42, but with simultaneous loss and gain of chromosomes), none was found to involve homeologous chromosomes. This finding is in contrast with the situations of both Brassica and Tragopogon, where loss and gain of chromosomes were found to predominantly involve homeologous chromosomes [e.g., compensatory monosomy/trisomy and nullisomy/tetrasomy (3)], explained by the gene-dosage balance hypothesis (5). One possible explanation for the absence of homeologous compensation in wheat is its hexaploid nature, in which the adverse effect of gene-dosage imbalance imposed by a given individual chromosome is less relative to that in tetraploidy. It should be noted that our results do not imply that chromosomal level genome balance (5) is less critical in wheat; it is likely the extent that makes the difference. Indeed, homeologous chromosome compensation is essential as well as sufficient for correcting the detrimental phenotypes of nullisomy (both copies of a homologous chromosome pair being lost) in natural common wheat, as established by the seminal work of Longwell and Sears (33). Why such compensatory aneuploidy rarely occurs in nascent allohexaploid wheats but the other types of aneuploidy are all prevalent remains unknown.
Reproductive Fitness Consequences of Aneuploidy in Nascent Allohexaploid Wheat and Their Evolutionary Implications.
It is commonly accepted that aneuploidy imposes adverse phenotypic consequences in general and compromised reproductive fitness in particular, a phenomenon collectively termed “aneuploidy syndrome” (34). For example, both pollen viability and seed yield were found to be significantly reduced by aneuploidy in resynthesized Brassica allotetraploid lines, and the greatest fertility was observed in euploid plants (3). It has been proposed that selection against aneuploid individuals with low fertility might be an important factor in the origin and establishment of natural B. napus, which is euploid (3). Although the exact selecting force under natural condition for euploidy remains to be identified in any plants, such a force must have been in action as evidenced clearly in the case of Brassica, in which it was found that without selection aneuploidy accrues rather than reduces with progressive generations (3).
We found that although whole-chromosome aneuploidy in the nascent allohexaploid wheats had a general negative effect on pollen viability in one of the two studied lines (960), only severe aneuploidy caused a significant reduction in seed production. Because pollen is abundant, an elevated percentage of less-viable pollens may not negatively impact seed fertility. This result should be especially so in self-pollinating plants like wheat, in which competition by pollen of different individuals or populations is limited. We suggest that if the original parental combinations leading to formation of the natural common wheat (T. aestivum) also met with similar prevalence and persistence of aneuploidy, then karyotype stabilization toward euploidy in the evolutionary process may have entailed novel genetic mutations or heritable epigenetic modifications. Indeed, it has been suggested that if aneuploid plant has little reduction in fitness, then selection against aneuploidy is difficult to predict, but such aneuploidy may be maintained to serve as a novel source of adaptive variations (4). This scenario has been experimentally demonstrated in yeast, in which aneuploidy alone represents a large-effect mutation that may confer fitness gains under specific conditions (35). Given the enhanced tolerance of aneuploidy types by an allohexaploid genome, it is clearly meaningful to investigate possible evolutionary roles of aneuploidy during the early generation postallopolyploidization in common wheat, particularly those types of aneuploidy that can be readily reverted back to euploidy.
Materials and Methods
Plant Materials.
Sixteen independently resynthesized allohexaploid wheat lines between different accessions of tetraploid wheat T. turgidum ssp. durum or carthlicum (genome BBAA) and diploid A. tauschii (genome DD) were used (for details, see SI Materials and Methods).
Karyotyping by Sequential GISH and FISH.
The protocols for GISH and FISH were essentially as described in Kato et al. (31) and Han et al. (36), with minor modifications. The procedure is elaborated in SI Materials and Methods.
Pyrosequencing.
The protocol was essentially as reported by Mochida et al. (37), and elaborated in SI Materials and Methods.
Profiling of Pollen Viability and Seed-Setting.
Glasshouse grown plants of two nascent allohexaploid wheat lines, 960 and AT5, were used to score pollen viability and seed-setting. For details of the procedure see SI Materials and Methods.
Statistical Analyses.
Statistics was performed using data analysis function of Microsoft Excel and SAS statistical software, version 9.2 (SAS). Details of data collection and statistical analyses are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Moshe Feldman, George Fedak, and Zhongfu Ni for providing the initial seeds of the resynthesized allohexaploid wheat lines and their parents, used in this study. This work was supported by the National Natural Science Foundation of China Grant 30120243 (to B.L.); the National High-Tech (863) Program of China Grant 2010AA1000686001 (to F.H.); and the Program for Introducing Talents to Universities B07017 (to B.L. and D.v.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300153110/-/DCSupplemental.
References
- 1.Feldman M, Lupton FGH, Miller T. Wheats. In: Smartt J, Simmonds NW, editors. Evolution of Crops. 2nd Ed. London: Longman Scientific; 1995. pp. 184–192. [Google Scholar]
- 2.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316(5833):1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA. 2011;108(19):7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chester M, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc Natl Acad Sci USA. 2012;109(4):1176–1181. doi: 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sears ER. Genetic control of chromosome pairing in wheat. Annu Rev Genet. 1976;10:31–51. doi: 10.1146/annurev.ge.10.120176.000335. [DOI] [PubMed] [Google Scholar]
- 7.Nicolas SD, Monod H, Eber F, Chèvre AM, Jenczewski E. Non-random distribution of extensive chromosome rearrangements in Brassica napus depends on genome organization. Plant J. 2012;70(4):691–703. doi: 10.1111/j.1365-313X.2012.04914.x. [DOI] [PubMed] [Google Scholar]
- 8.Mestiri I, et al. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010;186(1):86–101. doi: 10.1111/j.1469-8137.2010.03186.x. [DOI] [PubMed] [Google Scholar]
- 9.Matsushita SC, Tyagi AP, Thornton GM, Pires JC, Madlung A. Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. Genetics. 2012;191(2):535–547. doi: 10.1534/genetics.112.139295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buggs RJA, et al. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr Biol. 2012;22(3):248–252. doi: 10.1016/j.cub.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Feldman M, Levy AA. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192(3):763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy AA, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Bio J Linn Soci. 2004;82(4):607–613. [Google Scholar]
- 13.Qi B, et al. Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol. 2012;10:3. doi: 10.1186/1741-7007-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao N, et al. Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics. 2011;188(3):499–510. doi: 10.1534/genetics.111.127688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42(1):225–249. [PubMed] [Google Scholar]
- 16.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2010;13(2):153–159. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaakov B, Kashkush K. Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol Biol. 2012;80(4-5):419–427. doi: 10.1007/s11103-012-9957-3. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, Wendel JF. Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol. 2003;29(3):365–379. doi: 10.1016/s1055-7903(03)00213-6. [DOI] [PubMed] [Google Scholar]
- 20.Pires JC, et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Bio J Linn Soci. 2004;82(4):675–688. [Google Scholar]
- 21.Hegarty MJ, et al. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16(16):1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 22.Riley R, Kimber G. Aneuploids and the cytogenetic structure of wheat varietal populations. Heredity. 1961;16:275–290. [Google Scholar]
- 23.Gaeta RT, Chris Pires J. Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 2010;186(1):18–28. doi: 10.1111/j.1469-8137.2009.03089.x. [DOI] [PubMed] [Google Scholar]
- 24.von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 25.Storchová Z, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443(7111):541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, et al. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA. 2002;99(12):8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zohary D, Feldman M. Hybridization between amphidiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution. 1962;16(1):44–61. [Google Scholar]
- 28.Freeling M, et al. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol. 2012;15(2):131–139. doi: 10.1016/j.pbi.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Naranjo T. Chromosome structure of durum wheat. Theor Appl Genet. 1990;79(4):397–400. doi: 10.1007/BF01186085. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, Gill BS. Nonisotopic in situ hybridization and plant genome mapping: The first 10 years. Genome. 1994;37(5):717–725. doi: 10.1139/g94-102. [DOI] [PubMed] [Google Scholar]
- 31.Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA. 2004;101(37):13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayasaki M, Morikawa T, Tarumoto I. Intergenomic translocations of polyploid oats (genus Avena) revealed by genomic in situ hybridization. Genes Genet Syst. 2000;75(3):167–171. doi: 10.1266/ggs.75.167. [DOI] [PubMed] [Google Scholar]
- 33.Longwell J, Sears E. Nullisomics in tetraploid wheat. Am Nat. 1963;97(8):401–403. [Google Scholar]
- 34.Birchler JA. Insights from paleogenomic and population studies into the consequences of dosage sensitive gene expression in plants. Curr Opin Plant Biol. 2012;15(5):544–548. doi: 10.1016/j.pbi.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135(5):879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han F, Liu B, Fedak G, Liu Z. Genomic constitution and variation in five partial amphiploids of wheat—Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor Appl Genet. 2004;109(5):1070–1076. doi: 10.1007/s00122-004-1720-y. [DOI] [PubMed] [Google Scholar]
- 37.Mochida K, Yamazaki Y, Ogihara Y. Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol Genet Genomics. 2003;270(5):371–377. doi: 10.1007/s00438-003-0939-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.