Abstract
The maintenance of functional chloroplasts in photosynthetic eukaryotes requires real-time coordination of the nuclear and plastid genomes. Tetrapyrroles play a significant role in plastid-to-nucleus retrograde signaling in plants to ensure that nuclear gene expression is attuned to the needs of the chloroplast. Well-known sites of synthesis of chlorophyll for photosynthesis, plant chloroplasts also export heme and heme-derived linear tetrapyrroles (bilins), two critical metabolites respectively required for essential cellular activities and for light sensing by phytochromes. Here we establish that Chlamydomonas reinhardtii, one of many chlorophyte species that lack phytochromes, can synthesize bilins in both plastid and cytosol compartments. Genetic analyses show that both pathways contribute to iron acquisition from extracellular heme, whereas the plastid-localized pathway is essential for light-dependent greening and phototrophic growth. Our discovery of a bilin-dependent nuclear gene network implicates a widespread use of bilins as retrograde signals in oxygenic photosynthetic species. Our studies also suggest that bilins trigger critical metabolic pathways to detoxify molecular oxygen produced by photosynthesis, thereby permitting survival and phototrophic growth during the light period.
Keywords: biliverdin, heme oxygenase, iron homeostasis, oxidative stress, RNA-Seq analysis
The daily light–dark cycle requires all oxygenic photosynthetic species to survive the repeated transition from prolonged darkness to phototrophic metabolism at dawn. Most plants are unable to synthesize chlorophyll in darkness and therefore accumulate photosensitizing chlorophyll precursors at night (1). Sunrise induces an oxidative burst as photosynthesis resumes, so the transition to daylight requires careful coordination of many light-dependent processes. Multiple photoreceptors perform such roles in plants, the most notable being the red-sensing, linear tetrapyrrole (bilin)-based phytochromes and the blue-sensing, flavin-based cryptochromes and phototropins (2–5). Bilins are well-established plant retrograde signals, synthesized in plastids but enabling light sensing by cytosolic phytochromes. Phytochrome photoconversion then triggers nuclear translocation to positively regulate photosynthesis-associated nuclear gene (PhANG) expression (6, 7).
Genetic studies suggest that plastids also export negative retrograde signals, metabolites that suppress nuclear gene networks targeted by phytochromes (8–10). Among these metabolites are abscisic acid (ABA) (11), tetrapyrroles (12–14), 3′-phosphoadenosine 5′-phosphate (PAP) (15), β-cyclocitral (16), and methylerythritol cyclodiphosphate (MEcPP) (17). Although hypothetical export of a negative tetrapyrrole signal has received considerable support, biochemical evidence for such a retrograde signal remains equivocal in plants (18–20). Chlorophyte algae diverged from the streptophyte plant lineage over 500 million years ago but share a common chlorophyll a/b-based photosynthetic light-harvesting apparatus with plants. They thus might be expected to share similar mechanisms for light sensing and retrograde signaling. However, many chlorophyte genomes lack phytochromes (21–24), a deficiency offset by a larger complement of flavin- and retinal-based sensors (25–30).
Despite the absence of phytochromes, all known chlorophyte genomes retain cyanobacterial-derived genes for the two key enzymes required for bilin biosynthesis (Fig. 1A): a heme oxygenase (HMOX1) and a ferredoxin-dependent bilin reductase (PCYA). Both genes are found in the nuclear genome and encode proteins with apparent plastid targeting sequences (Fig. S1), which is consistent with plastid synthesis of the known bilin precursors of phytochromes, phycocyanobilin (PCB) and phytochromobilin (PΦB), in streptophytes (31). Chlorophytes also possess a second heme oxygenase gene (HMOX2). HMOX2 encodes an enzyme with a predicted C-terminal transmembrane endoplasmic reticulum-anchoring domain like those of mammalian heme oxygenases (32), suggesting a gene of eukaryotic origin but lost in the streptophyte lineage. Recent studies indicate that some chlorophyte species even possess a second plastid-targeted bilin reductase (33).
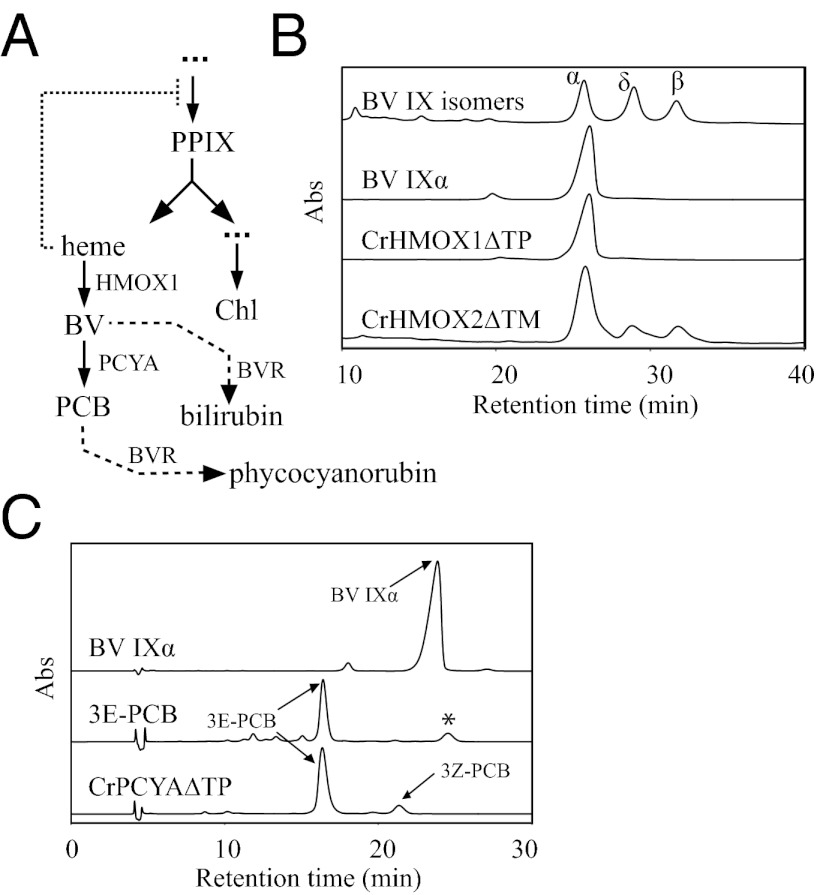
Fig. 1.
Both annotated C. reinhardtii heme oxygenases (HMOX1 and HMOX2) and PCYA bilin reductase genes encode catalytically active enzymes. (A) Tetrapyrrole pathways of C. reinhardtii and their perturbation by artificial introduction of mammalian biliverdin reductase (BVR). Feedback inhibition of tetrapyrrole synthesis by heme is indicated by a dotted line, and depletion of heme upon plastid expression of BVR is indicated by dashed lines. Chl, chlorophyll; PPIX, protoporphyrin IX. (B) HPLC elution profiles of HMOX metabolites and mixtures of BV isomers (α, δ, and β) obtained by chemical oxidative degradation of hemin. (C) HPLC analysis of PCYA-catalyzed formation of PCB from BV IXα and bilin standards (top traces). The asterisk indicates an impurity.
Here we exploit biochemical and reverse genetic approaches to explore the biological roles of bilin metabolism in the absence of phytochromes, using the representative chlorophyte species Chlamydomonas reinhardtii. We show that plastid-localized HMOX1 and PCYA yield bilin metabolites that regulate a unique nuclear gene network that potentially mitigates oxidative stress arising from photosynthetic oxygen evolution during daylight while also globally suppressing PhANG expression. Retention of the cytosolic HMOX2 confers the ability to scavenge extracellular heme as an iron source, which together with HMOX1 permits survival of C. reinhardtii under iron-limiting conditions. Our studies reveal bilins as a versatile negative retrograde signal needed for sustaining a functional chloroplast in light-grown C. reinhardtii cells.
Results
Chlamydomonas HMOX1, HMOX2, and PCYA Genes Encode Functional Enzymes.
To test their biochemical activities, we expressed and purified recombinant HMOX1∆TP (mature form lacking the predicted chloroplast transit peptide), HMOX2∆TM (soluble form lacking the C-terminal transmembrane helix), and ∆TP, ∆N, ∆C, and ∆NC truncations of PCYA in Escherichia coli (Fig. S2A). Both heme oxygenases were able to convert heme to biliverdin (BV) IXα, although the specific activity of HMOX2∆TM was approximately one-fifth that of HMOX1∆TP (Table S1). HPLC analysis of reaction products showed a lack of catalytic regiospecificity for HMOX2∆TM, explaining its apparent reduced activity. In contrast, HMOX1∆TP exclusively yielded the IXα isomer (Fig. 1B), consistent with high-throughput mammalian, cyanobacterial, and plant heme oxygenases that are coupled with an NADPH-dependent BV reductase (BVR) or with a ferredoxin-dependent bilin reductase (FDBR) such as PCYA or HY2 (31, 32). Heme oxygenases with relaxed regiospecificity similar to that of HMOX2 have been documented from insects, organisms that lack light-harvesting biliproteins, oxygen-carrying hemoproteins, and α-specific BV reductases (34). Recombinant PCYA was also active, with the core FDBR region of PCYA proving sufficient for conversion of BV IXα to PCB (Fig. 1C). These results implicate C. reinhardtii HMOX1 and HMOX2 as FDBR-coupled and FDBR-uncoupled heme oxygenases, respectively.
Subcellular Localization Studies Reveal Distinct Pathways for Bilin Synthesis in Chlamydomonas Cells.
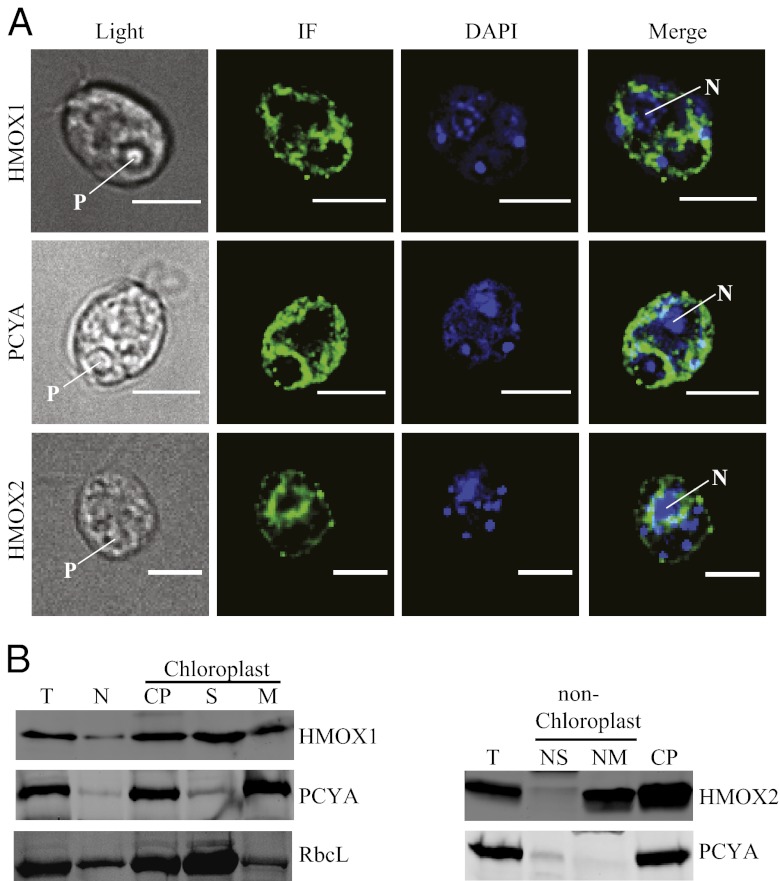
The plant-type heme oxygenase HMOX1 and bilin reductase PCYA contain putative chloroplast transit peptides, whereas the animal-type heme oxygenase HMOX2 possesses a predicted C-terminal transmembrane helix. We determined the subcellular localization of the three enzymes by confocal microscopy and by cell fractionation using monospecific antibodies to each protein. Immunofluorescence revealed that HMOX1 and PCYA were distributed within the chloroplast and were also present in whole-cell extracts of the cell-wall-deficient C. reinhardtii strain CC849 (Fig. 2 A and B). Biochemical fractionation studies confirmed that HMOX1 and PCYA were both present in plastids (Fig. 2B Left) but absent in mitochondria (Fig. S2B). Detection of these enzymes in the nonchloroplast fraction arises from partial plastid lysis during fractionation, because the Rubisco large subunit RbcL is encoded in the plastid genome yet was also seen in this fraction. Further fractionation revealed substantial association of PCYA with plastid membranes and HMOX1 distributed between stromal and membrane fractions (Fig. 2B Left). Due to low expression, HMOX2 could not be detected immunochemically in cell extracts. For this reason, full-length HMOX2 was overexpressed in strain CC849 (Fig. 2B Right), which permitted localization to perinuclear and cytoplasmic regions (Fig. 2A). Using a cell fractionation approach, we observed a distinct localization of HMOX2 to both cytoplasmic membranes and plastids (Fig. 2B Right). Although we cannot rule out some association of HMOX2 with the outer plastid envelope, these results indicate that HMOX2 is a nonspecific, low-throughput heme oxygenase with a cytosolic active site.
Fig. 2.
Subcellular distribution of heme oxygenases and bilin reductase. (A) Localization of HMOX1, PCYA, and HMOX2. DAPI, 4′,6-diamidino-2-phenylindole staining of the nucleus and chloroplast DNA; IF, indirect immunofluorescence; Light, bright field image; Merge, overlay of IF and DAPI. Pyrenoid (P) and nucleus (N) are shown with arrows. (Scale bars, 5 µm.) (B) Biochemical fractionation of HMOX1, PCYA,and HMOX2 as revealed by immunoblot analysis of total cellular proteins (T), nonchloroplast supernatant (N) and intact chloroplast (CP), and soluble (S) and membrane-associated (M) proteins from chloroplasts. The nonchloroplast supernatant was further separated into soluble (NS) and membrane (NM) fractions. Thirty micrograms of protein of each fraction was loaded for immunoblot.
Chloroplast HMOX1 and PCYA Are Responsible for Phycocyanobilin Synthesis from Heme in Vivo.
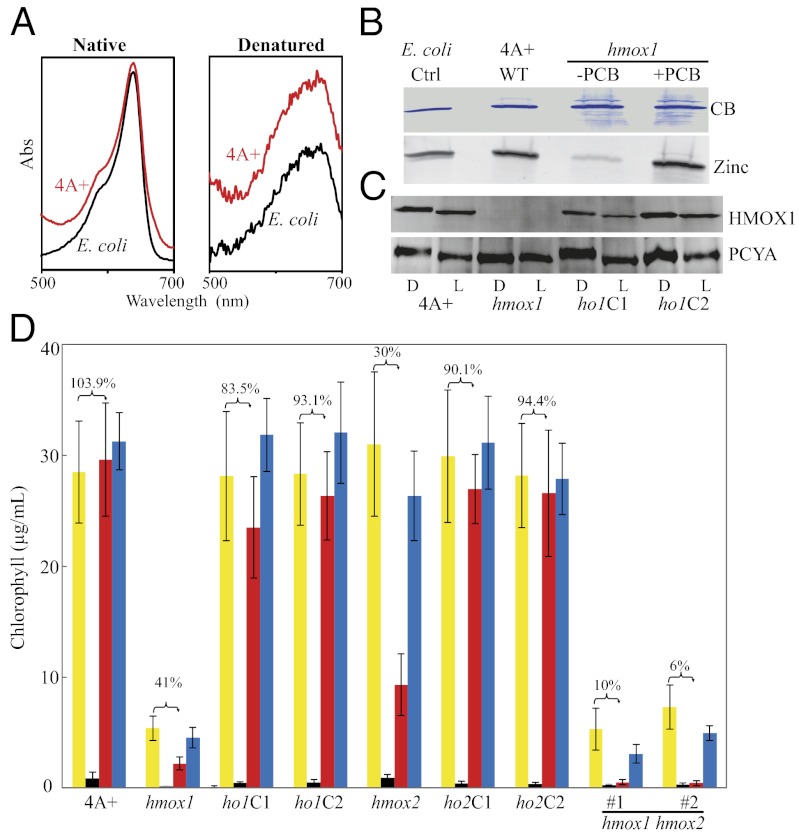
To establish that the chloroplast HMOX1 and PCYA are also functional in vivo, we transformed the bilin-binding cyanobacteriochrome (CBCR) reporter NpF2164g5 from the cyanobacterium Nostoc punctiforme (35) into the chloroplast genome of C. reinhardtii wild-type strain 4A+ (Fig. S2C). NpF2164g5 purified from 4A+ cells was shown to possess a covalently bound PCB chromophore with an absorption spectrum indistinguishable from that isolated from PCB-synthesizing E. coli cells (Fig. 3 A and B). HMOX1 and PCYA thus comprise a functional PCB biosynthetic pathway in the C. reinhardtii chloroplast. To assess the biological function of bilin biosynthesis in C. reinhardtii, we next screened insertion libraries for mutations in all three genes (36, 37). Insertional mutants were identified for the two heme oxygenases, hmox1 and hmox2 (Fig. S1A; see also Table S2 for primers used for genotyping). Unfortunately, we were able to obtain neither an insertional mutant nor an RNAi or artificial miRNA (38) knockdown mutant for PCYA. Immunoblot analysis confirmed that hmox1 was a null allele (Fig. 3C), but the low expression of HMOX2 in 4A+ wild type prevented immunochemical quantitation in the hmox2 mutant. NpF2164g5 isolated from the hmox1 mutant lacked a covalently bound chromophore but retained the ability to bind PCB in vitro (Fig. 3B). These results demonstrate that HMOX1, and not HMOX2, is required for PCB synthesis in vivo.
Fig. 3.
Chloroplast HMOX1 is responsible for synthesis of phycocyanobilin (PCB), and both heme oxygenases participate in heme iron assimilation in vivo. (A) Absorption spectra of native and denatured PCB-adducts of chloroplast-targeted cyanobacteriochrome NpF2164g5 purified from C. reinhardtii wild-type strain 4A+ or from PCB-producing E. coli. (B) NpF2164g5 purified from transgenic 4A+, hmox1, or E. coli cultures analyzed by Coomassie Blue (CB) staining (Upper) or by zinc-dependent fluorescence (Lower) for the presence of covalently bound PCB. Purified NpF2164g5 protein from hmox1 cells was still capable of binding PCB in vitro. (C) Immunoblot analysis of HMOX1 and PCYA in 4A+, hmox1, and two hmox1 complementing lines (ho1C1 and ho1C2) grown in suspension culture heterotrophically in darkness (D) or photoautotrophically under continuous light (L) at a fluence rate of 100 μmol photons⋅m−2⋅s−1. (D) Growth of C. reinhardtii with exogenous heme as iron source as measured by stationary-phase chlorophyll levels. All strains were grown in complete TAP medium (yellow columns), iron-free medium (black), or iron-free medium supplemented with either 10 µM hemin (red) or 20 µM Fe3+ (blue). The percent chlorophyll content of hemin-supplemented cultures relative to those grown on complete medium is shown for each genotype. Bars indicate the SD of three to four replicates.
Both HMOX1 and HMOX2 Genes Are Required for Heme Iron Acquisition Under Iron-Limiting Conditions.
To examine the phenotypic consequences of hmox1 and hmox2 mutations, we tested the hypothesis that one or both heme oxygenases were required for growth on heme as the sole iron source, a known function for heme oxygenases that do not generate the BV-IXα isomer (39). We compared growth as measured by chlorophyll accumulation in cultures of 4A+ wild type, hmox1 and hmox2 single mutants, complemented single mutants, and hmox1hmox2 double mutants. We used complete media (iron-replete), iron-deficient media, or iron-deficient media supplemented with 10 µM hemin or 20 µM Fe3+, concentrations chosen via pilot experiments testing growth of wild type on iron-deficient media. All cell lines harboring the hmox1 mutation accumulated significantly less chlorophyll than did other genotypes, and complementation studies show that the two heme oxygenases contributed additively to growth on extracellular heme as an iron source (Fig. 3D). Moreover, transcript levels of both heme oxygenases increased upon heme supplementation of iron-deficient 4A+ cultures (Table S3). Although both heme oxygenases contribute to iron homeostasis in C. reinhardtii, the retention of the two genes suggests nonredundant functions in extant chlorophyte species under specific environmental conditions.
Light-Dependent Chlorophyll Accumulation and Photoautotrophic Growth Are Severely Compromised by Perturbation of HMOX1 Function.
We next examined light dependence of the hmox1 phenotype in 4A+ wild-type cells. Strikingly, photoautotrophic growth of hmox1 cultures was strongly impaired, whereas heterotrophic growth of hmox1 cultures in darkness was indistinguishable from the wild type (Fig. 4 A and C and Fig. S3A). In wild-type cultures, chlorophyll content per cell was nearly three times higher in photoautotrophic cells than in heterotrophic dark-grown cells. This light-dependent increase was not seen for hmox1 mutant cultures (Fig. 4E). Both phenotypes were fully complemented by expression of the wild-type HMOX1 allele (Figs. 3C and 4 A, C, and E). By contrast, photoautotrophic growth and chlorophyll levels of light-grown hmox2 mutant cultures were indistinguishable from those of the light-grown wild type (Fig. S4). These results indicate that the plastid heme oxygenase HMOX1 is essential for light-dependent chlorophyll accumulation and for normal photoautotrophic growth.
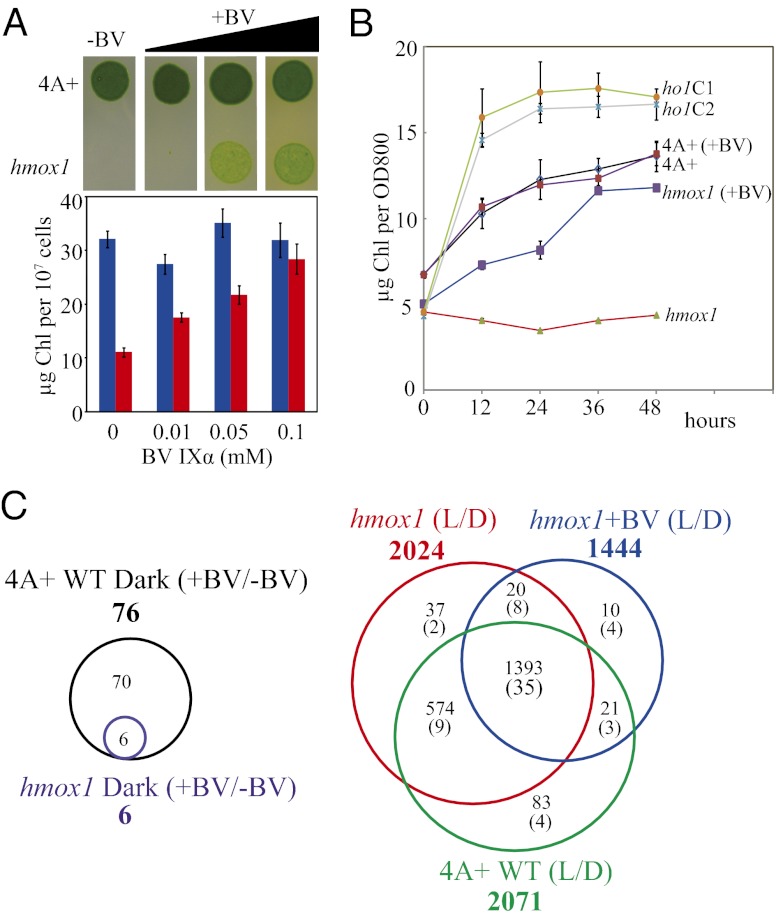
Fig. 4.
Photoautotrophic growth of both hmox1 mutant and BVR-expressing transgenic lines of C. reinhardtii is severely impaired. (A) Comparative growth on agar medium of 4A+, hmox1, and two complemented hmox1 lines (ho1C1 and ho1C2) in the dark (with acetate, heterotrophic) or under light (∼180 µmol photons⋅m−2⋅s−1, no acetate, photoautotrophic). (B) Comparative growth on agar medium of CC849 (wall-less wild type) and three BVR expression lines as in A. (C) Batch culture growth curves for hmox1 and complemented lines in continuous light (∼180 µmol photons⋅m−2⋅s−1) inoculated at the same optical density (OD800 = 0.001, ∼1.0 × 104 cells/mL). Bars indicate the SD of three to four replicates. (D) Suspension culture growth curves for the three BVR lines in continuous light as in C. (E) Normalized total chlorophyll (Chl a + Chl b) content per 107 cells in 4A+, hmox1, and complemented ho1C1 and ho1C2 cell lines. Black bars, heterotrophic (with acetate) dark-grown samples; gray bars, photoautotrophic (no acetate) light-grown samples. (F) Normalized total chlorophyll in BVR lines as in E.
The striking chlorophyll deficiency of hmox1 mutant cells is reminiscent of the phenotype of heme oxygenase-deficient plants, which arises from heme feedback inhibition of chlorophyll synthesis (Fig. 1A) and from the loss of phytochrome function (40, 41). To address a similar role for heme feedback effects, we introduced a codon-optimized rat BVR gene into the chloroplast genome of C. reinhardtii strain CC849. Mammalian BVR was previously used to impair phytochrome responses in plants by converting BV into bilirubin, thereby precluding phytochrome chromophore synthesis (42, 43). BVR is assumed to decrease heme levels by driving HMOX1-dependent heme turnover (Fig. 1A), alleviating product inhibition of heme oxygenase (32). Were hmox1 phenotypes due to feedback inhibition of chlorophyll synthesis by elevated heme, BVR expression should exert the opposite phenotype. Immunoblot analysis confirmed expression of BVR in three lines (Fig. S2D), all of which possessed chlorophyll-deficient, light-sensitive phenotypes similar to the hmox1 mutant (Fig. 4 B and D). The CC849 wild-type strain exhibited a smaller light-dependent increase in chlorophyll than did 4A+ wild type, and BVR expression actually reduced chlorophyll levels in the light. By comparison, BVR had no effect on chlorophyll accumulation of dark-grown cells (Fig. 4F). These results argue against the heme feedback hypothesis. We hypothesize that BV IXα, or its metabolite PCB, is directly responsible for the light-dependent accumulation of chlorophyll.
Biliverdin Feeding Rescues the Chlorophyll-Deficient Phenotype of the hmox1 Mutant.
To test the role of bilin in hmox1 phenotypes, we examined the ability of exogenous BV IXα to rescue the hmox1 mutant. Photoautotrophic growth of the hmox1 mutant on agar plates was partially rescued by increasing doses of BV IXα (Fig. 5A Upper). More significantly, chlorophyll content per cell was fully restored to that of wild type by addition of 0.1 mM BV IXα to liquid cultures (Fig. 5A Lower). Light-dependent chlorophyll accumulation of BV-treated hmox1 cultures also paralleled that of the wild type, but untreated hmox1 cells showed no increase in chlorophyll during the same time period (Fig. 5B). Control assays established that 0.1 mM BV IXα had no measurable influence on growth of 4A+ wild-type cultures and that there was no significant difference in chlorophyll content per cell for hmox1 cultures grown in darkness with or without 0.1 mM BV IXα (Fig. 5B). We also failed to observe a significant difference in cell-cycle progression for the two genotypes during the transition from dark to light (Table S4). The hmox1 cells were slightly larger than the wild type in darkness, which reflects a larger proportion of hmox1 cells arrested in the 4N stage. Based on these results, we conclude that BV supports a program of greening that the hmox1 mutant lacks.
Fig. 5.
Biliverdin rescues the light-sensitive and chlorophyll-deficient phenotypes of the hmox1 mutant. (A) Photoautotrophic (∼180 µmol photons⋅m−2⋅s−1) growth on agar media (Upper) and normalized total chlorophyll levels (Lower) of 4A+ and hmox1 suspension cultures in the presence of BV IXα. Normalized total chlorophyll levels of 4A+ (blue) and hmox1 (red) after 7 d of growth is presented as described in Fig. 4. (B) Time course of chlorophyll accumulation upon transfer to light. Exponential-phase cultures grown in TAP medium in darkness supplemented with or without 0.1 mM BV IXα were transferred to white light (∼180 µmol photons⋅m−2⋅s−1). Bars indicate the SD of three replicates. (C) RNA-seq reveals that BV regulates gene expression in darkness (Left) and also rescues a subset of light-regulated C. reinhardtii genes following the transfer from dark to light (Right). The diagrams show the number of nonoverlapping and shared differentially regulated transcripts with ≥twofold change and false discovery rate < 1% for each pairwise comparison. Numbers of genes that are also BV-regulated in the dark are shown in parentheses (Right). Genes of each subgroup are listed in Datasets S1 and S2.
Global Transcriptomic Analysis Reveals a Bilin-Specific Network of Gene Expression.
The increase in chlorophyll per cell following transfer of dark-grown C. reinhardtii cultures to light is somewhat analogous to deetiolation in flowering plants, a process known to be regulated by phytochromes and cryptochromes (44). Although chlorophytes retain the ability to synthesize chlorophyll in the absence of light and do not accumulate protochlorophyllide in darkness (45), light-dependent accumulation of chlorophyll is likely to be mediated by at least one photoreceptor. Indeed, cryptochromes and phototropins have been implicated in the light-regulated expression of various components of the C. reinhardtii photosynthetic apparatus (29, 30, 46). To test the hypothesis that bilins play a regulatory role during the dark–light transition in C. reinhardtii, we performed a multifactor transcriptome analysis using RNA sequencing (RNA-seq). We isolated RNA from heterotrophic suspension cultures of 4A+ wild type and the hmox1 mutant grown in darkness in the presence or absence of 0.1 mM BV IXα before and after transfer to light (∼150 µmol photons⋅m−2⋅s−1). Our analysis focused on the subset of transcripts whose expression was altered by more than a factor of two after 30 min of illumination (i.e., |log2R| > 1, where R = ratio of the expression estimates per gene for the light vs. dark samples). Using a twofold expression cutoff and keeping the false discovery rate below 1%, 2,024 and 2,071 differentially expressed transcripts were detected in dark–light transition cultures of hmox1 and wild type, respectively (Fig. 5C, Right, and Dataset S1). More than 92% of the genes whose expression was altered by light were shared by hmox1 and wild-type genotypes, that is, 1,967 genes out of 2,128 total in the two genotypes (Fig. 5C, Right, and Dataset S2). One hundred and four genes were no longer regulated by light without HMOX1. BV feeding restored light regulation to 21 of these (Dataset S2), including a CTR-type copper transporter, a glucosidase, a cation-transporting ATPase, ADP glucose pyrophosphorylase, and a number of unannotated genes.
The shared group of light-dependent genes lacked obvious homologs of early phytochrome-regulated genes and PhANGs found in similar transcriptomic experiments with plants (6). Of the 1,444 genes that were light-regulated in hmox1 mutant cells supplemented with BV, 1,393 (96%) overlapped with the light-responsive gene sets of both wild-type and mutant cells in the absence of BV (Fig. 5C, Right, and Datasets S1 and S2). This argues for a consistent ‟core group” of light-responsive transcripts in C. reinhardtii that is distinct from that of streptophyte plants. BV treatment damped light regulation for the vast majority of these core group genes: many of the RNAs that increased or decreased in abundance in the light changed to a lesser extent in the presence of BV (Dataset S2). Genes induced by light and suppressed by BV were enriched for protein families involved in high light stress (e.g., encoding LHCSR1, PSBS1, ELI3, HSP22, GPX, HSP70, and others). This suggests that the initial transcriptional response to light in C. reinhardtii is dominated by the response to oxidative stress, a combined effect of photosynthetic oxygen evolution and pigment photosensitization (47, 48). We therefore attribute the ‟global” attenuation of this response by BV to its ability to target a regulatory network needed to reduce or prevent oxidative stress. This hypothetical network is independent of the known SOR1 (Singlet Oxygen Resistant 1)-mediated response (49), because transcription of SOR1 and many SOR1-regulated genes is not affected in hmox1 cultures (Dataset S1). However, we cannot fully discount direct attenuation of light exposure by BV as responsible for some of the global suppression of PhANG expression in light.
We also identified a small number of BV-responsive genes in dark-grown cultures (Fig. 5C, Left, 6 and 76 in hmox1 and wild type, respectively; Dataset S1). All 6 of the BV-dependent genes in dark-grown hmox1 were present in the list of 76 BV-responsive genes for dark-grown wild type. The less robust response in the hmox1 mutant may be due to altered heme homeostasis in the hmox1 cells, which would not be efficiently rescued by BV feeding. Among the 76 BV-regulated genes are mono- and di-oxygenases, FAD-, heme- and iron-sulfur-containing redox proteins, a hemoglobin-like protein, and enzymes involved in oxidative amino acid catabolism. The majority of these genes, 70 out of 76 including all 6 HMOX1-independent genes, were up-regulated. These results indicate that C. reinhardtii possesses a light-independent signaling pathway using a plastid metabolite (bilin) to regulate nuclear gene expression: in other words, a bilin-based retrograde signaling pathway.
Discussion
The presence of cytosolic membrane-associated heme oxygenase in C. reinhardtii and other sequenced chlorophyte algae is unexpected, because such animal-type heme oxygenases are absent in both streptophyte algae and higher plants such as Arabidopsis thaliana. All heme oxygenases found in A. thaliana contain chloroplast translocation sequences that are sufficient for chloroplast targeting of the fusion proteins (50). Our data indicate that the animal-type HMOX2 of Chlamydomonas is a non-α–specific, FDBR-uncoupled enzyme involved in heme turnover and iron recycling. This suggests that, at the time of cyanobacterial capture, the eukaryote host already possessed the ability to detoxify heme, to scavenge iron from extracellular heme, and to make BV.
In response to environmental and developmental cues, multiple pathways and metabolites are involved in relaying information from plastids to nucleus to coordinate gene expression between the two genomes (10, 14, 15, 17). Identification of gun mutants (genome uncoupled) in A. thaliana using norflurazon implicates the tetrapyrrole pathway as a source of negative retrograde signals. A specific role of the heme branch (Fig. 1A) as a positive retrograde signal was proposed based on the ability of heme to induce PhANG expression in undeveloped Arabidopsis plastids (14). Heme was also found to regulate the expression of nuclear genes transiently in C. reinhardtii (51). Our data provide evidence that the heme catabolites BV and/or PCB specifically up-regulate a subset of nuclear genes in C. reinhardtii in darkness. Our data suggest that plastid-derived bilins function in the light as negative signals to suppress PhANG expression in C. reinhardtii, a network largely promoted by phytochromes in plants.
Based on the ability of exogenous BV to induce genes encoding oxygen-dependent redox enzymes in darkness, we propose that a bilin-responsive gene network in C. reinhardtii evolved to consume oxygen and to detoxify reactive oxygen species produced by light. We predict that a spike of PCB production occurs upon light exposure due to the increase in oxygen evolution and heme release from damaged hemoproteins, factors that both enhance HMOX1 enzymatic turnover. Heme turnover also will be facilitated by PCYA, which converts BV to PCB. We thus propose that PCB is the bilin signal responsible for induction of a photoprotective transcriptional program, because the genes regulated by exogenous heme treatment in darkness differ from those observed here (51–53). Our studies demonstrate that cells that lack HMOX1 are poorly equipped to deal with the transition from dark to light because they cannot produce bilins. This rationale accounts in part for the rescue of the hmox1 mutant by exogenous BV, whose uptake and metabolism by plants has been well documented (54, 55). Unlike plants, C. reinhardtii lacks phytochromes and other known bilin-dependent photosensors. The bilin retrograde signal network is thus consistent with a heme-derived retrograde signal to minimize light damage, while suppressing the activity of photoreceptors that function to up-regulate PhANG expression when light is present. Our inability to obtain a pcyA mutant may therefore reflect an essential function of this enzyme in C. reinhardtii, as has been shown in cyanobacteria (56).
Phytochromes are found in streptophyte algae (57, 58), at least one chlorophyte species (22), and the glaucophyte Cyanophora paradoxa (59), so we expect that PCB will serve as chromophore precursor for positive regulation of PhANG expression in these species. Our results raise the possibility that chlorophyte species lacking phytochromes possess complementary regulatory systems responsible for mediating the light-dependent increase in chlorophyll accumulation seen in C. reinhardtii. It is even conceivable that undiscovered bilin-based light sensors might be responsible for this response or for near UV- and blue-dependent responses of C. reinhardtii not ascribed to flavin-based sensors (26, 60), because PCB-based sensors can detect any wavelength of light from the near UV to the near IR (61, 62). Alternatively, bilin-based retrograde signaling may be the sole reason for retention of the bilin biosynthetic pathway in chlorophytes lacking phytochromes. In plants, retention of the bilin signaling pathway would confer significant adaptive advantage at the daily transition to photoautotrophic growth. Our RNA sequencing data implicate the bilin signaling pathway in suppression of reactive oxygen intermediates, a critical function for all oxygenic photosynthetic organisms that must cope with diurnal fluctuations of light fluence, and also for suppression of PhANG expression. Bilin biosynthesis thus produces an environmental integrating signal ideal for communicating the physiological status of the captured photosynthetic organelle. We therefore expect that bilin retrograde signaling will be widely distributed among oxygenic photosynthetic organisms both with and without phytochromes, including land plants.
Materials and Methods
Details are described in SI Materials and Methods. This includes information on protein expression and enzyme assays, hmox1 and hmox2 complementation, chloroplast transformation, antibody production and western analysis, immunofluorescence localization and cell fractionation, chlorophyll quantification, Chlamydomonas growth on heme, and RNA-seq protocols. Chlamydomonas strains were maintained at room temperature in TAP medium under continuous low intensity white light.
Supplementary Material
Acknowledgments
We thank Dr. Kempton M. Horken (University of Nebraska, Lincoln) for the carrier vector for construction of chloroplast transformation plasmids. The anti-RbcL antibody is a gift from Steven M. Theg (University of California, Davis). This work was supported by National Science Foundation Grant MCB-0843625 (to J.C.L.) and by National Institutes of Health Grant R24GM092473 (to S.S. Merchant) for RNA sequencing. Isolation of the hmox1 mutant, preparation of COX2 and AOX antibodies, and construction of the CBCR reporter NpF2164g5 was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, Office of Science, US Department of Energy Former Worker Program 449B (to K.K.N.) and Grants DE-FG02-04ER15529 (to S.S. Merchant) and DE-FG02-09ER16117 (to J.C.L.). K.K.N. is an investigator of the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation.
Footnotes
The authors declare no conflict of interest.
Database deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40031).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222375110/-/DCSupplemental.
See Commentary on page 3218.
References
- 1.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 4.Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6(5):220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaves I, et al. The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 6.Quail PH. Phytochrome-regulated gene expression. J Integr Plant Biol. 2007;49(1):11–20. [Google Scholar]
- 7.Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21(11):664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koussevitzky S, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316(5825):715–719. [PubMed] [Google Scholar]
- 9.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Leister D. Retrograde signaling in plants: From simple to complex scenarios. Front Plant Sci. 2012;3:135. doi: 10.3389/fpls.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasilewska A, et al. An update on abscisic acid signaling in plants and more…. Mol Plant. 2008;1(2):198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 12.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421(6918):79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 13.Voigt C, et al. In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol Plant. 2010;138(4):503–519. doi: 10.1111/j.1399-3054.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 14.Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol. 2011;21(10):897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estavillo GM, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23(11):3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109(14):5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Y, et al. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149(7):1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki N, et al. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010;15(9):488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Moulin M, McCormac AC, Terry MJ, Smith AG. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA. 2008;105(39):15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA. 2008;105(39):15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324(5924):268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 23.Prochnik SE, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329(5988):223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanc G, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22(9):2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang K, Beck CF. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2003;100(10):6269–6274. doi: 10.1073/pnas.0931459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman AR, Lohr M, Im CS. Chlamydomonas reinhardtii in the landscape of pigments. Annu Rev Genet. 2004;38:119–173. doi: 10.1146/annurev.genet.38.072902.092328. [DOI] [PubMed] [Google Scholar]
- 27.Mittag M, Kiaulehn S, Johnson CH. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 2005;137(2):399–409. doi: 10.1104/pp.104.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falciatore A, et al. The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev. 2005;19(1):176–187. doi: 10.1101/gad.321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Im CS, Eberhard S, Huang K, Beck CF, Grossman AR. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 2006;48(1):1–16. doi: 10.1111/j.1365-313X.2006.02852.x. [DOI] [PubMed] [Google Scholar]
- 30.Hegemann P. Algal sensory photoreceptors. Annu Rev Plant Biol. 2008;59:167–189. doi: 10.1146/annurev.arplant.59.032607.092847. [DOI] [PubMed] [Google Scholar]
- 31.Frankenberg N, Lagarias JC. In: The Porphyrin Handbook. Chlorophylls and Bilins: Biosynthesis Structure and Degradation. Kadish KM, Smith KM, Guilard R, editors. New York: Academic; 2003. pp. 211–235. [Google Scholar]
- 32.Ortiz de Montellano PR, Auclair K. In: The Porphyrin Handbook. The Iron and Cobalt Pigments: Biosynthesis, Structure and Degradation. Kadish KM, Smith KM, Guilard R, editors. New York: Academic; 2003. pp. 183–210. [Google Scholar]
- 33.Chen YR, Su YS, Tu SL. Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc Natl Acad Sci USA. 2012;109(21):8310–8315. doi: 10.1073/pnas.1201744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Sato M, Sasahara M, Migita CT, Yoshida T. Unique features of recombinant heme oxygenase of Drosophila melanogaster compared with those of other heme oxygenases studied. Eur J Biochem. 2004;271(9):1713–1724. doi: 10.1111/j.1432-1033.2004.04077.x. [DOI] [PubMed] [Google Scholar]
- 35.Rockwell NC, Martin SS, Lagarias JC. Red/green cyanobacteriochromes: Sensors of color and power. Biochemistry. 2012;51(48):9667–9677. doi: 10.1021/bi3013565. [DOI] [PubMed] [Google Scholar]
- 36.Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137(2):545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Ballester D, et al. Reverse genetics in Chlamydomonas: A platform for isolating insertional mutants. Plant Methods. 2011;7:24. doi: 10.1186/1746-4811-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molnar A, et al. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58(1):165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- 39.Caignan GA, et al. Oxidation of heme to beta- and delta-biliverdin by Pseudomonas aeruginosa heme oxygenase as a consequence of an unusual seating of the heme. J Am Chem Soc. 2002;124(50):14879–14892. doi: 10.1021/ja0274960. [DOI] [PubMed] [Google Scholar]
- 40.Terry MJ, Linley PJ, Kohchi T. Making light of it: The role of plant haem oxygenases in phytochrome chromophore synthesis. Biochem Soc Trans. 2002;30(4):604–609. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 41.Cornah JE, Terry MJ, Smith AG. Green or red: What stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 2003;8(5):224–230. doi: 10.1016/S1360-1385(03)00064-5. [DOI] [PubMed] [Google Scholar]
- 42.Lagarias DM, Crepeau MW, Maines MD, Lagarias JC. Regulation of photomorphogenesis by expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Cell. 1997;9(5):675–688. doi: 10.1105/tpc.9.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montgomery BL, et al. Biliverdin reductase-induced phytochrome chromophore deficiency in transgenic tobacco. Plant Physiol. 2001;125(1):266–277. doi: 10.1104/pp.125.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 45.Fujita Y, Bauer CE. In: The Porphyrin Handbook. Chlorophylls and Bilins: Biosynthesis Structure and Degradation. Kadish KM, Smith KM, Guilard R, editors. New York: Academic; 2003. pp. 109–156. [Google Scholar]
- 46.Beel B, et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012;24(7):2992–3008. doi: 10.1105/tpc.112.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledford HK, Chin BL, Niyogi KK. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot Cell. 2007;6(6):919–930. doi: 10.1128/EC.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer BB, Wiesendanger M, Eggen RI. Growth condition-dependent sensitivity, photodamage and stress response of Chlamydomonas reinhardtii exposed to high light conditions. Plant Cell Physiol. 2006;47(8):1135–1145. doi: 10.1093/pcp/pcj085. [DOI] [PubMed] [Google Scholar]
- 49.Fischer BB, et al. SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2012;109(20):E1302–E1311. doi: 10.1073/pnas.1116843109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem J. 2010;425(2):425–434. doi: 10.1042/BJ20090775. [DOI] [PubMed] [Google Scholar]
- 51.Voss B, et al. Hemin and magnesium-protoporphyrin IX induce global changes in gene expression in Chlamydomonas reinhardtii. Plant Physiol. 2011;155(2):892–905. doi: 10.1104/pp.110.158683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Gromoff ED, Treier U, Beck CF. Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol Cell Biol. 1989;9(9):3911–3918. doi: 10.1128/mcb.9.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell. 2008;20(3):552–567. doi: 10.1105/tpc.107.054650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elich TD, McDonagh AF, Palma LA, Lagarias JC. Phytochrome chromophore biosynthesis. Treatment of tetrapyrrole-deficient Avena explants with natural and non-natural bilatrienes leads to formation of spectrally active holoproteins. J Biol Chem. 1989;264(1):183–189. [PubMed] [Google Scholar]
- 55.Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3(11):1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvey RM, Biswas A, Schluchter WM, Bryant DA. Effects of modified Phycobilin biosynthesis in the Cyanobacterium Synechococcus sp. Strain PCC 7002. J Bacteriol. 2011;193(7):1663–1671. doi: 10.1128/JB.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kidd DG, Lagarias JC. Phytochrome from the green alga Mesotaenium caldariorum. Purification and preliminary characterization. J Biol Chem. 1990;265(12):7029–7035. [PubMed] [Google Scholar]
- 58.Winands A, Wagner G. Phytochrome of the green alga Mougeotia: cDNA sequence, autoregulation and phylogenetic position. Plant Mol Biol. 1996;32(4):589–597. doi: 10.1007/BF00020200. [DOI] [PubMed] [Google Scholar]
- 59.Price DC, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335(6070):843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- 60.Teramoto H, et al. Action spectrum for expression of the high intensity light-inducible Lhc-like gene Lhl4 in the green alga Chlamydomonas reinhardtii. Plant Cell Physiol. 2006;47(3):419–425. doi: 10.1093/pcp/pcj009. [DOI] [PubMed] [Google Scholar]
- 61.Rockwell NC, Lagarias JC. A brief history of phytochromes. ChemPhysChem. 2010;11(6):1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA. 2011;108(29):11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.