Abstract
Objective
Enamel matrix derivative (EMD), is an extract of porcine developing enamel matrix. Its commercialised form Emdogain, is claimed to stimulate periodontal regeneration by recapitulating original developmental processes, although the mechanism remains unclear. Our objective was to investigate interactions between EMD and human periodontal ligament (HPDL) fibroblasts in vitro.
Design
HPDL fibroblasts were cultured in the presence of fluorescently labelled EMD and cellular EMD uptake was monitored using confocal laser scanning microscopy and immunohistochemistry. Internalised EMD proteins were characterised using SDS-PAGE.
Results
EMD was internalised by HPDL fibroblasts leading to the appearance of multiple, vesicle-like structure in the cytoplasm. The internalised protein was composed mainly of the major 20 kDa amelogenin component of EMD which was subsequently processed with time to generate a cumulative 5 kDa component.
Conclusions
Cellular uptake and subsequent intracellular processing of EMD components by dental mesenchymal cells may play a role in EMD bioactivity and in part explain the turnover of Emdogain when placed clinically.
Keywords: Emdogain®, Periodontal ligament fibroblasts, Endocytosis, Processing, Cytoplasmic vesicles
1. Introduction
Emdogain is a commercialised extract of porcine developing enamel derived matrix (EMD) used clinically to regenerate periodontal tissues reportedly by mimicking the developmental processes associated with tooth development.1 The molecular mechanism(s) responsible for EMD’s efficacy are not fully known. Since early immunoassay studies were unable to detect the presence of growth-factors within EMD the effects of EMD were largely attributed to amelogenins2 which comprise >90% of the proteins in EMD.3 Full-length amelogenin4 and the alternatively spliced variant, leucine rich amelogenin peptide (LRAP), appear capable of controlling gene expression.5–8 However, other studies have revealed that EMD does, indeed, contain growth factor like activity e.g. TGF-β1 and BMP.9, 10 Matters are complicated by reports indicating that EMD also stimulates autocrine production of growth factors, TGF-β and PDGF.11
Whatever the mechanism, interaction of EMD with cells is critical. Therefore, the aim of the present work was to investigate the interaction of fluorescently labelled EMD with cultured human periodontal ligament fibroblasts using confocal laser scanning microscopy and immunocytochemistry, while internalised proteins were characterised using SDS-PAGE.
2. Materials and methods
2.1. Source of material
EMD was supplied by Biora (Malmö, Scania, Sweden) as a lyophilised preparation without the PGA vehicle used to deliver it clinically.
2.2. Fluorescein isothiocyanate (FITC) conjugation
Lyopholised proteins (i.e. whole EMD and BSA) were conjugated to FITC (Sigma, Poole, Dorset, UK) by dissolving in 0.1 M NaHCO3 buffer, pH 9, at a concentration of 4 mg/ml. FITC was dissolved in anhydrous DMSO at 1 mg/ml and 50 µl of FITC solution was slowly added in 5 µl aliquots to each millilitre of EMD solution with gentle stirring. The solution was then incubated in the dark for 8 h at 4 °C. NH4Cl was added to a final concentration of 50 mM and the solution incubated for a further 2 h at 4 °C.
The solution was then dialysed (Slide-A-Lyzer® dialysis cassettes Pierce Cramlington, Northumberland, UK) for 36 h against PBS to remove free FITC. For use in cell culture, 1.25 ml of dialysed FITC labelled proteins (in PBS) were added to 8.75 ml of DMEM culture medium to give a final concentration of 0.5 mg/ml (assuming no change in volume during dialysis).
2.3. Size exclusion chromatography of EMD–FITC
Size exclusion fractionation of EMD–FITC was carried out to obtain an EMD–FITC fraction free of the 5 kDa component and a fraction containing just the FITC labelled 5 kDa component itself. These two fractions were used to better understand the origin of a 5 kDa EMD component that was found to accumulate in cells incubated with EMD–FITC. Fractionation was carried out using a formic acid mobile phase. Briefly, 20 mg of lyophilized EMD was dissolved in 0.75 ml of 0.125 mol/l formic acid and subjected to size exclusion chromatography using a 90 cm × 1.6 cm column of Bio Gel P10 (Bio-Rad, Hemel Hempstead, Hertfordshire, UK) eluted with 0.125 mol/l formic acid at 0.3 ml/min. The eluant was monitored at 280 nm and 5 ml fractions were collected. Fractions containing proteins above 5 kDa were pooled to generate a “5 kDa free fraction”. This pooled fraction and the remaining 5 kDa fraction were lyophilised.
2.4. Cell culture
Premolars extracted from consenting healthy individuals for orthodontic reasons were used as a source of HPDL fibroblasts as described previously.12 Only HPDL fibroblasts at passages 2–5 were used in this study. Cells were grown in DMEM supplemented with 10% FCS, 100 units/ml penicillin and 100 µg/ml streptomycin (Sigma, Poole, Dorset, UK), in a humidified atmosphere of 5% CO2 in air at 37 °C.
2.5. EMD–FITC treated cells viewed by confocal laser scanning microscopy in monolayer morphology
Cells were grown to confluence on sterile glass coverslips placed in six-well culture flasks and cultured for 17 h in DMEM containing 0.5 mg/ml FITC labelled EMD as described above. The coverslip bound cells were then washed (3 × 5 min) with ice-cold PBS and fixed for 10 min at room temperature in 3.7% formaldehyde. The coverslip bound cells were then washed (3 × 5 min) in PBS and mounted onto glass microscope slides with Vectashield, an aqueous-mountant containing anti-fade reagent (Vector Laboratories Ltd., Peterborough, Cambridgeshire, UK) and viewed using confocal laser scanning microscopy (FITC absorption and fluorescence emission maxima: 496 nm and 520 nm respectively; excitation light source: Green Ar/ArKr laser setting 488 nm).
2.6. Immunocytochemistry
Cells were grown to confluence in T75 culture flasks and then cultured for 17 h in DMEM containing either 0.5 mg/ml EMD–FITC or 0.5 mg/ml unlabelled EMD (or 0.5 mg/ml BSA–FITC or 0.5 mg/ml unlabelled BSA in controls) in a humidified atmosphere of 5% CO2 in air at 37 °C. The monolayer was then washed (3 × 5 min) with ice-cold PBS and the cells detached by trypsinization. The cell suspension was centrifuged for 10 min at 1200 rpm to produce a pellet. The supernatant was discarded and the pelleted cells were washed 3 times by repeated resuspension and centrifugation in 3 ml of PBS (3 min at 1200 rpm with final spin for 10 min at 1200 rpm). The final supernatant (PBS) was discarded. The pellet was resuspended in 500 µl of 4% gelatin in PBS, and stored at 4 °C for approximately 4 h to set the gelatin. The cells in gelatin were then fixed overnight in 10 ml of 10% neutral buffered formalin at 4 °C before being subjected to paraffin embedding and sectioning for analysis. Paraffin sections were mounted on 3-aminopropyltriethoxysilane (APES) coated glass microscope slides and probed with rabbit anti-20 kDa-amelogenin primary antibodies (a kind gift from Biora, Malmö , Sweden). Cross-reactivity was detected by using a DAKO StreptABComplex/HRP Duet, Mouse/Rabbit Immunoperoxidase kit (Dako UK Ltd., Ely, Cambridgeshire) used according to manufacturer’s instructions. Controls were run in parallel in which the primary antibody was omitted.
2.7. Retrieval of FITC labelled protein internalised by cells in culture
Cells were grown to confluence in T75 culture flasks and then cultured in DMEM containing 0.5 mg/ml EMD–FITC in a humidified atmosphere of 5% CO2 in air at 37 °C for various lengths of time (1 h, 3 h, 6 h and 17 h). In order to recover any internalised EMD–FITC conjugate for analysis, a substantial amount of contaminating extracellular material present in an aggregated form first had to be removed. This was achieved by washing with a low pH buffer which dissolved and removed extracellular EMD–FITC (as determined by fluorescence microscopy) while leaving cells intact. Following cell treatment with the EMD–FITC conjugate the culture medium was aspirated off and the cells were washed (3 × 2 min) with PBS and then washed (3 × 5 min) with low pH wash buffer (0.1 M formic acid (pH 2.2), 0.69% NaCl). The cells were washed again (3 × 2 min) with PBS, then any residual PBS was aspirated off and the cells lysed by adding 1 ml of SDS sample loading buffer to the culture flask followed by vigorous shaking for 30 min. The lysate was heated at ~90 °C for 5 min, centrifuged at 20,000 × g for 10 min and the supernatant removed for SDS-PAGE.
Cells were also incubated in cultured in DMEM containing either an EMD–FITC fraction devoid of any FITC labelled 5 kDa material or a fraction containing the FITC labelled 5 kDa material itself (concentration of both fractions equivalent to the relative amount in present in 0.5 mg/ml EMD–FITC (assuming 100% recovery of protein following chromatographic preparation of fractions)) in a humidified atmosphere of 5% CO2 in air at 37 °C for various lengths of time (3 h, 6 h and 17 h).
2.8. SDS-PAGE
Lysates of EMD–FITC treated cells were subjected to SDS-PAGE according to Laemmli13 using 15% mini gels. Samples were loaded at 10 µl per lane along with 10 µg of the original EMD–FITC conjugate. Gels were viewed using UV transillumination to visualise the fluorescently labelled EMD.
3. Results
3.1. Interaction of EMD–FITC with HPDL fibroblasts as revealed by confocal laser scanning microscopy
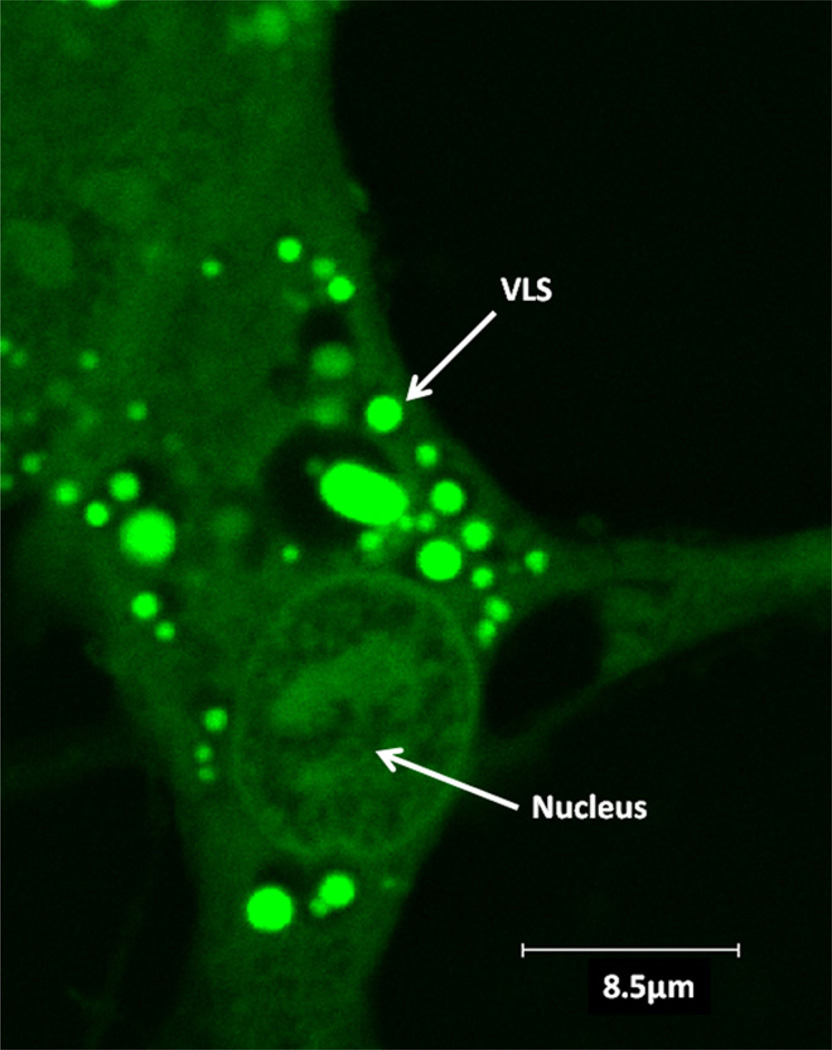
Fig. 1 shows a confocal laser scanning microscopy image of a HPDL fibroblast cultured with EMD–FITC conjugate. Strongly fluorescent VLSs were present throughout the cytoplasm but were absent from the nucleus. Some VLSs contained a centralised fluorescent region surrounded by a dark nonfluorescent region. Cells incubated with BSA–FITC conjugate showed no fluorescence (data not shown).
Fig. 1.
Periodontal fibroblasts treated with EMD–FITC and viewed by confocal laser scanning microscopy. A typical image of confluent HPDL fibroblasts incubated in culture for 17 h with 0.5 mg/ml EMD–FITC and viewed in monolayer by confocal laser scanning microscopy. Multiple, strongly fluorescent vesicle like structures (VLSs) were observed within the cytoplasm of the cells. Some vesicles exhibited a dark non florescent area surrounding a fluorescent central region.
3.2. Interaction of EMD–FITC with HPDL fibroblasts as revealed by immunocytochemistry
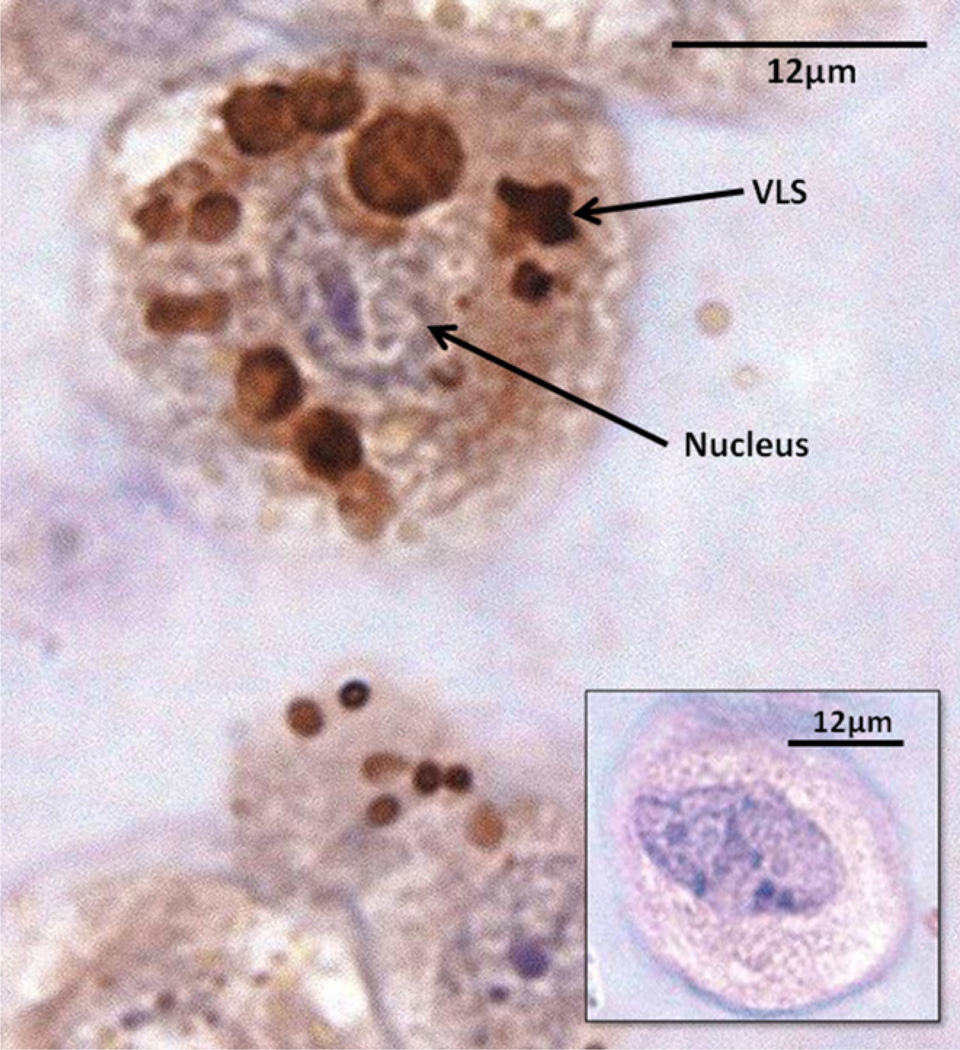
HPDL fibroblasts previously incubated with EMD–FITC conjugate were subjected to immunocytochemistry using antibodies raised against 20 kDa pig amelogenin. Fig. 2 shows amelogenin cross reactivity concentrated in globules throughout the cell cytoplasm with no obvious nuclear staining. The immunostained VLSs appeared generally larger than fluorescently stained VLSs in cells derived from the same donor. Inset shows a negative control section with no primary antiamelogenin antibody. Cells treated with unlabelled EMD gave identical results (data not shown).
Fig. 2.
Paraffin sections of EMD–FITC treated HPDL fibroblasts probed with anti-20 kDa-amelogenin antibodies. Cells were counterstained with haematoxylin and eosin. Multiple, strongly cross-reactive VLSs were evident within the cytoplasm (arrowed). Inset shows negative control (no primary antibody). No significant cross-reactivity observed.
3.3. Biochemical characterisation of intracellular EMD–FITC conjugate recovered following its uptake by HPDL fibroblasts
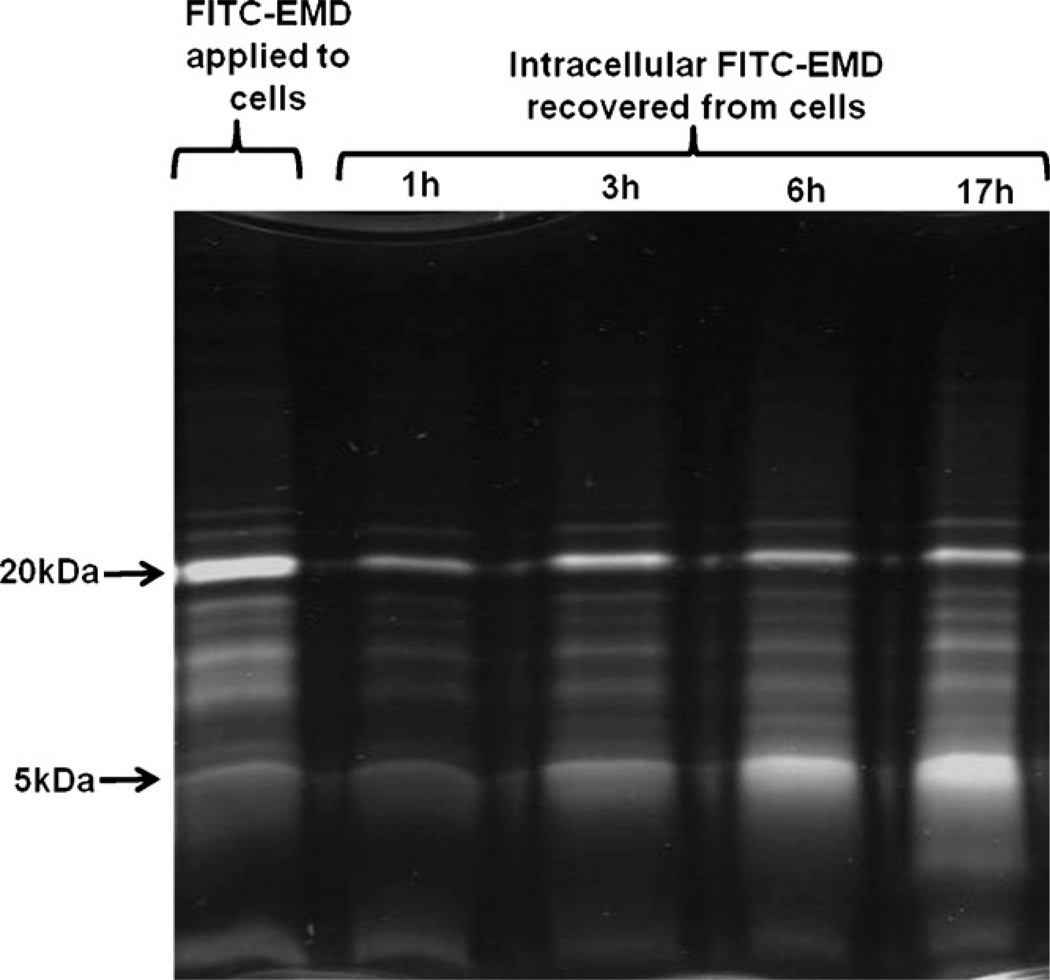
Intracellular material recovered from HPDL fibroblasts that had been incubated with EMD–FITC conjugate for either 1, 3, 6 or 17 h was analysed by SDS-PAGE. Fig. 3 shows the whole EMD–FITC conjugate as applied to the cells (lane 1) compared to the intracellular proteins retrieved after culturing the cells with EMD–FITC conjugate for either 1, 3, 6 or 17 h (lanes 2–5). The composition of the intracellular material recovered after 1 h incubation with EMD–FITC conjugate (lane 2) reflected the composition of the applied EMD–FITC (lane 1) with the 20 kDa band being most prominent. However, over 17 h there was a gradual accumulation of protein at 5 kDa which accumulated with time to become the dominant band present at later time points (lanes 3–5).
Fig. 3.
SDS-PAGE of whole EMD–FITC (as applied to the cells) and lysates of cells exposed to EMD–FITC for 1–17 h (viewed by UV transillumination). The composition of the intracellular material recovered after 1 h incubation with EMD–FITC conjugate (lane 2) reflected the composition of the applied EMD–FITC (lane 1) with the 20 kDa band being most prominent. Over 17 h there was a gradual accumulation of proteins, especially the 5 kDa protein which accrued to become the dominant band present at later time points (lanes 3–5).
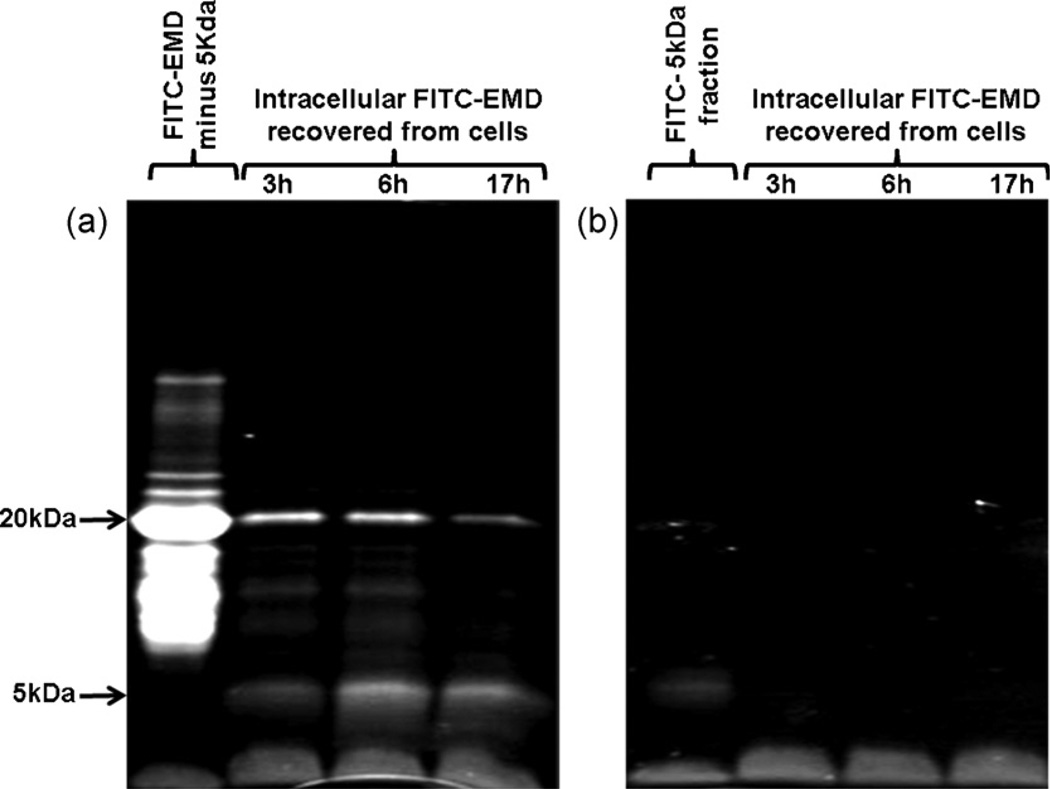
To investigate the origin of the accumulating 5 kDa protein, cells were incubated either with EMD–FITC containing no 5 kDa material or an isolated fraction of the FITC labelled 5 kDa protein itself. Fig. 4a shows the proteins recovered following incubation with EMD–FITC lacking the 5 kDa material. Although no fluorescent 5 kDa material was applied to the cells (lane 1), 5 kDa material clearly accumulated intracellularly with time (lanes 2–5) even though protein at this molecular weight was absent from the EMD applied to the cells.
Fig. 4.
(a) SDS-PAGE of retrieved intracellular protein following incubation of the cells with EMD–FITC containing no 5 kDa protein. Although no 5 kDa protein was applied to the cells, material at this molecular weight still accumulated with time presumably due to intracellular degradation of a larger precursor (e.g. the 20 kDa protein). (b) SDS-PAGE of retrieved intracellular proteins following incubation of the cells with FITC labelled 5 kDa EMD fraction. No uptake was detected.
Fig. 4b shows the protein recovered following incubation of cells with a protein preparation containing the FITC labelled 5 kDa fraction alone. The FITC labelled 5 kDa fraction applied to the cells (lane 1) was not taken up by cells to any great extent as evidenced by the lack of any 5 kDa fluorescent intracellular material retrieved from the cells at any time point (lanes 2–5).
It appears therefore, that the intracellular 5 kDa material retrieved from cells following their incubation with EMD is generated by intracellular degradation of higher molecular weight EMD components e.g. the 20 kDa component.
4. Discussion
The present work, based on fluorescently labelled EMD and immunocytochemistry, demonstrated that EMD components, particularly the 20 kDa EMD component (corresponding to the major 20 kDa amelogenin processing product P148) were selectively taken up by cultured HPDL fibroblasts. The absence of any obvious gross aggregates at the cell membrane suggests that 20 kDa amelogenin was recognised at the cell membrane in its monomeric form or possibly as small homogeneous complexes similar to previously reported nanospheres.14 Once internalised, the protein was concentrated in vesicle like structures, present in the cytoplasm. With time, internalized 20 kDa amelogenin appeared to be degraded, generating material at 5 kDa. Earlier studies have reported the apparent uptake of amelogenin by various cell types. For example, amelogenin was immunolocalised to odontoblast lysosomes15; FITC labelled recombinant mouse amelogenin was shown to be taken up by dental epithelial cells (HAT-7 cells) from culture medium and localised to the perinuclear region16; fluorescently labelled EMD and recombinant full length mouse amelogenin-DsRed hybrid protein were taken up by mouse pre-osteoblast MC3T3-E1 cells directly into LAMP-1 and CD63 coated vesicles (late endosomes or lysosomes)17; EMD was shown to be internalised by primary osteoblasts through a pathway mediated via clathrin coated pits18 and amelogenin was shown to be internalised by normal human dermal fibroblasts possibly via integrin binding.19 Van der Pauw et al.20 showed that HPDL fibroblasts phagocytised 2 µm beads coated with either EMD or BSA. 13% of cells phagocytosed EMD coated beads compared with ≤6% for BSA. Using fluorescently labelled proteins in solution, the present study indicated almost no uptake of BSA compared with the 20 kDa amelogenin component of EMD (data not shown). This suggests the presence of a highly selective receptor mediated endocytotic mechanism of for example the 20 kDa amelogenin uptake rather than non specific phagocytosis or pinocytosis. LAMP-1 and CD63 have been reported to bind amelogenin6, 21 and to a limited extent, CD63 and LAMP-1 cycle from the Golgi via the plasma membrane to late endosomes and lysosomes22, 23 and may therefore act as the specific cell surface receptors involved in capturing amelogenin and triggering amelogenin endocytosis.
The role of CD63 as an amelogenin receptor does not invalidate the previously mentioned report18 that amelogenin uptake is mediated via clathrin coated pits as CD63 can itself be endocytosed via clathrin coated pits.22 Likewise, LAMP-1 can also be internalised via clathrin coated pits.24 In other words, amelogenin may be captured at the cell surface by CD63 and/or LAMP-1 and the resulting complexes internalized through clathrin coated pits.
CD63 also interacts with integrins25 and a model explaining how ameloblastin regulates osteogenic differentiation has been suggested wherein CD63 interactions with integrins are mediated by the initial formation of CD63-ameloblastin complexes.26 Given this, the previously described report, suggesting that amelogenin uptake may involve integrin binding19 may actually involve integrin binding with CD63–amelogenin complexes.
Both CD63 and integrins are implicated in signal transduction and it is tempting to speculate that the bioactivity associated with EMD/amelogenin may be mediated via amelogenin binding to CD63 and/or integrins resulting in specific signal transduction events. Since both CD63 and integrin can be subject to clathrin mediated endoytosis22, 27 endoyctosis of CD63–amelogenin and/or integrin–amelogenin complexes would provide a means of attenuating amelogenin signalling.
In the present study, fluorescence did not completely fill the vesicle-like structures and a dark zone sometimes separated a florescent core from the vesicular boundary. This separation was not apparent with immunohistochemistry where the anti-amelogenin positive material extended to the vesicular boundary (hence the perception that immunostained vesicles were often bigger than fluorescently stained vesicles). This supports the previously published notion that these vesicles are lysosomal,17 since the observed loss of fluorescence adjacent to the lysosomal membrane may be due to lysosomal membrane proton pumps acidifying the lysosomal lumen to <pH 5 which abolishes fluorescein fluorescence. The use of fluorescein labelled probes and pH dependent fluorescence is a well documented technique for studying the acidification of intracellular compartments.28
Published work on the cellular uptake of amelogenins has not revealed whether amelogenin uptake is selective i.e. whether certain amelogenin isoforms are taken up preferentially compared to others or what happens to the amelogenin in the intracellular environment. Xu et al.16 used recombinant full length amelogenin but SDS-PAGE and Western blotting showed it contained multiple amelogenin derived molecules and it was not clear which of these molecules were being internalised by cells. Shapiro et al.17 used a crude cell extract of their amelogenin-DsRed hybrid protein and it is unclear if the amelogenin part of this construct remained intact or whether a multitude of hybrid proteins were present exhibiting amelogenin that had been processed to varying degrees.
The results showed that HPDL fibroblasts take up EMD components. The dominant protein internalised corresponds to the 20 kDa amelogenin (P148). This, with time, was degraded to a 5 kDa molecule. When EMD free of any 5 kDa protein was applied to cells, there was still an intracellular accumulation of 5 kDa material indicating that this 5 kDa material is an accumulating intracellular degradation product of larger EMD components taken up (notably the 20 kDa amelogenin). When the FITC labelled 5 kDa fraction from FITC-EMD was added to cells no uptake was observed within the limits of detection (Fig. 3). Although this does not rule out the possibility that some 5 kDa component can be taken up directly, it supports the notion that the intracellular levels of the 5 kDa protein can in part at least be due to degradation of a larger precursor. This intracellular 5 kDa degradation product is most likely to be TRAP (tyrosine rich amelogenin peptide – corresponding to residues 1–45 of the 20 kDa amelogenin P148). We base this assumption on the fact that the intracellular 5 kDa protein was fluorescent and must therefore contain the N-terminal of its precursor protein and/or the single lysine residue (residue 24) present in the 20 kDa amelogenin (since fluorescein labelling is via primary amines either at the N-terminal or on the side chains of internal lysine residues). The 20 kDa amelogenin is well known to be sensitive to cleavage between residues 44 and 45.29 This particular cleavage occurs in the developing enamel matrix during amelogenesis and it appears that this processing step is mimicked here in the intracellular environment of the HPDL fibroblast.
The question regarding the mechanism of EMD action, particularly with regard to the role of specific molecules, is still unresolved. It is clear that, albeit poorly characterised, amelogenins, derived by proteolytic processing of recombinant amelogenin, are taken up by HAT-7 cells in culture and can influence amelogenin gene expression by stabilising amelogenin mRNA.16 However, this would require amelogenin to be released from the vesicles described here and elsewhere17 or by amelogenin being taken up and released directly into the cytoplasm. EMD has been shown to affect the expression of multiple genes30 and it remains unclear to what extent cell surface receptor (e.g. CD63) activation and/or internalisation of EMD proteins influence gene expression. Interestingly, CD63 is capable of transducing signals across the cell membrane in association with serine protein kinase activity.31 Amelogenin binding to cell surface CD6317 may trigger a kinase based signal transduction mechanism leading to specific intracellular events that give rise to EMD’s bioactivity.
In summary, HPDL cells selectively take up amelogenin from EMD and internalise it in cytoplasmic vesicle like structures. With time, a 5 kDa peptide (which may be TRAP) accumulates in the cells following degradation of EMD initially taken up. The effect of internalised amelogenin on cell function remains to be clarified but it may be important in terms of the bioactivity associated with EMD.
Acknowledgments
Funding: This work was supported by the European Commission Fifth Framework Programme (Grant Number QLK3-CT-2001-00090) and The Wellcome Trust (Grant Number 082448).
Ethical approval: Teeth extracted for orthodontic reasons at Leeds Dental were used in this work. Patients gave consent for teeth to be used in research in accordance with the Institute’s regulations relating to consent and ethics that were in force at the time.
Footnotes
Competing interests: There are no conflicts of interest to declare from the authors.
References
- 1.Straumann The regenerative approach. Available from: http://www.straumann.co.uk/gb-index/products/products-biologics/products-emdogain.htm [cited 13.04.12]
- 2.Gestrelius S, Andersson C, Lidstrom D, Hammarstrom L, Somerman M. In vitro studies on periodontal ligament cells and enamel matrix derivative. Journal of Clinical Periodontology. 1997;24(9 Pt 2):685–692. doi: 10.1111/j.1600-051x.1997.tb00250.x. [DOI] [PubMed] [Google Scholar]
- 3.Maycock J, Wood SR, Brookes SJ, Shore RC, Robinson C, Kirkham J. Characterization of a porcine amelogenin preparation EMDOGAIN, a biological treatment for periodontal disease. Connective Tissue Research. 2002;43(2–3):472–476. doi: 10.1080/03008200290000880. [DOI] [PubMed] [Google Scholar]
- 4.Viswanathan HL, Berry JE, Foster BL, Gibson CW, Li Y, Kulkarni AB, et al. Amelogenin: a potential regulator of cementum-associated genes. Journal of Periodontology. 2003;74(10):1423–1431. doi: 10.1902/jop.2003.74.10.1423. [DOI] [PubMed] [Google Scholar]
- 5.Tompkins K, Alvares K, George A, Veis A. Two related low molecular mass polypeptide isoforms of amelogenin have distinct activities in mouse tooth germ differentiation in vitro. Journal of Bone and Mineral Research. 2005;20(2):341–349. doi: 10.1359/JBMR.041107. [DOI] [PubMed] [Google Scholar]
- 6.Tompkins K, George A, Veis A. Characterization of a mouse amelogenin [A-4]/M59 cell surface receptor. Bone. 2006;38(2):172–180. doi: 10.1016/j.bone.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Tompkins K, Veis A. Polypeptides translated from alternatively spliced transcripts of the amelogenin gene, devoid of the exon 6a, b, c region, have specific effects on tooth germ development in culture. Connective Tissue Research. 2002;43(2–3):224–231. doi: 10.1080/03008200290001096. [DOI] [PubMed] [Google Scholar]
- 8.Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, et al. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. Journal of Biological Chemistry. 2000;275(52):41263–41272. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- 9.Kawase T, Okuda K, Momose M, Kato Y, Yoshie H, Burns DM. Enamel matrix derivative (EMDOGAIN) rapidly stimulates phosphorylation of the MAP kinase family and nuclear accumulation of smad2 in both oral epithelial and fibroblastic human cells. Journal of Periodontal Research. 2001;36(6):367–376. doi: 10.1034/j.1600-0765.2001.360604.x. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki S, Nagano T, Yamakoshi Y, Gomi K, Arai T, Fukae M, et al. Enamel matrix derivative gel stimulates signal transduction of BMP and TGF-{beta} Journal of Dental Research. 2005;84(6):510–514. doi: 10.1177/154405910508400605. [DOI] [PubMed] [Google Scholar]
- 11.Lyngstadaas SP, Lundberg E, Ekdahl H, Andersson C, Gestrelius S. Autocrine growth factors in human periodontal ligament cells cultured on enamel matrix derivative. Journal of Clinical Periodontology. 2001;28(2):181–188. doi: 10.1034/j.1600-051x.2001.028002181.x. [DOI] [PubMed] [Google Scholar]
- 12.Somerman MJ, Archer SY, Imm GR, Foster RA. A comparative study of human periodontal ligament cells and gingival fibroblasts in vitro. Journal of Dental Research. 1988;67(1):66–70. doi: 10.1177/00220345880670011301. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Moradian-Oldak J, Iijima M, Bouropoulos N, Wen HB. Assembly of amelogenin proteolytic products and control of octacalcium phosphate crystal morphology. Connective Tissue Research. 2003;44(Suppl 1):58–64. [PubMed] [Google Scholar]
- 15.Inai T, Kukita T, Ohsaki Y, Nagata K, Kukita A, Kurisu K. Immunohistochemical demonstration of amelogenin penetration toward the dental pulp in the early stages of ameloblast development in rat molar tooth germs. Anatomical Record. 1991;229(2):259–270. doi: 10.1002/ar.1092290213. [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Harada H, Yokohama-Tamaki T, Matsumoto S, Tanaka J, Taniguchi A. Reuptake of extracellular amelogenin by dental epithelial cells results in increased levels of amelogenin mRNA through enhanced mRNA stabilization. Journal of Biological Chemistry. 2006;281(4):2257–2262. doi: 10.1074/jbc.M507695200. [DOI] [PubMed] [Google Scholar]
- 17.Shapiro JL, Wen X, Okamoto CT, Wang HJ, Lyngstadaas SP, Goldberg M, et al. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cellular and Molecular Life Sciences. 2007;64(2):244–256. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reseland JE, Reppe S, Larsen AM, Berner HS, Reinholt FP, Gautvik KM, et al. The effect of enamel matrix derivative on gene expression in osteoblasts. European Journal of Oral Sciences. 2006;114(Suppl 1):205–211. doi: 10.1111/j.1600-0722.2006.00333.x. [discussion 254–206, 381–202] [DOI] [PubMed] [Google Scholar]
- 19.Almqvist S, Werthen M, Johansson A, Agren MS, Thomsen P, Lyngstadaas SP. Amelogenin is phagocytized and induces changes in integrin configuration, gene expression and proliferation of cultured normal human dermal fibroblasts. Journal of Materials Science. 2010;21(3):947–954. doi: 10.1007/s10856-009-3952-5. [DOI] [PubMed] [Google Scholar]
- 20.van der Pauw MT, Van den Bos T, Everts V, Beertsen W. Phagocytosis of fibronectin and collagens type I, III, and V by human gingival and periodontal ligament fibroblasts in vitro. Journal of Periodontology. 2001;72(10):1340–1347. doi: 10.1902/jop.2001.72.10.1340. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Tannukit S, Zhu D, Snead ML, Paine ML. Enamel matrix protein interactions. Journal of Bone and Mineral Research. 2005;20(6):1032–1040. doi: 10.1359/JBMR.050111. [DOI] [PubMed] [Google Scholar]
- 22.Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Experimental Cell Research. 2009;315(9):1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 23.Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. Journal of Cell Biology. 1996;132(4):565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honing S, Griffith J, Geuze HJ, Hunziker W. The tyrosine-based lysosomal targeting signal in lamp-1 mediates sorting into Golgi-derived clathrin-coated vesicles. The EMBO Journal. 1996;15(19):5230–5239. [PMC free article] [PubMed] [Google Scholar]
- 25.Berditchevski F, Bazzoni G, Hemler ME. Specific association of CD63 with the VLA-3 and VLA-6 integrins. Journal of Biological Chemistry. 1995;270(30):17784–17790. doi: 10.1074/jbc.270.30.17784. [DOI] [PubMed] [Google Scholar]
- 26.Iizuka S, Kudo Y, Yoshida M, Tsunematsu T, Yoshiko Y, Uchida T, et al. Ameloblastin regulates osteogenic differentiation by inhibiting Src kinase via cross talk between integrin beta1 and CD63. Molecular and Cellular Biology. 2011;31(4):783–792. doi: 10.1128/MCB.00912-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. Journal of Cell Biology. 2009;187(5):733–747. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(7):3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brookes SJ, Robinson C, Kirkham J, Bonass WA. Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Archives of Oral Biology. 1995;40(1):1–14. doi: 10.1016/0003-9969(94)00135-x. [DOI] [PubMed] [Google Scholar]
- 30.Parkar MH, Tonetti M. Gene expression profiles of periodontal ligament cells treated with enamel matrix proteins in vitro: analysis using cDNA arrays. Journal of Periodontology. 2004;75(11):1539–1546. doi: 10.1902/jop.2004.75.11.1539. [DOI] [PubMed] [Google Scholar]
- 31.Iida J, Skubitz AP, McCarthy JB, Skubitz KM. Protein kinase activity is associated with CD63 in melanoma cells. Journal of Translational Medicine. 2005;3:42. doi: 10.1186/1479-5876-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]