Abstract
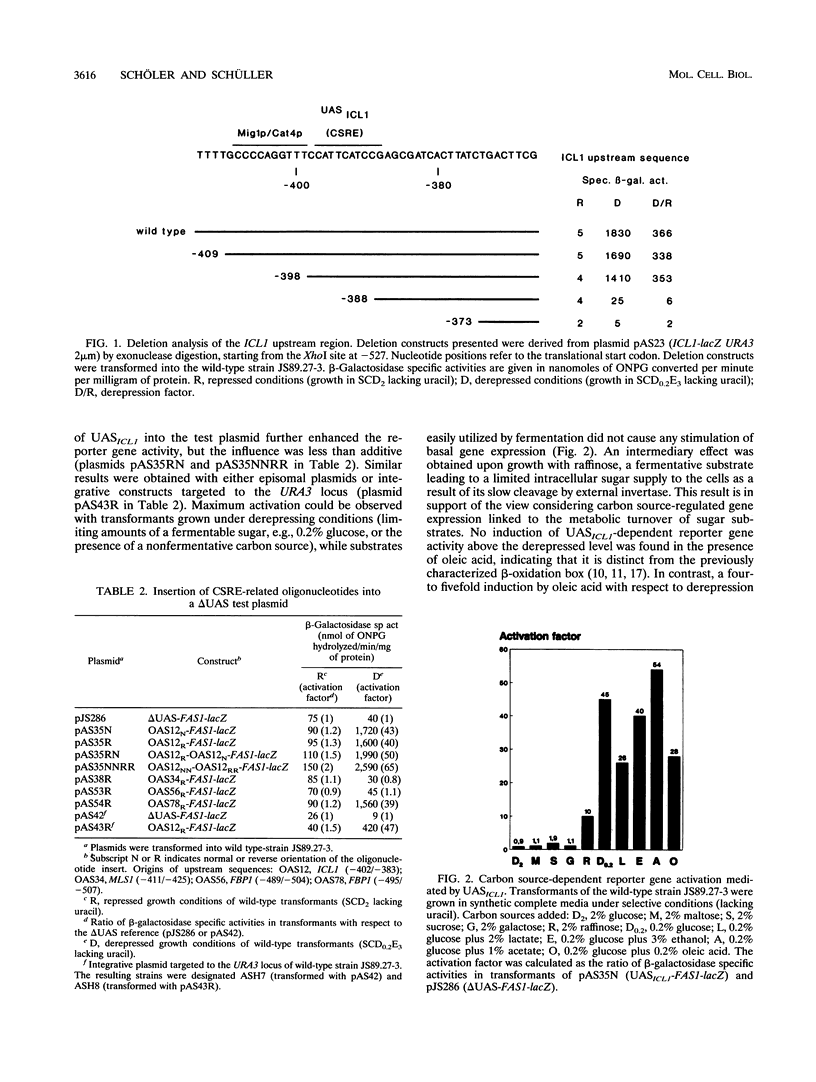
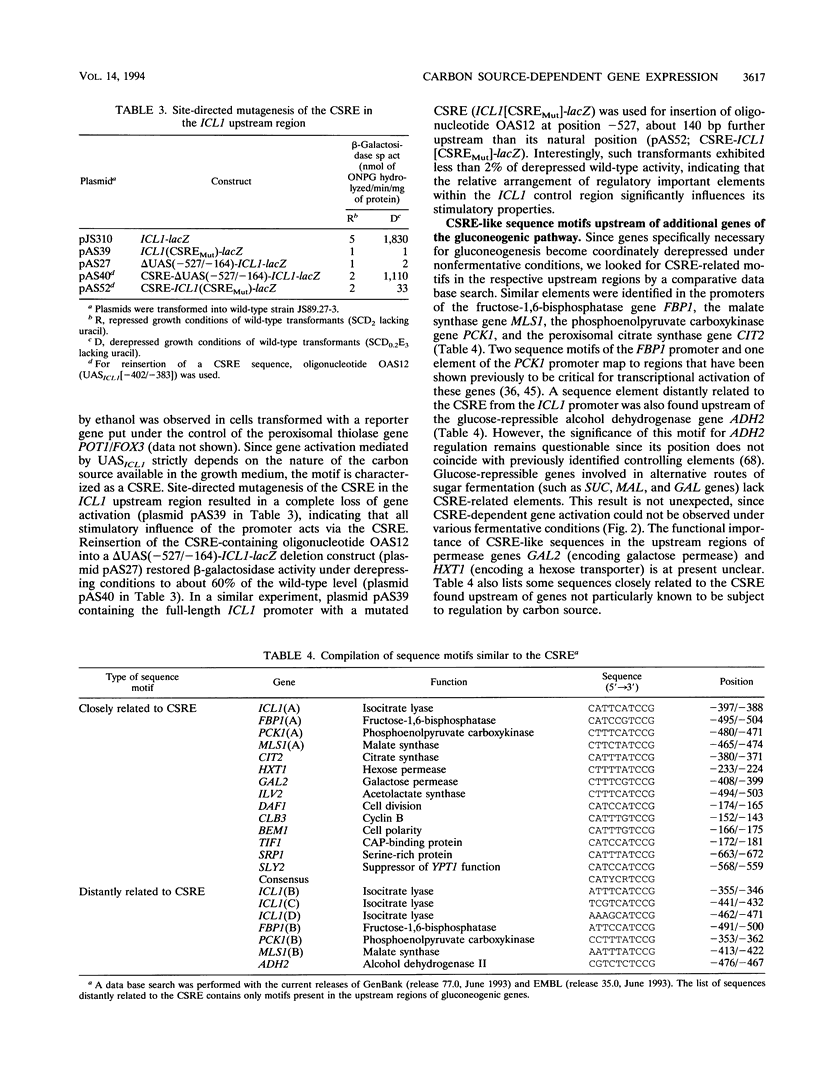
The expression of yeast genes encoding gluconeogenic enzymes depends strictly on the carbon source available in the growth medium. We have characterized the control region of the isocitrate lyase gene ICL1, which is derepressed more than 200-fold after transfer of cells from fermentative to nonfermentative growth conditions. Deletion analysis of the ICL1 promoter led to the identification of an upstream activating sequence element, UASICL1 (5' CATTCATCCG 3'), necessary and sufficient for conferring carbon source-dependent regulation on a heterologous reporter gene. Similar sequence motifs were also found in the upstream regions of coregulated genes involved in gluconeogenesis. This carbon source-responsive element (CSRE) interacts with a protein factor, designated Ang1 (activator of nonfermentative growth), detectable only in extracts derived from derepressed cells. Gene activation mediated by the CSRE requires the positively acting derepression genes CAT1 (= SNF1 and CCR1) and CAT3 (= SNF4). In the respective mutants, Ang1-CSRE interaction was no longer observed under repressing or derepressing conditions. Since binding of Ang1 factor to the CSRE could be competed for by an upstream sequence derived from the fructose-1,6-bisphosphatase gene FBP1, we propose that the CSRE functions as a UAS element common to genes of the gluconeogenic pathway.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achstetter T., Wolf D. H. Proteinases, proteolysis and biological control in the yeast Saccharomyces cerevisiae. Yeast. 1985 Dec;1(2):139–157. doi: 10.1002/yea.320010203. [DOI] [PubMed] [Google Scholar]
- Bailey R. B., Woodword A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193(3):507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Neigeborn L., Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984 May;107(1):19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol. 1989 Nov;9(11):5034–5044. doi: 10.1128/mcb.9.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M., Freidel K., Löhning C. Characterization of trans-acting mutations affecting Ty and Ty-mediated transcription in Saccharomyces cerevisiae. Curr Genet. 1991 Dec;20(6):441–448. doi: 10.1007/BF00334769. [DOI] [PubMed] [Google Scholar]
- Dombek K. M., Camier S., Young E. T. ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol Cell Biol. 1993 Jul;13(7):4391–4399. doi: 10.1128/mcb.13.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einerhand A. W., Kos W. T., Distel B., Tabak H. F. Characterization of a transcriptional control element involved in proliferation of peroxisomes in yeast in response to oleate. Eur J Biochem. 1993 May 15;214(1):323–331. doi: 10.1111/j.1432-1033.1993.tb17927.x. [DOI] [PubMed] [Google Scholar]
- Einerhand A. W., Voorn-Brouwer T. M., Erdmann R., Kunau W. H., Tabak H. F. Regulation of transcription of the gene coding for peroxisomal 3-oxoacyl-CoA thiolase of Saccharomyces cerevisiae. Eur J Biochem. 1991 Aug 15;200(1):113–122. doi: 10.1111/j.1432-1033.1991.tb21056.x. [DOI] [PubMed] [Google Scholar]
- Eisen A., Taylor W. E., Blumberg H., Young E. T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988 Oct;8(10):4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol Gen Genet. 1980;178(3):633–637. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. Glycolytic enzymes and intermediates in carbon catabolite repression mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1980 Jan;177(2):345–350. doi: 10.1007/BF00267449. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982 Sep;151(3):1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E., Moreno F., Rodicio R. The ICL1 gene from Saccharomyces cerevisiae. Eur J Biochem. 1992 Mar 15;204(3):983–990. doi: 10.1111/j.1432-1033.1992.tb16720.x. [DOI] [PubMed] [Google Scholar]
- Filipits M., Simon M. M., Rapatz W., Hamilton B., Ruis H. A Saccharomyces cerevisiae upstream activating sequence mediates induction of peroxisome proliferation by fatty acids. Gene. 1993 Sep 30;132(1):49–55. doi: 10.1016/0378-1119(93)90513-3. [DOI] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991 Oct;11(10):5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol. 1988 Feb;8(2):647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M. Carbon catabolite repression in yeast. Eur J Biochem. 1992 Jun 1;206(2):297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Promoter elements determining weak expression of the GAL4 regulatory gene of Saccharomyces cerevisiae. Mol Cell Biol. 1993 Aug;13(8):4999–5009. doi: 10.1128/mcb.13.8.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hohmann S., Gozalbo D. Structural analysis of the 5' regions of yeast SUC genes revealed analogous palindromes in SUC, MAL and GAL. Mol Gen Genet. 1988 Mar;211(3):446–454. doi: 10.1007/BF00425699. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kuchin S. V., Kartasheva N. N., Benevolensky S. V. Genes required for derepression of an extracellular glucoamylase gene, STA2, in the yeast Saccharomyces. Yeast. 1993 May;9(5):533–541. doi: 10.1002/yea.320090510. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992 Sep;6(9):1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F., Fugit D. R., Mackay V. L. Pleiotropic Mutations at the TUP1 Locus That Affect the Expression of Mating-Type-Dependent Functions in SACCHAROMYCES CEREVISIAE. Genetics. 1980 Apr;94(4):899–920. doi: 10.1093/genetics/94.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J., Tanouye L., Michels C. A. The UAS(MAL) is a bidirectional promotor element required for the expression of both the MAL61 and MAL62 genes of the Saccharomyces MAL6 locus. Curr Genet. 1992 Sep;22(3):181–189. doi: 10.1007/BF00351724. [DOI] [PubMed] [Google Scholar]
- Lombardo A., Cereghino G. P., Scheffler I. E. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Jul;12(7):2941–2948. doi: 10.1128/mcb.12.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M., Nehlin J. O., Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994 Mar;14(3):1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Yoshimatsu T., Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K., Entian K. D. Genetic analysis of serine biosynthesis and glucose repression in yeast. Curr Genet. 1992 Apr;21(4-5):295–300. doi: 10.1007/BF00351686. [DOI] [PubMed] [Google Scholar]
- Mercado J. J., Gancedo J. M. Regulatory regions in the yeast FBP1 and PCK1 genes. FEBS Lett. 1992 Oct 19;311(2):110–114. doi: 10.1016/0014-5793(92)81379-z. [DOI] [PubMed] [Google Scholar]
- Mukai Y., Harashima S., Oshima Y. AAR1/TUP1 protein, with a structure similar to that of the beta subunit of G proteins, is required for a1-alpha 2 and alpha 2 repression in cell type control of Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jul;11(7):3773–3779. doi: 10.1128/mcb.11.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992 Oct 25;20(20):5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984 Dec;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics. 1987 Feb;115(2):247–253. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B. F., Needleman R. B. Identification of the upstream activating sequence of MAL and the binding sites for the MAL63 activator of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Jul;10(7):3797–3800. doi: 10.1128/mcb.10.7.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederacher D., Entian K. D. Characterization of Hex2 protein, a negative regulatory element necessary for glucose repression in yeast. Eur J Biochem. 1991 Sep 1;200(2):311–319. doi: 10.1111/j.1432-1033.1991.tb16187.x. [DOI] [PubMed] [Google Scholar]
- Niederacher D., Schüller H. J., Grzesitza D., Gütlich H., Hauser H. P., Wagner T., Entian K. D. Identification of UAS elements and binding proteins necessary for derepression of Saccharomyces cerevisiae fructose-1,6-bisphosphatase. Curr Genet. 1992 Nov;22(5):363–370. doi: 10.1007/BF00352437. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992 Feb 7;68(3):573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Rose M., Albig W., Entian K. D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by hexokinases PI and PII. Eur J Biochem. 1991 Aug 1;199(3):511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x. [DOI] [PubMed] [Google Scholar]
- Sarokin L., Carlson M. Short repeated elements in the upstream regulatory region of the SUC2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2324–2333. doi: 10.1128/mcb.6.7.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schamhart D. H., Ten Berge A. M., Van De Poll K. W. Isolation of a catabolite repression mutant of yeast as a revertant of a strain that is maltose negative in the respiratory-deficient state. J Bacteriol. 1975 Mar;121(3):747–752. doi: 10.1128/jb.121.3.747-752.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler A., Schüller H. J. Structure and regulation of the isocitrate lyase gene ICL1 from the yeast Saccharomyces cerevisiae. Curr Genet. 1993 May-Jun;23(5-6):375–381. doi: 10.1007/BF00312621. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J Bacteriol. 1991 Mar;173(6):2045–2052. doi: 10.1128/jb.173.6.2045-2052.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Isolation and expression analysis of two yeast regulatory genes involved in the derepression of glucose-repressible enzymes. Mol Gen Genet. 1987 Sep;209(2):366–373. doi: 10.1007/BF00329667. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Molecular characterization of yeast regulatory gene CAT3 necessary for glucose derepression and nuclear localization of its product. Gene. 1988 Jul 30;67(2):247–257. doi: 10.1016/0378-1119(88)90401-5. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Hahn A., Tröster F., Schütz A., Schweizer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992 Jan;11(1):107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy J. M., Fraenkel D. G. Fructose bisphosphatase of Saccharomyces cerevisiae. Cloning, disruption and regulation of the FBP1 structural gene. J Mol Biol. 1985 Nov 20;186(2):307–319. doi: 10.1016/0022-2836(85)90107-x. [DOI] [PubMed] [Google Scholar]
- Shah H. C., Carlson G. P. Alteration by phenobarbital and 3-methyl-cholanthrene of functional and structural changes in rat liver due to carbon tetrachloride inhalation. J Pharmacol Exp Ther. 1975 Apr;193(1):281–292. [PubMed] [Google Scholar]
- Sikorski R. S., Boguski M. S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990 Jan 26;60(2):307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Simon M., Adam G., Rapatz W., Spevak W., Ruis H. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol Cell Biol. 1991 Feb;11(2):699–704. doi: 10.1128/mcb.11.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Thrash-Bingham C., Fangman W. L. A yeast mutation that stabilizes a plasmid bearing a mutated ARS1 element. Mol Cell Biol. 1989 Feb;9(2):809–816. doi: 10.1128/mcb.9.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly R. J. Cloning and characterization of the CYC8 gene mediating glucose repression in yeast. Gene. 1988 Dec 15;73(1):97–111. doi: 10.1016/0378-1119(88)90316-2. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Isolation of Saccharomyces cerevisiae mutants constitutive for invertase synthesis. J Bacteriol. 1986 Jun;166(3):1123–1127. doi: 10.1128/jb.166.3.1123-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K. S., Norbeck L. L., Nolan S. L., Atkinson N. S., Hopper A. K. SRN1, a yeast gene involved in RNA processing, is identical to HEX2/REG1, a negative regulator in glucose repression. Mol Cell Biol. 1992 Jun;12(6):2673–2680. doi: 10.1128/mcb.12.6.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992 Nov;8(11):387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Yang X., Hubbard E. J., Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992 Jul 31;257(5070):680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Yamashita I. The GAM1/SNF2 gene of Saccharomyces cerevisiae encodes a highly charged nuclear protein required for transcription of the STA1 gene. Mol Gen Genet. 1991 Aug;228(1-2):270–280. doi: 10.1007/BF00282476. [DOI] [PubMed] [Google Scholar]
- Yu J., Donoviel M. S., Young E. T. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jan;9(1):34–42. doi: 10.1128/mcb.9.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K., Kaufmann I., Rasenberger H., Haubetamann P. Genetics of carbon catabolite repression in Saccharomycess cerevisiae: genes involved in the derepression process. Mol Gen Genet. 1977 Feb 28;151(1):95–103. doi: 10.1007/BF00446918. [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Montgomery D. L., Nichols D. L., Hall B. D. Transcriptional regulation of the yeast cytochrome c gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3627–3631. doi: 10.1073/pnas.76.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]