Abstract
The Synechococcus elongatus KaiA, KaiB and KaiC proteins in the presence of ATP generate a post-translational oscillator (PTO) that runs in a temperature-compensated manner with a period of 24 hours. KaiA dimer stimulates phosphorylation of KaiC hexamer at two sites per subunit, T432 and S431, and KaiB dimers antagonize KaiA action and induce KaiC subunit exchange. Neither the mechanism of KaiA-stimulated KaiC phosphorylation nor that of KaiB-mediated KaiC dephosphorylation is understood in detail at the present time. We demonstrate here that the A422V KaiC mutant sheds light on the former mechanism. It was previously reported that A422V is less sensitive to dark pulse-induced phase resetting and has a reduced amplitude of the KaiC phosphorylation rhythm in vivo. A422 maps to a loop (422-loop) that continues toward the phosphorylation sites. By pulling on the C-terminal peptide of KaiC (A-loop), KaiA removes restraints from the adjacent 422-loop whose increased flexibility indirectly promotes kinase activity. We found in the crystal structure that A422V KaiC lacks phosphorylation at S431 and exhibits a subtle, local conformational change relative to wild-type KaiC. MD simulations indicate higher mobility of the 422-loop in the absence of the A-loop and mobility differences in other areas associated with phosphorylation activity between wild-type and mutant KaiCs. The A-loop···422-loop relay that informs KaiC phosphorylation sites of KaiA dimer binding propagates to loops from neighboring KaiC subunits, thus providing support for a concerted allosteric mechanism of phosphorylation.
The Role of Phosphorylation in the KaiABC Circadian Oscillator from the Cyano-bacterium Synechococcus elongatus
Cyanobacteria are the simplest organisms known to possess a circadian clock.1,2 In Synechococcus elongatus (S. elongatus) the KaiA, KaiB and KaiC proteins3 constitute a post-translational oscillator (PTO)4 that coupled to a transcription/translation oscillatory feedback loop (TTFL)5,6 controls protein expression genome-wide in a non-promoter specific fashion.7,8 The discovery that the KaiA, KaiB and KaiC proteins from S. elongatus in the presence of ATP generate a temperature-compensated oscillator with a 24-hour period in a test tube4 has now been replicated with the coresponding proteins from the thermophile Thermosynechococcus elongatus (T. elongatus).9 The three Kai proteins interact with each other in vitro and in vivo to form heteromultimeric complexes (Figure 1).10–21 The KaiC homo-hexamer,13,22 the central component of the clock, is the result of a gene duplication23 and possesses ATPase,24,25 kinase26,27 and ATP synthase/phosphotransferase (dephosphorylation)28,29 activities. KaiA stimulates KaiC phosphorylation30,31 and KaiB antagonizes KaiA action,32 whereby KaiB binding to KaiC coincides with exchange of subunits among hexamers.33 The KaiC subunit shuffling process appears to be crucial for maintaining a robust amplitude.16
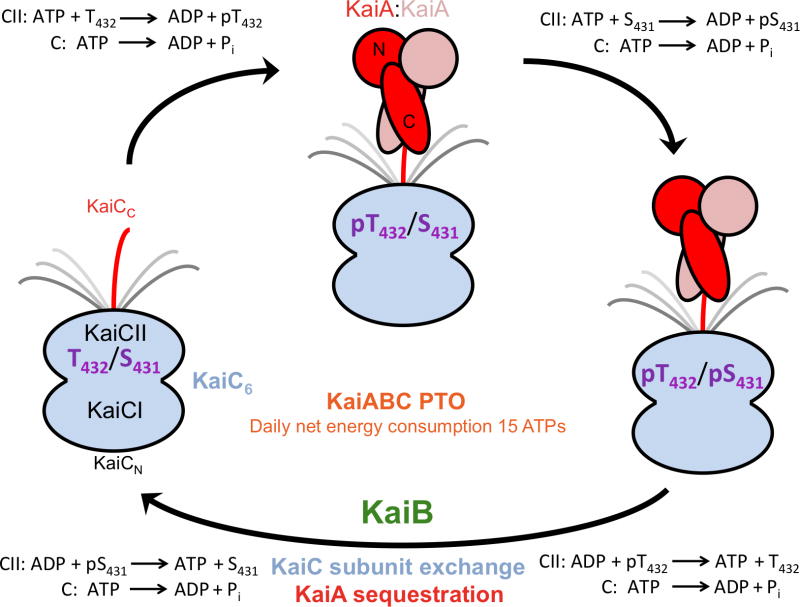
Figure 1.
Phosphorylation and dephosphorylation of KaiC during the daily cycle of the KaiABC PTO. KaiC in the hypo-phosphorylated T432/S431 (TS) state, upon interacting with KaiA dimer via a CII C-terminal peptide is first phosphorylated at T432 residues (pTS) and then also at S431 residues (pTpS). ATPase activity fuels kinase activity in the CII ring. KaiB dimers associate with the KaiC hexamer in the hyper-phosphorylated state. Unlike the C-terminal alpha-helical bundle domains of KaiA dimer (C-KaiA), KaiB displays no affinity for the C-terminal peptide.18 Dephosphorylation first of pT432 residues via an ATP synthase mechanism (TpS) and then of pS431 residues leads back to the hypo-phosphorylated state and is accompanied by KaiC subunit exchange and the appearance of a ternary KaiABC complex in which KaiA is prevented from stimulating KaiC phosphorylation (KaiA sequestration).16,21,42
Phosphorylation and dephosphorylation of KaiC are limited to the C-terminal KaiCII domain,34,35 but both the N-terminal KaiCI ring and KaiCII have ATPase activity.25,28 Phosphorylation occurs first at T432 and then at S431 and dephosphorylation proceeds in the same order: TS → pTS → pTpS → TpS → TS.36,37 The strict order is most likely the result of kinetic control as T432 lies consistently closer to S431 in structures of KaiC.13,38 An increasing number of interactions across the subunit interface as a result of phosphorylation renders unfavorable the reverse reaction, forming the basis for of ratcheting mechanism that drives forward the oscillator.17 In addition to T432 and S431, phosphorylation was also observed at T426 in mutant KaiCs38 and the T426A mutant is arhythmic.34 Although T426 has to be phosophorylatable for the clock to function properly,39 S431 and T426 cannot both carry a phosphate at the same time (the side chain of T426 is H-bonded to pS431)38 and the most likely role of T426 is in the stabilization of pS431 and the latter’s dephosphorylation.28
A hybrid structural approach involving X-ray crystallography, negative-stain and cryo EM, small angle X-ray scattering (SAXS) and NMR has yielded insight into the protein-protein interactions over the daily cycle in the KaiABC PTO.8,13–21,40 Binding between the C-terminal domains of a KaiA dimer to the C-terminal peptide(s) from the hypo-phosphorylated form of KaiC hexamer13–15 triggers phosphorylation first at T432 and then at S431, ultimately resulting in the hyper-phosphorylated form with 12 pT432 and pS431 residues per KaiCII hexamer (Figure 1). KaiB dimers do not compete with KaiA for the same binding site18 and exhibit the highest affinity for the TpS form.41 KaiBC complex formation and dephosphorylation coincide with the appearance of another Kai protein complex form of distinct shape that contains all three proteins.16 Native PAGE experiments using full-length KaiC and a mutant lacking the C-terminal peptide tail indicate that binding of KaiB to KaiC hexamer creates a new binding site for KaiA at the KaiBC interface.42 The EM envelope of the ternary complex is consistent with binding of KaiA monomers to the KaiBC particle and native PAGE confirmed that the N-terminal KaiA pseudoreceiver domain is used for binding.21 The KaiBC complex thus sequesters KaiA dimer and prevents it from binding to C-terminal KaiC peptide(s), an event that is crucial for proper functioning of the clock,37,43,44 ultimately bringing about the hypo-phosphorylated state before the PTO cycle starts anew (Figure 1).
Unraveling of the C-terminal KaiC A-Loop by KaiA Stimulates KaiC Kinase Activity
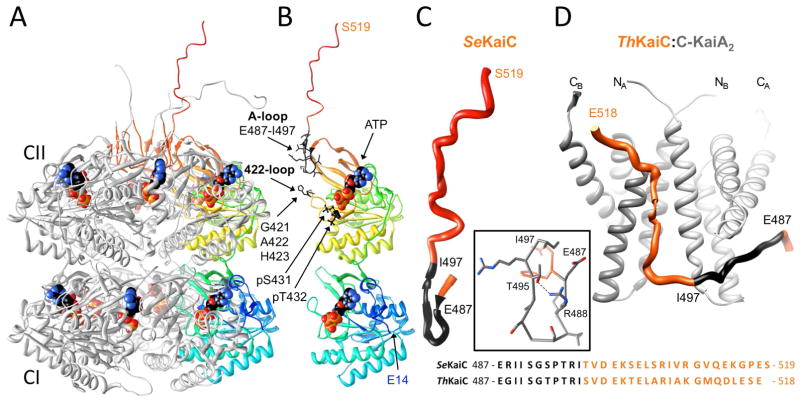
The KaiC homo-hexamer takes on the shape of a double-doughnut with overall dimensions of ca. 100×100×100 Å.13 A constricted waist in the KaiC barrel marks the boundary between the rings formed by N-terminal KaiCI subunit domains and C-terminal KaiCII subunit domains (Figure 2A). ATP molecules occupy clefts between subunits in both halves such that phosphorylation at T432 and S431 proceeds across the subunit interface (Figures 2A, 3).34 With the exception of the C-terminal region, the CI and CII domains adopt very similar folds (see Figure 2B in ref. 13). Residues 245–260 at the C-terminal end of CI act as a linker that adopts an extended conformation between CI and CII. Residues 483–497 near the C-terminus of CII are folded into an S-shaped loop and the last 22 residues (498–519) protrude from the CII dome and exhibit dynamic conformational behavior (Figure 2). The A-loop is made up of residues 487–497 with the hydrogen-bonded R488 and T495 acting as a clamp (Figure 2C). Deletion of the last 30 residues in KaiC (Δ30C-KaiC mutant) abolishes circadian rhythmicity and the R488A mutant displays enhanced damping of rhythmicity.15
Figure 2.
Three-dimensional structures of S. elongatus KaiC and the T. elongatus KaiA-KaiC peptide complex. (A) Crystal structure of full-length KaiC (PDB ID 3DVL; http://www.rcsb.org).13,17 Subunits B, C, D, E and F of the hexamer are colored in gray (except for A-loop residues 487–497 that are highlighted in red), and subunit A is shown in rainbow coloring. ATP molecules bound between subunits are shown in space filling mode with carbon atoms colored in black. At the N-terminal end of CI (cyan), residues 1–13 are missing in the crystallographic model. C-terminal peptide tails of CII (red) were only traced completely for subunits A and F.15 (B) The A subunit depicted separately in the same orientation and coloring as in panel A. N- and C-terminal residues are labeled along with ATPs, the two phosphorylation sites (T432 and S431), and A-loop residues 487–497 and A422-loop residues 421–423 (amino acids in both loops are highlighted in black). (C) Close-up view of the conformation of the A-loop (black) and CII-terminal region in the crystal structure. Inset: The A-loop conformation is stabilized by a hydrogen bond between R488 and T495. (D) NMR solution structure of the complex between KaiC C-terminal peptide (see panels B and C for coloring) and KaiA dimer (C-terminal domains) from T. elongatus.14 Each KaiA dimer can bind two KaiC peptides although only one peptide is depicted here. Note the stretched conformation of the former A-loop in the structure of the complex compared to the curled arrangement in the crystal structure (C). An alignment of the sequences of C-terminal regions of KaiCs from S. elongatus and T. elongatus below panels C and D illustrates that they are highly similar.
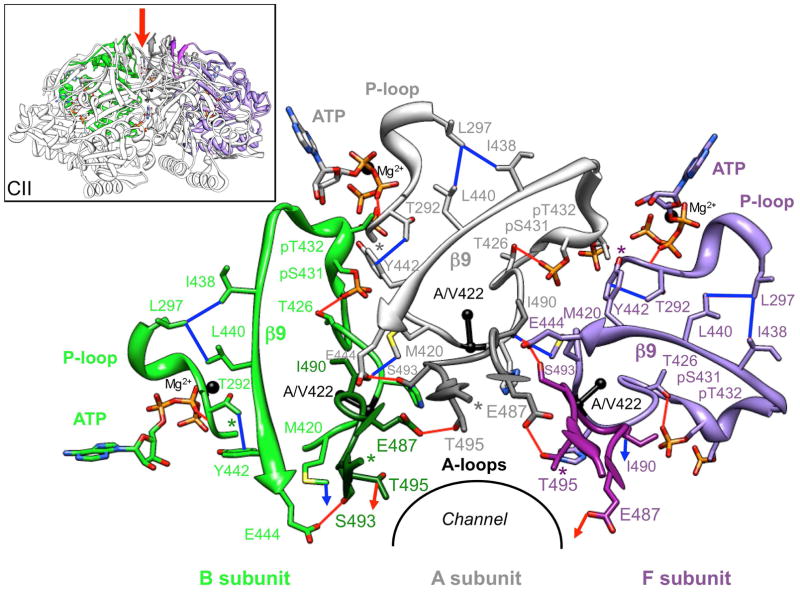
Figure 3.
Components of C-terminal KaiCII subunits crucial for relaying A-loop binding by KaiA to the KaiC kinase active site. The view is down the central channel (red arrow in inset) with N- and C-terminal residues marked by asterisks. A-loops, 422-loops (Cα and Cβ of A422 are shown as black spheres) and adjacent phosphorylation loop regions, including residues T426, pS431 and pT432, β9 strands, P-loops and ATPs (black spheres represent Mg2+ ions) are depicted for three neighboring subunits, A (gray), B (green) and F (purple). A-loops are shown in darker shades, amino acids are labeled and important hydrogen bonds and hydrophobic contacts linking key structural elements within or across subunits are shown as red and blue lines, respectively. Intra-subunit interactions between the A-loop and the 422-loop are illustrated in more detail in Figures 5 and 6.
LiWang and coworkers demonstrated using NMR that the C-terminal domains of KaiA dimer bind to the C-terminal region of KaiC comprising residues I490 to Q513 (Figure 2D).14 In the structure of the C-KaiA:KaiC-peptide complex the A-loop region adopts an extended conformation. Therefore, binding of KaiA unravels the A-loop, thus severing the hydrogen bond between the side chains of R488 and T495 (Figure 2C, D). Interestingly, the KaiC I497 deletion mutant (residues 498–519 deleted) is constitutively hypo-phosphorylated and the KaiC E487 deletion mutant (residues 488–519, including the A-loop deleted) is constitutively hyper-phosphorylated.45 Moreover, the E487A mutant is arhythmic and also constitutively hyper-phosphorylated. Therefore, the conformation of the KaiC A-loop (extended vs. curled) constitutes a circadian phase “switch” that is flipped by KaiA.45 Like the E487A mutation T495A also confers arhythmicity,3 and the crystal structure of KaiC provides a rationalization for the more severe consequences of the E487A and T495A mutations compared to R488A.13 In particular, hydrogen bonds between E487 and T495 residues from adjacent subunits connect A-loops around the entire CII ring (see the illustration with three subunits depicted in Figure 3). Therefore, KaiA binding to a C-terminal KaiC peptide and concomitant stretching of the A-loop from one subunit can be expected to destabilize the A-loops from neighboring subunits.45 Abrogation of this inter-subunit hydrogen bonding network as a result of either E487A or T495A mutation has drastic consequences in terms of function. In comparison to the T495A mutant that lacks both the intra-subunit hydrogen bond to R488 and the inter-subunit hydrogen bond to E487, the R488A mutation results only in the loss of an intra-subunit hydrogen bond and might not alter anything beyond facilitating the transition from the curled to the extended conformation of the A-loop region upon KaiA binding.
Although the conformational changes in the KaiC C-terminal region as a result of docking of KaiA are now hypothetically understood, it is unclear how the unfolding of the A-loop is transmitted to the KaiCII kinase active site beyond a model45 that invoked a potential change in ATP orientation triggered by a relay involving A-loop, the I438-E444 region of the β9 strand and the P-loop (Figure 3). The E444D mutation is thought to disrupt this relay and the mutant was found to be constitutively hyper-phosphorylated.45
Phenotype of the KaiC A422V Mutant
Some 40,000 ethylmethanesulfonate-mutagenized S. elongatus strains were screened in terms of their reaction to a 5-hour dark pulse. Among many arhythmic and period-altered mutants the so-called pr1 mutant was the only one that retained a normal period but displayed an abnormal response to the dark pulse.46 The pr1 mutation corresponds to an alanine to valine substitution at residue 422 in KaiCII (Figure 2B). KaiC acts as the main repressor of kaiBC expression such that overexpresssion of KaiC represses activity of its own promoter and temporal overexpression triggers a shift in the phase of circadian rhythm in a phase-dependent manner. Both functions were affected in the pr1 mutant. In addition to the inability to shift phase following a dark pulse at any time of the day, the phenotype of the A422V mutant includes: (i) abolished rhythm of KaiC accumulation, (ii) reduced amplitude of the KaiC phosphorylation rhythm and (iii) reduced phase shift compared to wild-type S. elongatus when KaiC and pr1-KaiC were co-expressed in wild-type cells.46 A rationale for the drastic changes in clock function as a result of a conservative Ala→Val substitution is currently missing. The A422 residue lies in proximity to the T432 and S431 phosphorylation sites and also maps to one of the so-called KaiA-binding domains,11 i.e. the dome-shaped KaiCII surface and residues located beneath it13 along with C-terminal A-loops and tentacles (Figure 2).14 However, a mechanism linking residue 422 to KaiA binding and stimulation of phosphorylation has not yet been proposed.
We hypothesized that the effects displayed by the KaiC A422V mutant on key properties of the cyanobacterial clock, such as the ability to phase shift, the amplitude of KaiC phosphorylation and rhythmic accumulation of KaiC should provide key insights into the interaction between KaiA and KaiC and the mechanism of KaiA-stimulated KaiC phosphorylation. Therefore, we undertook a thorough investigation of the structure, stability and dynamic behavior of the A422V KaiC mutant protein.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Site-directed mutagenesis of S. elongatus KaiC was performed by a modified method of Papworth and coworkers.47 The His6-tagged (C-terminus) KaiC A422V mutant protein was expressed in E. coli (BL21 cell line, Novagen/EMD Biochemicals) following earlier protocols for wild-type KaiC.13,22 The protein was first purified by metal affinity chromatography (TALON IMAC resin, Takara Bio Clontech) and then by gel filtration chromatography (Sephacryl S-300 HR resin, GE Healthcare). The purity of the protein was assessed by SDS-PAGE and its identity established by trypsin digestion and subsequent analysis by MALDI-TOF mass spectrometry.
Crystal Structure of the S. elongatus KaiC A422V Mutant
The His6-tagged KaiC A422V mutant protein was crystallized as previously described for wild-type KaiC.13 Crystals were flash-frozen and X-ray diffraction data collected on the 21-ID-F beamline of the LS-CAT at the Advanced Photon Source (Argonne, IL) and processed with HKL2000.48 Selected crystal data and data collection parameters are listed in Table 1. The structure was determined by molecular replacement with the program CNS49 and using the native KaiC coordinates (PDB ID 3DVL)15,17 without solvent molecules as the search model. Model building was performed with the program Coot50 and refinement was carried out with the program Phenix51 using all data to 3.3 Å resolution. Final refinement parameters are listed in Table 1.
Table 1.
Crystal Data and Refinement Statistics for KaiC A422V Mutant
| Data Collection | |
|---|---|
| space group | P212121 |
| unit cell constants [Å] | a=131.36, b=134.47, c=203.91 |
| X-ray source | APS, LS-CAT, 21-ID-F |
| detector | MAR300 |
| wavelength [Å] | 0.97872 |
| resolution range [Å] (last shell) | 30-3.35 (3.41-3.35) |
| redundancy (last shell) | 6.6 (6.7) |
| R-merge [%] | 5.6 (50.3) |
| no. of unique reflections (last shell) | 52,438 (2,564) |
| completeness [%] (last shell) | 100 (100) |
| I/σ(I) (last shell) | 31.1 (2.6) |
| Refinement | |
| R-work [%] | 21.7 |
| R-free [%] | 26.0 |
| reflections used to calculate R-free [%] | 5.0 |
| overall average B-factor [Å2] | 68.5 |
| no. of protein and ATP atoms | 23,796 |
| no. of ATP molecules | 12 |
| no. of solvent mol., Mg2+, phosphate ions | 397, 7, 8 |
| r.m.s.d. from ideal geometry | |
| bonds [Å] | 0.007 |
| angles [°] | 0.7 |
| Ramachandran plot | |
| favored region [%] | 90.9 |
| outliers [%] | 2.4 |
Crystallographic Data Deposition
Final coordinates and structure factors for the crystallographic model of the S. elongatus KaiC A422V mutant protein have been deposited in the Protein Data Bank (http://www.rcsb.org): PDB ID 4IJM.
CD Melting Experiments
Melting experiments with wt and A422V mutant KaiC were conducted in a quartz cuvette with 1 mm light path on a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller. The protein concentration was 0.25 mg/mL (4.3 μM) in a solution containing 20 mM HEPES-NaOH pH 8, 100 mM NaCl, 5% glycerol, 1 mM ATP and 2 mM 2-mercaptoethanol. Thermal denaturation was monitored by recording the ellipticity at 222 nm while the sample was heated from 15°C to 95°C at 1°/min.
Molecular Dynamics Simulations
Starting coordinates for the KaiC simulations were taken from the crystal structure with PDB ID 3DVL.15 Four variations of the protein were generated manually: wildtype sequence versus A422V mutant, combined with making the C-terminus E487 versus I497. The LEaP program in AmberTools 12 was used to parameterize the systems with the current AMBER ff12SB force field52 for the protein, the Meagher et al. parameters53 for the twelve ATP molecules, and the SPC/E water model54 and compatible ions. LEaP was used to add missing hydrogen atoms, 90 Na+ ions for charge neutralization, and to solvate the proteins using 66,063 (C-terminal E487) or 66,273 water molecules (C-terminal I497) in an orthorhombic box, with an 11 Å clearance to the box edges.
Simulations were performed in AMBER’s PMEMD on GPU55 using periodic boundary conditions, a real-space cutoff of 9 Å, a Langevin thermostat set to 293K with gamma_ln = 3.0/ps, constant pressure of 1.0 bar with tau_p set to 3.0, SHAKE and SETTLE algorithms56,57 for bonds involving hydrogen, and a timestep of 1.5 fs. Before simulation, the systems were minimized in three stages: first by allowing solvent to relax while the protein was held fixed with strong 1,000 kcal/mol/Å2 restraints, then by allowing the protein atoms to relax while the solvent was held fixed with weaker 100 kcal/mol/Å2 restraints, and finally by relaxing the entire system without restraints. After minimization, the systems were equilibrated using seven stages of decreasing restraints on the solute atoms from 16 to 0 kcal/mol/Å2, over the course of 450 ps starting in constant volume conditions. The last stage of equilibration using weak 0.0625 kcal/mol/Å2 restraints was switched to constant pressure conditions using isotropic position scaling that were continued for 25 ns with no restraints for production dynamics.
RESULTS
Crystal Structure of KaiC A422V Mutant
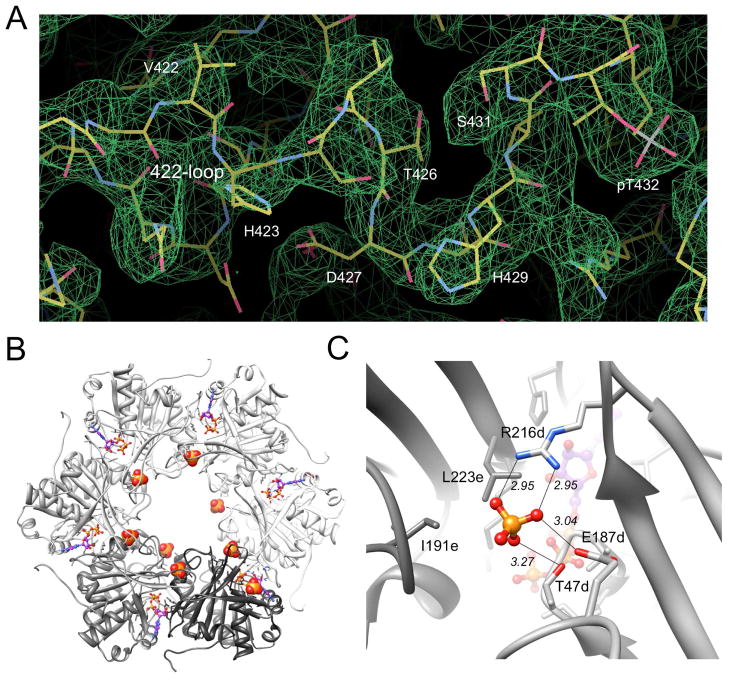
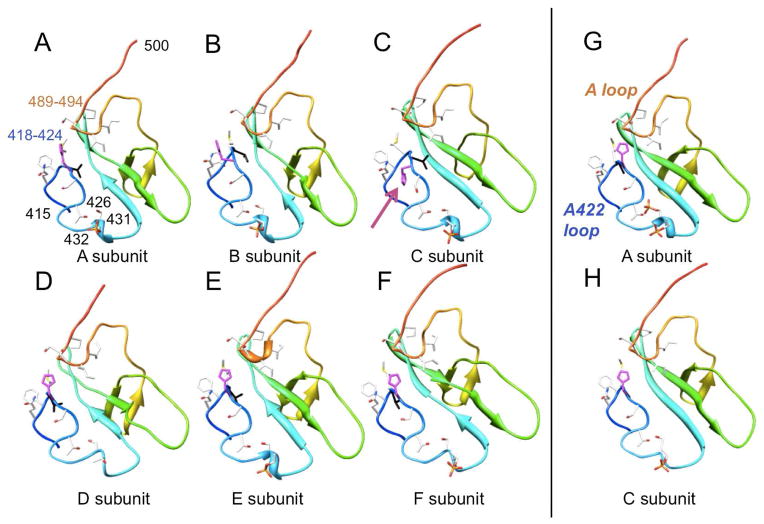
To analyze the effects of replacing alanine by valine at position 422 on the local conformation and the phosphorylation state of KaiC, we determined the crystal structure of the A422V KaiC mutant at 3.3 Å resolution. Crystals of the mutant protein were obtained under conditions very similar to those previously used for wt-KaiC and other mutants.13,21,28,38 Crystal data, X-ray diffraction data and refinement statistics are summarized in Table 1 and a representative example of the quality of the final electron density is depicted in Figure 4A. Inspection of the T432 and S431 phosphorylation sites in CII domains from subunits of the A422V mutant reveals that T432 carries a phosphate in all six and that S431 is not phosphorylated (Figure 5A–F). Thus, the A422V mutant displays phosphorylation that amounts to 60% of the level seen in the crystal structure of wt-KaiC [10 phosphates with S431 residues in subunits C (Figure 5H) and D not being phosphorylated].13,34 Interestingly, eight phosphate ions are visible in the CI half (Figure 4B, C), undoubtedly the result of hydrolysis of ATP by the ATPase there. Previous structures of the KaiC hexamer did not reveal phosphate ions in the lower ring.
Figure 4.
Quality of the KaiC A422V mutant crystal structure and phosphate ion coordination in the CI half. (A) Final 2Fo-Fc Fourier sum electron density (1σ threshold) in the region of V422 to pT432 from subunit C. (B) The KaiCI ring viewed from the C-terminal side with phosphate ions mostly coordinated near the central channel. ATP molecules bound between subunits of the hexamer (A-subunit, white → F-subunit, charcoal) are drawn in ball-and-stick mode. (C) Close-up view of the phosphate ion located between subunits D and E. Selected amino acids are depicted in stick mode and are labeled and the coordination geometry is indicated by thin solid lines with distances in Å.
Figure 5.
Conformations of the KaiCII C-terminal halves (residues 415–500), including A-loop and V422-loop in (A-F) all six subunits in the crystal structure of the KaiC A422V mutant. The protein backbone is shown in secondary structure cartoon mode and rainbow coloring. Side chains of selected amino acids are depicted in stick mode and labeled, with carbon, oxygen, nitrogen, sulfur and phosphorus atoms colored in gray, red, blue, yellow and orange, respectively. The V422 side chain is highlighted in black and the H423 side chain is highlighted in magenta. The conformations of the corresponding regions from subunits (G) A and (H) C in the wild-type KaiC crystal structure (PDB ID 3DVL) are shown for comparison (A422 highlighted in black). In subunit C of A422V KaiC, the orientation of H423 is flipped relative to all other subunits in both the wt and mutant KaiC crystal structures (arrow in panel C). Note the absence of phosphorylation at S431 in the entire A422V KaiC structure.
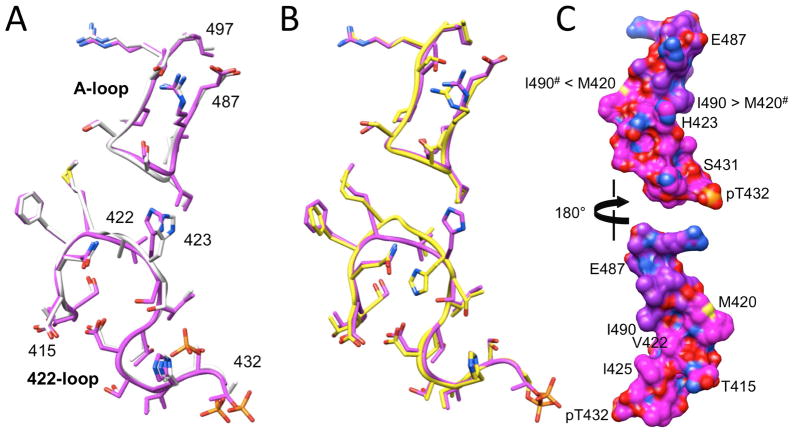
At the structural level, the switch from alanine to valine leads to a subtle conformational adjustment at the apex of the 422-loop as the more bulky valine side chain can no longer be accommodated inside the loop (Figure 5). Thus, the isopropyl moiety swings outwards and away from the loop such that its orientation is more or less perpendicular to that of loop main chain atoms. This effect is particularly obvious in the C subunit (Figure 5C). By comparison, the methyl side chain of alanine is accommodated in the interior of the loop and therefore lies roughly in the best plane defined by loop residue Cα atoms. Despite the different orientations of the alanine and valine side chains, the conformation of the 422-loop exhibits only very minor deviations between the wt and mutant KaiC crystal structures (Figure 6A). As the superimposition of the loops from subunits A in the two structures illustrates, the most significant adjustments occur with the backbone and side chains of residue 422.
Figure 6.
Comparison between the conformations of the A-loop and A/V422-loop regions from selected subunits in the KaiC wt and A422V mutant crystal structures. (A) Superimposition of the loops from subunits A in wt KaiC (gray chain) and A422V KaiC (magenta chain). (B) Superimposition of the loops from subunits A (magenta chain) and C (yellow chain) in A422V KaiC. (C) Surface model of the A-loop···V422-loop van der Waals interaction in KaiCII with pT432 in the foreground (top) and rotated around the vertical by 180° (bottom). Carbon atoms of A-loop and V422-loop residues are colored in purple and magenta, respectively, and oxygen, nitrogen, sulfur and phosphorus atoms are colored in red, blue, yellow and orange, respectively. Selected residues are labeled and arrows between I490 (A-loop) and M420 (V422-loop) residues indicate hydrophobic contacts between side chains from adjacent subunits (marked by #).
In addition to the local rotation of the loop backbone that helps push out the valine side chain in all six subunits of the mutant crystal structure, subunit C displays a conformational change for H423 (Figure 5). Unlike in the rest of the A422V KaiC hexamer, the side chain of H423 there is flipped and thus directed away from the A-loop, resulting in a slight swelling of the 422-loop (Figure 6B). Whereas the change in the loop conformation as a consequence of the Ala → Val mutation is essentially limited to the immediate vicinity of residue 422 (Figure 6A), the altered orientation of the H423 side chain in subunit C affects the conformation of the peptide chain for residues S420, G421, V422 and H423 (Figure 6B).
A-Loop and A422-Loop are in Direct Contact
As illustrated in Figures 2B, 3, 4A, 5 and 6, the A-loop and the 422-loop are paired and line the KaiC central channel at its C-terminal end. Examination of the distances between residues of the two loops, i.e. S420, G421, A/V422 and H423 (422-loop) and I490, S491 and G492 (A-loop) indicates that the two loops are only weakly associated. In the crystal structure of wt-KaiC hexamer, distances between the amide nitrogen of G421 and the carbonyl oxygen of S491 in the six subunits range from 3.27 Å to 3.77 Å (average 3.52 Å). For atom pairs at the shorter end of this distance range it is reasonable to postulate the existence of a weak hydrogen bond. Moreover, the terminal methyl group of the M420 side chain (Cε) forms a hydrophobic interaction with Cα of G492 (distances range from 3.52 to 4.03 Å; average 3.77 Å). On the other side of the loop, H423 is too far removed from I490 to engage in a hydrogen bond or a hydrophobic contact.
Looking at the loop-loop interaction in the crystal structure of the A422V KaiC mutant, it becomes clear that there are only minor changes in the distances between paired residues relative to wt-KaiC. Distances between N(G421) and O(S491) range from 3.37 to 4.23 Å (average 3.76 Å) in the six subunits and those between Cε(M420) and Cα(G492) range from 3.49 to 4.37 Å (average 3.96 Å). The hydrophobic contact involving methionine is therefore not disrupted by the switch from alanine to valine at position 422 and the concomitant local rotation of the loop backbone. However, except for subunit C (Figure 5C), distances between N(G421) and O(S491) (>3.65 Å) are all outside the hydrogen bonding range. The shorter distance there (3.37 Å) may account for a weak stabilization and is related to the flipped orientation of H423 that pushes up the 422-loop, thus moving it closer toward the A-loop (Figure 6B). As this illustration shows, the A-loop maintains an almost identical conformation and orientation despite the closer approach of the 422-loop relative to a subunit with the more common orientation of the histidine side chain in the KaiC A422V mutant structure (Figure 6A). In the C subunit, the distance between Cε(M420) and Cα(G492) is 4.17 Å, N(V422) and O(490) are separated by 3.67 Å, and one of the outer methyl carbons of the V422 side chain (Cγ1) lies at 3.72 Å from the side chain of I490 (Cγ2), consistent with a hydrophobic interaction.
The closer spacing between the side chains of V422 and I490 from the 422- and A-loops, respectively, is clearly related to the altered backbone conformation of the 422-loop in the C subunit with the flipped orientation of the H423 side chain. However, the valine side chain jutting away from the loop in the crystal structure of the mutant also aids in the formation of a closer approach between residues 422 and 490. In subunit A, the distance between Cγ1(V422) and Cγ2(I490) is 4.33 Å, indicative of a hydrophobic interaction. However, in the remaining subunits, this distance is increased and the contacts can no longer be considered to be of importance for the loop-loop interaction. In the crystal structure of wt-KaiC, the orientation of the A422 side chain precludes a hydrophobic contact to side chain atoms of I490 and all distances exceed 7 Å.
The above comparison between the wt and A422V KaiC crystal structures of the juxtaposition of the A-loop and the 422-loop demonstrates first, that the two loops are not engaged in a tight interaction and, second, that the Ala → Val mutation does not significantly alter this weak association. Although the spacing of loop backbones and side chains may result in slight electrostatic and hydrophobic contributions, it may be more appropriate to think of the two loops as essentially engaging in van der Waals interactions. Inspection of a molecular surface model of the A-loop…V422-loop interface illustrates this point (Figure 6C). Much like an upper lip, the A-loop touches the lower lip - the 422-loop - and neatly seals the gap between them that is apparent in the wire models (Figures 2B, 4A, 5 and 6). However, KaiA binding to the C-terminal KaiCII peptide opens up the lips by unraveling the A-loop and thus freeing the 422-loop from its constraints.
Thermodynamic Stability of the KaiC A422V Mutant
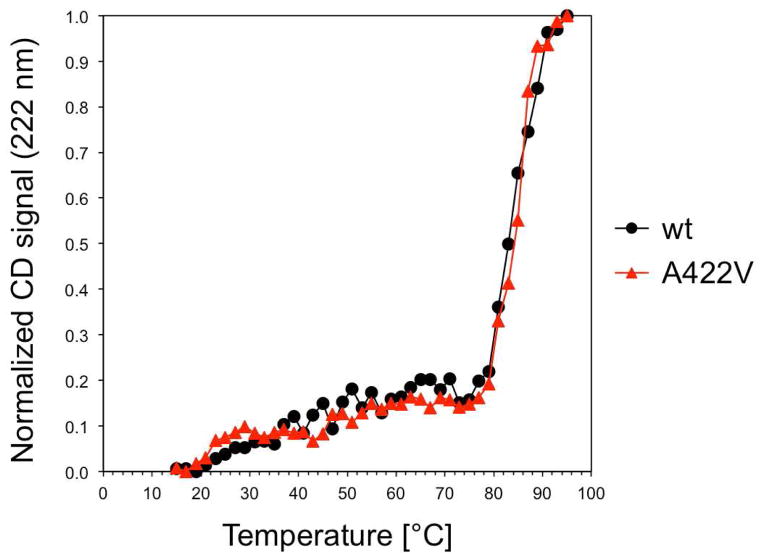
To assess a potential effect of the Ala → Val mutation at residue 422 on the stability of KaiC, we conducted CD melting experiments with both the wt and mutant proteins. As shown in Figure 7, the two proteins exhibit virtually identical melting temperatures of around 85°C. Therefore, the manifold changes upon mutation at the functional level cannot be attributed to an altered stability. The similar stabilities of wt KaiC and the A422V KaiC mutant are in line with the outcome of the above comparison between the conformations of the two proteins. At the structural level the A422V mutation leads to only minor adjustments at the apex of the 422-loop and its orientation relative to the A-loop in the six subunits. Therefore, changes in conformation and/or stability of A422V KaiC relative to wt KaiC are unlikely to account for the phenotype of the pr1 mutant46 and the reduced phosphorylation of the mutant protein in the crystal structure (Figure 5).
Figure 7.
CD-melting curves for wild-type and A422V mutant KaiC indicate virtually identical thermodynamic stability.
Altered Dynamics of the A422 Loop Upon Valine Mutation
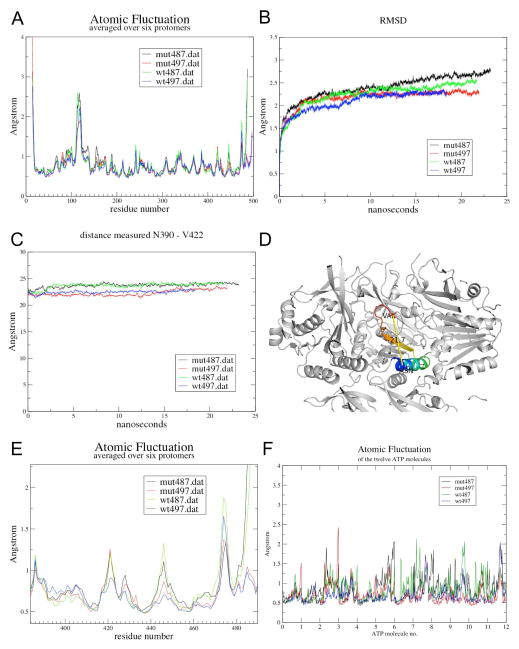
The observations that the A422V mutation appears not to affect the thermodynamic stability of KaiC and does not change the 422-loop conformation in a substantial way, but does attenuate phosphorylation and abrogates phase shifts in vivo could indicate that valine at position 422 in the KaiC sequence alters the flexibility of that loop. To examine this hypothesis and whether presence or absence of the adjacent A-loop residues affects the dynamics of the 422-loop, we conducted molecular dynamics (MD) simulations of hexameric, hyperphosphorylated KaiC (residues 14–497, 12 phosphates on T432 and S431) with either A (wt) or V (mutant) at position 422 (A-loop present) and KaiC (residues 14–487) with either A or V at position 422 (A-loop absent, i.e. a mimic of KaiC with KaiA bound). The current analysis is based on simulations over 25 nanoseconds and focuses on three parameters that represent averages of the six subunits: (i) atomic fluctuations, (ii) root mean square (rms) deviations relative to the initial conformation, and (iii) distances between Cα (N390) and Cα (A/V422) over the duration of the simulation (Figure 8A, B and C, respectively). We chose residue N390 as a reference point in the KaiCII subunit to evaluate the degree of mobility for 422-loop residues as a function of the presence (497) or absence (487) of the A-loop and the identity of the residue at position 422 (A = wt versus V = mut; Figure 8).
Figure 8.
Molecular dynamics (MD) simulations over 25 nanoseconds for S. elongatus KaiC hexamers with either alanine or valine at position 422 and including A-loop residues (E14 to I497; blue and red trace, respectively) or lacking A-loop residues (E14 to E487; green and black trace, respectively). Panels depict (A) atomic fluctuations for KaiC residues, (B) root mean square deviation (rmsd), (C) variations over the duration of the simulation in (D) the distance between N390 and A/V422 (Cα positions), (E) Zoomed atomic fluctuations for the KaiCII region entailing residues 380–490, and (F) atomic fluctuations for ATP molecules.
Inspection of the graphs depicting the three above parameters readily discloses that wt487 and mut487 proteins exhibit higher mobility than their wt497 and mut497 counterparts (Figure 8). Interestingly, in terms of a potential effect of the A422V mutation, the relative fluctuations between wt497 and wt487 hexamers on the one hand and mut497 and mut487 hexamers on the other are quite different. For example, the 422-loop in wt487 gains significantly in flexibility relative to wt497, whereas mut497 and mut487 display similar mobilities. Further along the chain, the region around the phosphorylation sites appears somewhat more mobile in the mut497 and mut487 proteins relative to both wt497 and wt487. On the other hand, around E444 (see also Figure 3), wt497 and wt487 exhibit the lowest and highest fluctuations, respectively, whereas mut497 and mut487 behave in a more similar fashion and the latter’s flexibility is reduced relative to wt487. Deviating relative fluctuations in these areas could account for altered trends in terms of the phosphorylation of S431 seen in the wt and A422V mutant KaiCs. Consistent with the higher average mobility of protein residues seen in the simulations of wt487 and mut487 KaiCs relative to the 497 constructs with the A-loop present, ATP molecules in wt487 and mut487 KaiC exhibit elevated mobility relative to wt497 and mut497 (Figure 8F). The calculated average fluctuations for 12 ATPs in the four simulated KaiC hexamers are: wt487 0.86 Å, mut487 0.84 Å, wt497 0.70 Å, and mut497 0.68 Å.
Perhaps more surprising than the above changes in mobility of residues in relative proximity of the A- and A/V-422 loops is the implication from the MD simulations that the removal of the A-loop appears to alter the dynamic behavior of the hexamer in regions that are relatively distant from residue 422. For example, residues of the KaiCI-CII linker region (245–260) in the wt487 hexamer show higher atomic fluctuations than in the mut487 hexamer (Figure 8A). It is tempting to speculate that these differences might affect the transfer of energy from ATP hydrolysis generated in CI to the CII ring, or serve the propagation of conformational changes in CII as a result of KaiA and/or KaiB binding there to CI. A further surprise revealed by the MD simulations concerns the S109 to G120 loop regions of subunits at the bottom of the CI ring that exhibit by far the highest mobility in the hexamer (excluding the N- and C-terminal residues; Figure 8A). The mobility of these loops appears to be also influenced somewhat by the presence or absence of the A-loops at the opposite end of the KaiC hexamer. To some extent, the higher mobility of residues in this region is related to their surface location. This is consistent with the observation in the crystal structure of KaiC that the loops formed by residues 110–120 display the highest temperature factors apart from the C-terminal tails (not shown). Overall, the MD simulations clearly reveal that the A422V mutation and unraveling of the A-loop affect the relative dynamics of KaiCII residues, whereby the latter event in general leads to a significant mobility increase.
Clearly, it would be desirable to conduct MD simulations for this system on a much longer timescale. However, there is a practical tradeoff between the size of the system we simulate, and the timescale to which we have access. In order to try to conserve resources, our first attempt comprised a much smaller system: only three CII subunits, which proved unstable in the simulation, as did the second attempt, using six CII subunits. Only the full hexameric system remained stable, and only with the use of a gentle seven-stage thermal equilibration protocol described in the methods. We have used the short-timescale data that we are able to collect, to generate mechanistic interpretations of the molecular system. Also, we have taken advantage of the six-fold symmetry of the system (which is not enforced during the simulation) to effectively explore conformational space in an ensemble that is six times greater than that of a single subunit. To some extent, this should mitigate the limited time duration of the simulation. At the current stage of our MD simulations (that have only been carried out for the hyper-phosphorylated form of KaiC), we cannot satisfactorily answer the question how phosphorylation (i.e. ST, SpT, pSpT, pST) affects KaiC dynamics? Clearly, to gain insight into this aspect, we would need to simulate all phosphorylation states and then conduct a detailed comparison between them.
DISCUSSION
KaiA stimulates KaiC phosphorylation and is thus crucial for initiating the rhythmic cycle between the KaiC hypo-, hyper-, and hypo-phosphorylated states with a period of 24 hours that lies at the heart of the cyanobacterial circadian clock. We know that the KaiA dimer binds to KaiC C-terminal peptide(s)14,15 and that the conformation of the A-loop region near the C-terminal end of KaiC is altered upon KaiA binding (Figure 2). The observation that a KaiC deletion mutant with the A-loop sequence and the attached C-terminal residues missing (Δ487-KaiC) is constitutively hyper-phosphorylated without KaiA demonstrates the important role played by the A-loop vis-a-vis KaiC phosphorylation.45 However, how the A-loop fold observed in the KaiC crystal structure13 and its unraveling by KaiA suppresses and activates, respectively, the KaiCII kinase, has remained an enigma. Our concept of the mechanism of KaiA-stimulated KaiC phosphorylation assigns a central role to a loop with residue A422 at its apex (termed 422-loop here) in the activation of KaiC kinase activity at subunit interfaces and phosphorylation of T432 and S431 as a result of KaiA contacting a KaiC C-terminal tail. The A422-loop has so far not been considered in the context of the activation of the KaiC kinase. We provide evidence here based on analyses of the 3D-structure, phosphorylation state and dynamics of the A422V KaiC mutant that together with previous data regarding its phenotype46 strongly implicate the 422-loop in the activation of KaiC phosphorylation by KaiA.
Relaying KaiA Binding to the KaiCII Kinase Active Sites
Because the KaiC phosphorylation/dephosphorylation cycle is so critical for proper functioning of the S. elongatus clock, the significant suppression of phosphorylation displayed by the conservative A422V mutation, in addition to its disruption of clock hallmarks such as phase shifting and rhythm of protein abundance, are particularly striking. The proximity of A422 and the T432 and S431 phosphorylation sites is certainly noteworthy (Figures 2B, 3, 5), but the fact that the A-loop and the 422-loop are directly associated in the structure of the KaiCII half is particularly interesting in regard to the question how A-loop unfolding is relayed to the kinase active site. Detailed examination of structural elements in the vicinity of ATP and the two phosphorylation sites reveals a network of interactions that ties together, either directly or indirectly, A-loops, 422-loops (that continue to S431 and T432), β9 strands, P-loops and therefore ATP, from individual subunits (Figure 3). A-loops from adjacent subunits are tethered together by hydrogen bonds between T495 and E487 that are severed upon KaiA binding. A-loops also engage in interactions with A422-loops from their own (G492…M420) and an adjacent subunit (i.e. I490…M420) and with β9 strands from the same subunit (S493…E444). From this branch point (A422-loop ← A-loop → β9), absence or presence of the A-loop is then relayed both to the phosphorylation region (T432, S431 and T426), via differential flexibility of the A422-loop, and to the P-loop and therefore ATP, via the β9 strand (i.e. Y442…E292, L440…L297, and I438…L297; Figure 3). Thus, KaiA binding to KaiC C-terminal peptide does not just unravel the A-loop but removes a host of constraints that link the above structural elements in individual subunits and across the entire KaiCII hexamer, ultimately resulting in closer proximity between T432 and S431 hydroxyl groups and the γ-phosphate of ATP and phosphoryl transfer. It is clear that the order of phosphorylation in KaiCII, T432 first and S431 second,36,37 is kinetically controlled and related to the tighter spacing between T432 and γP relative to S431 throughout the KaiC structure.13,34,38
Not surprisingly, mutations that alter the interactions among substructures of this network lead to severely distorted periods (e.g. G421R and Y442H) of the clock or loss of rhythmicity (e.g. T495A, E487A and T426A). Both the T495A3 and E487A45 KaiC mutations that disrupt hydrogen bond formation between neighboring A-loops show arhythmic behavior and E487A was also found to be constitutively hyper-phosphorylated. We have described the consequences of the A422V mutation46 for clock function and how it relates to KaiA-stimulated KaiC phosphorylation in detail in the present contribution. In addition, the G421R mutation that can be expected to alter the A-loop/A422-loop interface exhibits a clock period of 44 h.3 Closer to the phosphorylation sites, the T426A mutant has long been known to be arhythmic.34,38,39 The side chain of T426 engages in a direct interaction with the phosphate on S43113,34 and since we envision an important role of the short peptide stretch between A422 and S431/T432 in transmitting the increased flexibility of the A422-loop as a result of the removal of the A-loop by KaiA to the phosphorylation sites, it is not surprising that T426 mutations would be detrimental to clock function. In the context of changed network interactions as a result of KaiA binding to the A-loop region that affect β9, P-loop and ultimately ATP phosphates, it is noteworthy that the Y442H mutation lengthens the clock period to 60 h.3 Overall, the available point mutation data are consistent with a key role of these structural elements in the regulation of phosphorylation.
Concerted Allostery of KaiA Stimulation
“Pulling” by KaiA on the C-terminal tail and the attached A-loop from a single KaiC subunit will affect the subunits to its left and right as well. This is because of the aforementioned network of interactions that links adjacent subunits at the level of the A- and 422-loops. From the first affected subunit, there will be a spiraling cascade of phosphorylations around the KaiCII hexamer on T432 residues which progress to spiraling phosphorylations of S431 residues. This network of intramolecular interactions results in a “domino effect” of phosphorylations jumping from one subunit to the next. Therefore it seems unnecessary to invoke a model with a KaiA dimer migrating from one KaiC subunit to the next58 or two or more KaiA dimers simultaneously binding to multiple KaiC tails. In fact, it was found early on in the study of the cyanobacterial clock that a single KaiA dimer is sufficient to upregulate phosphorylation of a KaiC hexamer to the maximum level.59 Consequently, an allosteric mechanism of KaiA-stimulated KaiC auto-phosphorylation is more appealing.60 Concerted allosteric control is not just in line with the hexameric nature of KaiC and the intriguing network of interactions among subunits in the region of KaiA-KaiC interactions (Figure 3) and elsewhere in the KaiC hexamer, but is also consistent with the relative concentrations/stoichiometries of Kai proteins in the cell33 and in the in vitro reaction.4 Thus, the cellular concentrations of the KaiB and KaiC proteins are estimated at 10–30 μM and the concentration of KaiA is estimated at 0.5–1 μM. Further support for the concerted allosteric model comes from the KaiA-KaiC binding mode. Although Figure 2D depicts only a single KaiC peptide chain bound between the C-terminal domains from a KaiA dimer, each KaiA dimer can in principle bind two KaiC peptides.14 The three-dimensional model of the KaiA-KaiC complex based on electron microscopy15,20 is not of sufficient resolution to determine whether KaiA is tethered to two KaiC C-terminal peptides in all cases. However, the illustrations in Figure 3 make it clear that unfolding by KaiA of A-loops from two KaiC subunits that are not direct neighbors in the hexamer will loosen the association among all six subunits.
The mechanism proposed here in regard to the stimulation of KaiC phosphorylation by KaiA is compatible with the ratcheting concept for KaiC phosphorylation,17 i.e. why time in the in vitro oscillator is unidirectional.8 This is because the stimulation mechanism does not affect the order of phosphorylation that is kinetically controlled and the fact that phosphorylation of T432 results in an increased number of interactions between residues across the subunit interface, thus promoting the next step, phosphorylation of S431.
Structural Basis of the A422V Phenotype
Recent research has provided crucial insights into two sources of metabolic input to the circadian oscillator that allow cyanobacteria to sense the daily light/dark cycle. Thus, KaiC phosphorylation is directly affected by the cellular ATP/ADP ratio which itself depends on photophosphorylation.61 In fact, changes in the ATP/ADP ratio were also demonstrated to reset the phase of KaiC phosphorylation in the in vitro oscillation reaction constituted by KaiA, KaiB and KaiC in the presence of ATP. A second metabolic signal that influences the phase of the KaiABC PTO is related to the oxidation state of the plastoquinone (PQ) pool.62 Oxidized PQ causes KaiA aggregation62 and interferes with KaiA binding to KaiC C-terminal peptide,63 thus limiting stimulation of KaiC phosphorylation by KaiA. These two cues, oxidized PQ and ATP/ADP ratio, appear to act in tandem to signal the onset of darkness (when the PQ pool becomes oxidized rapidly) and to measure the duration of darkness (triggering a slow decline on the ATP/ADP ratio that induces a phase shift), respectively.64
Both metabolic input signals serving entrainment of the cyanobacterial oscillator therefore affect KaiC phosphorylation. This suggests that the inability of the KaiC A422V mutant to shift phase could be directly related to its distorted phosphorylation [i.e. the low amplitude of the phosphorylation cycle46 and the inability to phosphorylate S431 (this paper; Figure 4A)]. We have shown here that the A422V mutant’s defective phosphorylation may be the result of an altered mobility of its 422-loops and adjacent regions relative to wt KaiC, brought on by KaiA binding and concomitant unraveling of A-loops. Relaying by the 422-loop the removal of restraints exerted by A-loops to the phosphorylation sites and P-loops/ATP involves numerous hydrophobic contacts (A-loop ··· 422-loop, β9 ··· P-loop, etc.; Figure 3). Unlike other non-covalent interactions, such as hydrogen bonds or van der Waals forces, hydrophobic interactions become weaker as the temperature is lowerered.65 That hydrophobics play a crucial role in the transmission of KaiA binding to the KaiC kinase active site may be supported by the observation that KaiC becomes hyper-phosphorylated at low temperature (4°C) relative to ambient conditions.42 It is also possible that the A422V mutation directly affects KaiA-KaiC binding and the ability of KaiA to pull on the KaiC tail and unravel the A-loop.
In summary, an apparently minor change in residue from alanine to valine at position 422 in the cyanobacterial master clock protein KaiC gives rise to drastic changes at the functional level that include an inability to reset the phase and to distorted phosphorylation. The present analysis of the structure, phosphorylation pattern, stability and dynamics of the KaiC A422V mutant provides a much-improved understanding of the mechanism of KaiA-stimulated KaiC phosphorylation.
Acknowledgments
Funding Sources
We are grateful to the National Institutes of Health for supporting clock research in our labs (R01 GM073845 to ME and R01 GM067152 to CHJ). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
We would like to thank Dr. Spencer Anderson, LS-CAT, Advanced Photon Source, Argonne National Laboratory (Argonne, IL) for help with X-ray data collection and Prof. Terry Lybrand, Vanderbilt University, for helpful discussions.
References
- 1.Johnson CH. Precise circadian clocks in prokaryotic cyanobacteria. Curr Issues Molec Biol. 2004;6:103–110. [PubMed] [Google Scholar]
- 2.Ditty JL, Mackey SR, Johnson CH, editors. Bacterial Circadian Programs. Springer Publishers Inc; Heidelberg, Germany: 2009. [Google Scholar]
- 3.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 5.Tomita J, Nakajima M, Kondo T, Iwasaki H. Circadian rhythm of KaiC phosphorylation without transcription-translation feedback. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 6.Qin X, Byrne M, Yu X, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami R, Mutoh R, Iwase R, Furukawa Y, Imada K, Onai K, Morishita M, Yasui S, Ishii K, Valencia Swain JO, Uzumaki T, Namba K, Ishiura M. The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J Biol Chem. 2012;287:29506–29015. doi: 10.1074/jbc.M112.349092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki H, Taniguchi Y, Kondo T, Ishiura M. Physical interactions among circadian clock proteins, KaiA, KaiB and KaiC, in cyanobacteria. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taniguchi Y, Yamaguchi A, Hijikata A, Iwasaki H, Kamagata K, Ishiura M, Go M, Kondo T. Two KaiA-binding domains of cyanobacterial circadian clock protein KaiC. FEBS Lett. 2001;496:86–90. doi: 10.1016/s0014-5793(01)02408-5. [DOI] [PubMed] [Google Scholar]
- 12.Kageyama H, Kondo T, Iwasaki H. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J Biol Chem. 2003;278:2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- 13.Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell. 2004;15:375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA. 2004;101:10925–10930. doi: 10.1073/pnas.0403037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M. Analysis of KaiA-KaiC protein interactions in the cyanobacterial circadian clock using hybrid structural methods. EMBO J. 2006;25:2017–2038. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori T, Williams DR, Byrn M, Qin X, Egli M, Mchaourab H, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson CH, Egli M, Stewart PL. Structural insights into a circadian oscillator. Science. 2008;322:697–701. doi: 10.1126/science.1150451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pattanayek R, Williams DR, Pattanayek S, Mori T, Johnson CH, Stewart PL, Egli M. Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J. 2008;27:1767–1778. doi: 10.1038/emboj.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akiyama S, Nohara A, Ito K, Maéda Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell. 2008;29:703–716. doi: 10.1016/j.molcel.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Egli M, Stewart PL. Structural aspects of the cyanobacterial KaiABC circadian clock. In: Ditty JL, Mackey SR, Johnson CH, editors. Bacterial Circadian Programs. Springer Publishers Inc; Heidelberg, Germany: 2009. pp. 121–140. [Google Scholar]
- 21.Pattanayek R, Williams DR, Rossi G, Weigand S, Mori T, Johnson CH, Stewart PL, Egli M. Combined SAXS/EM based models of the S. elongatus posttranslational oscillator and its interactions with the output His-kinase SasA. PLoS ONE. 2011;6:e23697. doi: 10.1371/journal.pone.0023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA. 2002;99:17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 24.Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. The ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells. 2008;13:387–395. doi: 10.1111/j.1365-2443.2008.01174.x. [DOI] [PubMed] [Google Scholar]
- 26.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egli M, Mori T, Pattanayek R, Xu Y, Qin X, Johnson CH. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51:1547–1558. doi: 10.1021/bi201525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287:18030–18035. doi: 10.1074/jbc.M112.350660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB, and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:1–8. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA. 2004;101:13933–13938. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA. 2004;101:13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattanayek R, Mori T, Xu Y, Pattanayek S, Johnson CH, Egli M. Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS ONE. 2009;4:e7529. doi: 10.1371/journal.pone.0007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Mori T, Qin X, Yan H, Egli M, Johnson CH. Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLoS ONE. 2009;4:e7509. doi: 10.1371/journal.pone.0007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang YG, Tseng R, Kuo NW, LiWang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci USA. 2012;109:16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murayama Y, Mukaiyama A, Imai K, Onoue Y, Tsunoda A, Nohara A, Ishida T, Maéda Y, Terauchi K, Kondo T, Akiyama S. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 2011;30:68–78. doi: 10.1038/emboj.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin X, Byrne M, Mori T, Zou P, Williams DR, Mchaourab H, Johnson CH. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA. 2010;107:14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brettschneider C, Rose RJ, Hertel S, Axmann IM, Heck AJR, Kollmann M. A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Molec Sys Biol. 2010;6:389. doi: 10.1038/msb.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markson JS, O’Shea EK. The molecular clockwork of a protein-based circadian oscillator. FEBS Lett. 2009;583:3938–3947. doi: 10.1016/j.febslet.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YI, Dong G, Carruthers C, Golden SS, LiWang A. A day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105:12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacteiral circadian clock. J Bacteriol. 2005;187:2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papworth C, Bauer JC, Braman J, Wright DA. Site-directed mutagenesis using double-stranded plasmid DNA templates. Strategies. 1996;9:3–4. doi: 10.1385/0-89603-332-5:31. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossvari I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 53.Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 54.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 55.Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1 Generalized Born. J Chem Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryckaert JP, Ciccotti G, Berendsen HJC, Hirasawa K. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 57.Miyamoto S, Kollman PA. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 58.Egli M, Pattanayek R, Pattanayek S. Protein-protein interactions in the cyanobacterial KaiABC circadian clock. In: Boeyens JCA, Ogilvie JF, editors. Models, Mysteries and Magic of Molecules. Springer Publishers; Dordrecht, The Netherlands: 2007. pp. 283–299. [Google Scholar]
- 59.Hayashi F, Ito H, Fujita M, Iwase R, Uzumaki T, Ishiura M. Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem Biophys Res Comm. 2004;316:195–202. doi: 10.1016/j.bbrc.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 60.van Zon JS, Lubensky DK, Altena PRH, ten Wolde PR. An allosteric model of circadian KaiC phosphorylation. Proc Natl Acad Sci USA. 2007;104:7420–7425. doi: 10.1073/pnas.0608665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood TL, Bridwell-Rabb J, Kim YI, Gao T, Chang YG, LiWang A, Barondeau DP, Golden SS. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Si USA. 2010;107:5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pattanayek R, Sidiqi SK, Egli M. Crystal structure of the redox-active cofactor DBMIB bound to circadian clock protein KaiA and structural basis for DBMIB’s ability to prevent stimulation of KaiC phosphorylation by KaiA. Biochemistry. 2012;51:8050–8052. doi: 10.1021/bi301222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim YI, Vinyard DJ, Ananyev G, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial oscillator. Proc Natl Acad Sci USA. 2012;109:17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hochachka PW, Somero GN. Biochemical adaptation: mechanism and process in physiological evolution. Oxford University Press; Oxford, UK: 1973. [Google Scholar]