Abstract
Addition of poly(A) to the 3′ ends of cleaved pre-mRNA is essential for mRNA maturation and is catalyzed by Pap1 in yeast. We have previously shown that a non-viable Pap1 mutant lacking the first 18 amino acids is fully active for polyadenylation of oligoA, but defective for pre-mRNA polyadenylation, suggesting that interactions at the N-terminus are important for enzyme function in the processing complex. We have now identified proteins that interact specifically with this region. Cft1 and Pta1 are subunits of the cleavage/polyadenylation factor, in which Pap1 resides, and Nab6 and Sub1 are nucleic-acid binding proteins with known links to 3′ end processing. Our results suggest a novel mechanism for controlling Pap1 activity, and possible models invoking these newly-discovered interactions are discussed.
Keywords: Poly(A) polymerase, Polyadenylation
1. Introduction
Polyadenylation of eukaryotic mRNA involves cleavage of precursor and addition of adenosines onto the new 3′ end, and is catalyzed by a conserved complex of over 20 proteins [1]. Acquisition of the poly(A) tail promotes release from the site of transcription and efficient mRNA export, and the length affects turnover rate and translation [2,3].
The poly(A) polymerase (PAP) that adds the tail is tightly controlled during a cycle of 3′ end processing so that initiation of poly(A) synthesis occurs promptly after cleavage yet terminates once the tail is sufficiently long. Because of this central role, PAPs have become targets for regulatory proteins and post-translational modifications such as acetylation, phosphorylation and sumoylation [1,4] that potentially alter the network of interactions made by PAP within the processing complex. Mammalian PAP interacts with the CPSF-160 and hFip1 subunits, which direct the enzyme to the correct RNA substrates by interacting with signal sequences that define the poly(A) site [5,6]. Several proteins that are not part of the basic processing complex, such U1A, U1-70K and hnRNP H, can regulate polyadenylation by direct interaction with PAP [7–9].
Less is known about contacts made with yeast Pap1. Fip1 acts as a flexible tether to incorporate the enzyme into the cleavage/polyadenylation factor (CPF) [10–12] through a Fip1 binding site in the Pap1 C-terminal domain [13]. Deletion of the first 18 amino acids of Pap1 is lethal in yeast and causes defective polyadenylation of pre-mRNA when assayed using purified factors [14]. Surprisingly, this mutation had no effect on the catalytic activity of the enzyme when it is separated from CPF [11,14]. The truncated Pap1 was also normal for interaction with Fip1. The same region of vertebrate enzymes has 56% similarity to this yeast sequence, but has not been correlated with a particular function. These findings suggest that contacts between the N-terminus of Pap1 and unknown subunits of the mRNA 3′ end processing complex are critical for its essential function in mRNA maturation. In this study, we report the identification of proteins that interact with this part of Pap1 and possibly modulate its activity.
2. Materials and methods
2.1. Yeast strains
The yeast strains used in this study were W303 (MATa leu2–3, 112 trp1–1 can1–100 ura3-1 ade2-1 his3–11, 15) and PJ69-4A (MAT a leu2–3, 112 ura3–52 trp1–901 his3–200 gal4Δ gal80Δ GAL-ADE2 lys2::GAL1-HIS3 met2::GAL7-LacZ). KANMX or HIS3 replacements of the SUB1 and NAB6 genes were made in W303.
2.2. Two-hybrid analysis
Pap1-GBD and ΔN-Pap1-GBD [15] and gal4 activation domain (GAD)-Pta1 constructs [16] were described previously. Two-hybrid analysis was performed by transforming plasmid pairs into the PJ69-4A strain [15,16]. Transformants were selected on medium lacking leucine and tryptophan to ensure that both the GAD and GBD plasmids were present. Protein–protein interactions were scored by the ability of cells to grow in the absence of histidine.
2.3. Peptide columns
A Pap1 peptide, MSSQKVFGITGPVSTVGA, or an Npl3 peptide, RGGYDSPRGGY, was coupled to column matrix (Pierce UltraLink EDC/DADPA Immobilization Kit), incubated for 2 h at 25 °C with nuclear extract prepared as described [17] except using IPP-150 buffer (10 mM Tris, pH 7.9, 150 mM NaCl, 1 mM MgOAc, 2 mM CaCl2, 0.1% NP-40, 1 mM DTT and the protease inhibitors PMSF, leupeptin, antipain and pepstatin-A), washed using 12 column volumes of IPP-150, and eluted using 100 mM glycine, pH 3. Eluted proteins were analyzed by mass spectroscopy through the Tufts Core Facility.
2.4. In vitro protein–protein interaction assays
For protein expression in Escherichia coli Rosetta (DE3), the Pap1-GST, ΔN-Pap1-GST, and Nab6-V5-His6 plasmids were made by modifying pJPAP1 [14]. Other expression plasmids were described previously: Pap1-His6 and ΔN-Pap1-His6 [14], Fip1(1–206) [11], Sub1-V5-His6 [18], Pta1 and Pta1 truncations [16], and Cft1-GST [19]. Expression and purification was performed as described [14]. GST pull-downs followed a modification of a previous protocol [16], using 50 µl glutathione-Sepharose beads in 200 µl of IP-150 for 2 h at 4 °C with gentle shaking, followed by four washes with IP-150, and elution with IP-150 containing 50 mM glutathione at 4 °C for 1 h with gentle shaking. Protein–protein interactions were analyzed by resolving proteins on an SDS–10% polyacrylamide gel, followed by Western blotting with the following antibodies: monoclonal Pap1 antibody at 1:100 dilution, V5 antibody (Invitrogen) at 1:5000 dilution, and polyclonal Fip1 antibody at 1:7500 dilution.
2.5. In vitro 3′ end processing
Yeast extract was prepared and used for processing assays as described [11,16], except that the final dialysis was twice against 2 l of buffer D for 2 h, and then overnight.
3. Results
3.1. The Pap1 N-terminus interacts with the first 300 amino acids of Pta1 and with Cft1
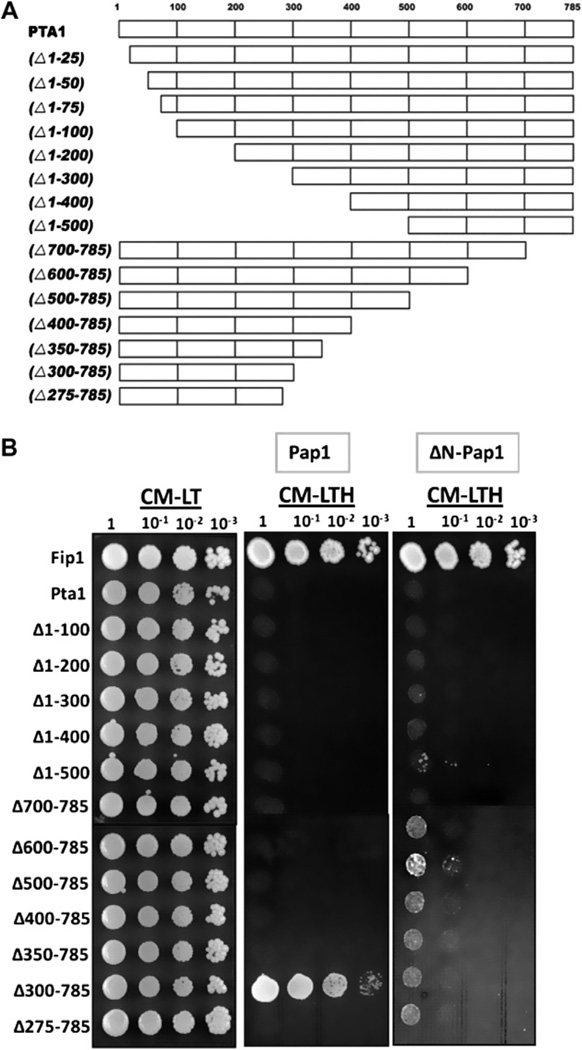
Pta1 has been shown to interact with in vitro translated Pap1 [16]. We used two-hybrid analysis to confirm this interaction in vivo and identify interaction domains by pairing full-length and truncations of Pta1 (Fig. 1A) with full-length Pap1 or Pap1 lacking the N-terminus (ΔN-Pap1). The Pta1 constructs have been previously shown to express protein and interact with other proteins [16]. Consistent with the known C-terminal Fip1 binding site, both Pap1 and ΔN-Pap1 interact with Fip1, but no interaction was seen between Pap1 and full-length Pta1 (Fig. 1B). Analysis using Pta1 truncations revealed an interaction between Pta1 (Δ300–785) and full-length Pap1 that was lost with ΔN-Pap1 or when another 25 amino acids of Pta1 were removed (Δ275–785). The lack of interaction between full-length Pta1 and Pap1 may be due to steric constraints that prevent a two-hybrid signal when both proteins are part of CPF.
Fig. 1.
Two-hybrid analysis of interactions between Pta1 and Pap1. (A) Full-length Pta1 and Pta1 truncations used for two-hybrid analysis [16]. (B) The first 300 amino acids of Pta1 interact with the Pap1 N-terminus. Pta1 truncations were fused to GAD, while Pap1 and ΔNPap1 were fused to GBD. The GAD-Fip1 interaction confirms that ΔNPap1 is expressed. Tenfold serial dilutions of liquid cultures were spotted onto medium lacking leucine and tryptophan (CM-LT) to select for the two-hybrid plasmids. Activation of HIS3 expression (indicative of protein interaction) was tested by spotting the same strains onto medium lacking leucine, tryptophan and histidine (CM-LTH), followed by incubation for 2–3 days at 30 °C.
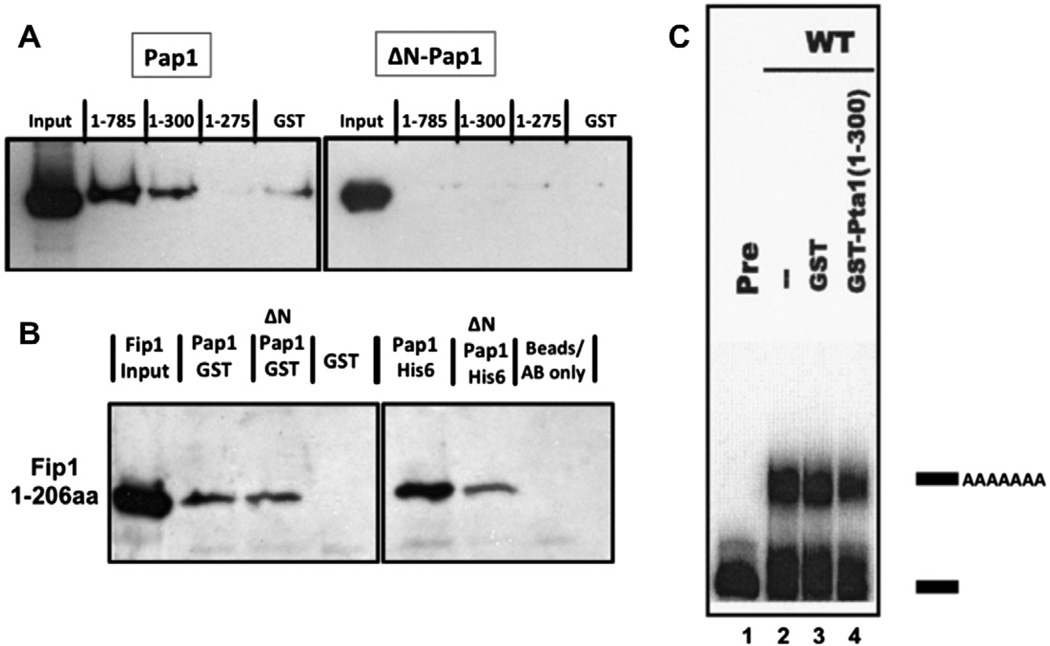
We also assessed physical contact using recombinant GST-Pta1 and His6-tagged Pap1 or ΔN-Pap1. Pap1 interacted with full-length Pta1 and the 1–300 amino acid fragment of Pta1, but not with the 1–275 fragment of Pta1 (Fig. 2A). ΔN-Pap1 did not interact with any forms of Pta1, but bound to Fip1 (Fig. 2B), indicating that unfolding of the protein does not explain a lack of interaction. These findings extend the two-hybrid results by showing a direct interaction between Pap1 and Pta1 mediated by amino acids 1–300 of Pta1 and the N-terminus of Pap1.
Fig. 2.
Pta1 interacts directly with the Pap1 N-terminus. (A) Western blot showing the amount of Pap1-His6 or ΔN-Pap1-His6 pulled down with full-length GST-Pta1 or the indicated Pta1 fragments. (B) Western blot showing that full-length Pap1 and ΔNPap1 interact with amino acids 1–206 of Fip1 [11]. Left panel: Fip1 pull-downs with the Pap1-GST, ΔNPap1-GST, or GST proteins used in Figs. 2A and 4. Right panel: co-immunoprecipitations of Fip1 using Pap1-His6 or ΔNPap1-His6 immobilized on protein A beads by Pap1 antibody, or using beads and antibody without Pap1. (C) Addition of the 1–300 amino-acid fragment of Pta1 inhibits poly(A) addition performed with yeast extract and radioactive GAL7 RNA substrate ending at the poly(A) site. Lane 1, unreacted precursor (Pre); lane 2, extract alone; lane 3, extract plus GST; lane 4, extract plus GST-Pta1 (1–300). Positions of precursor and polyadenylated RNAs are indicated on the right.
To study the function of the Pta1/Pap1 interaction in 3′ end processing, the 1–300 Pta1 fragment was added to a poly(A) addition assay using yeast extract and radiolabeled GAL7 RNA substrate ending at the poly(A) site. Extract supplemented with the Pta1 fragment showed reduced accumulation of polyadenylated product compared to extract receiving an equal amount of GST (Fig. 2C), suggesting a role for this Pta1 domain in regulating Pap1 activity.
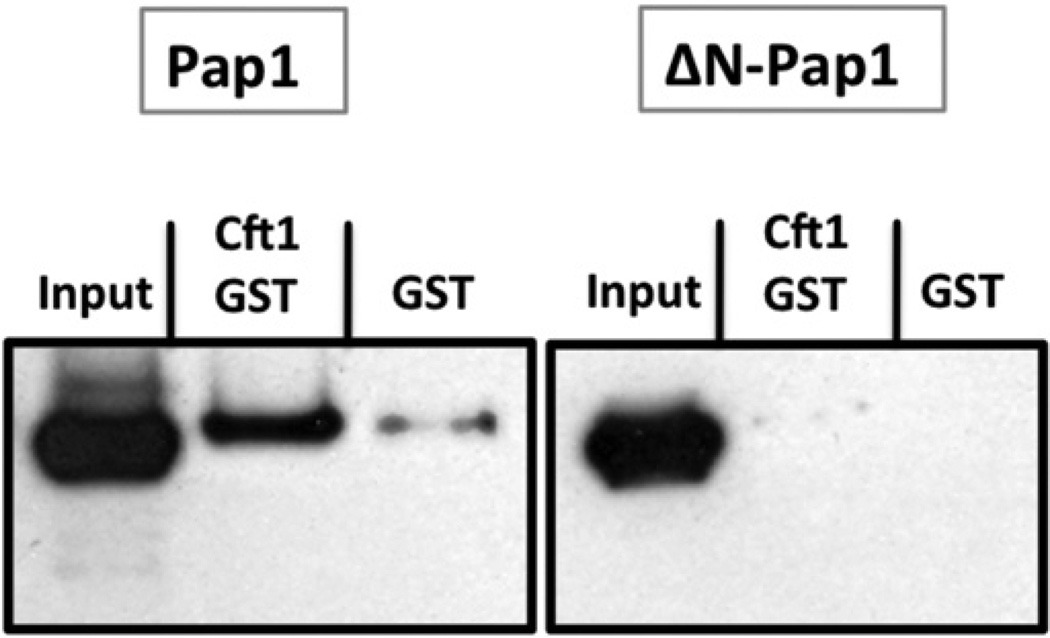
To ask if the interaction of CPSF160, the mammalian Cft1 homologue, with poly(A) polymerase [6] is conserved in yeast, pull-downs were performed using GST-Cft1 and His6-tagged Pap1 or ΔN-Pap1. Cft1 bound full-length Pap1, but not Pap1 lacking the N-terminus (Fig. 3). Thus, like its mammalian homologue, Cft1 interacts with Pap1, but in yeast, this interaction is localized to the Pap1 N-terminus.
Fig. 3.
Cft1 interacts directly with the Pap1 N-terminus. Pull-downs showing interaction between GST-Cft1 and Pap1-His6 but not with GST-Cft1 and ΔNPap1-His6. Pap1 was detected using Pap1 antibody. Pap1-His6 and DNPap1-His6 both interact with Fip1 (Fig. 2B).
3.2. Sub1 and Nab6 interact with the Pap1 N-terminus
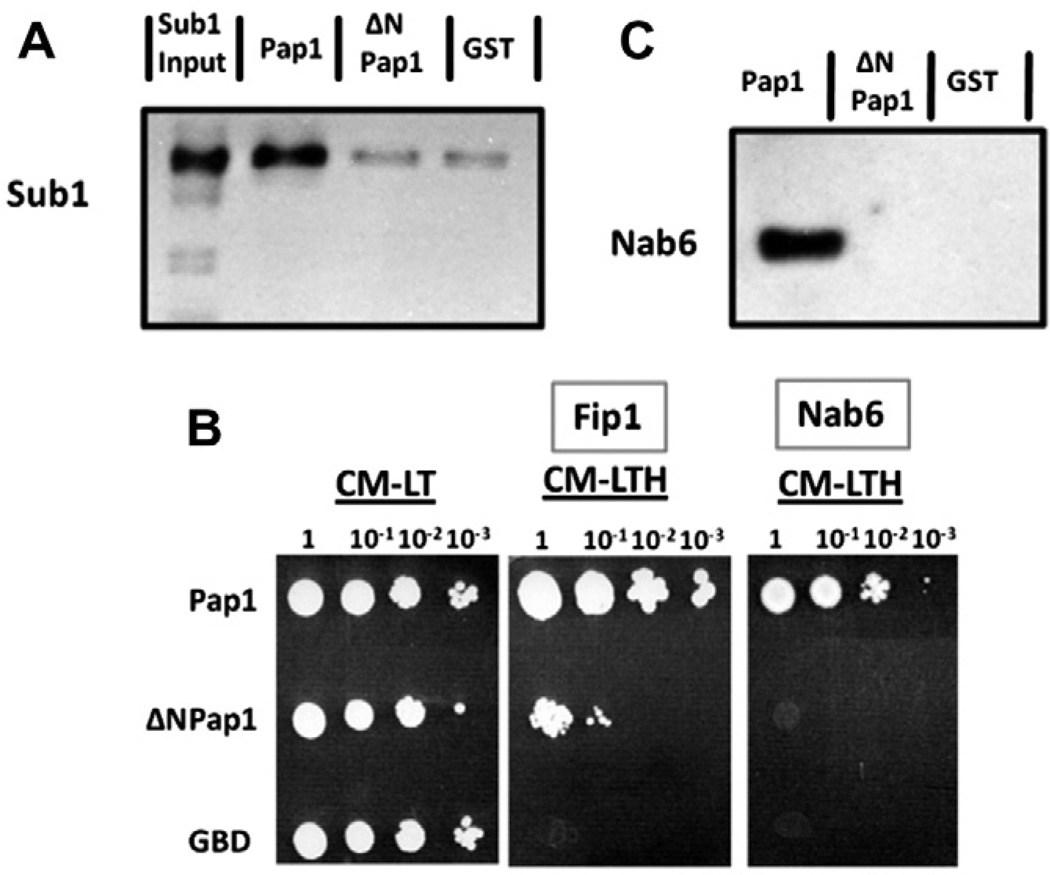
To find other proteins that interact with the Pap1 N-terminus, a column coupled to a peptide comprising the first 18 amino acids of Pap1 was used to pull down proteins from yeast nuclear extract. Sub1, a protein implicated in both transcription and 3′ end processing [20], and references therein), was one of the proteins identified from mass spectroscopy analysis of bound proteins. Sub1 was not found on a control column using a peptide from a different yeast protein, Npl3. Pull-downs showed that Sub1 interacts directly with full-length Pap1-GST, and this interaction is lost in the absence of the Pap1 N-terminus (Fig. 4A).
Fig. 4.
Sub1 and Nab6 interact with the Pap1 N-terminus. (A) Western blot of pull-downs using Sub1-V5-His6 and Pap1-GST or ΔN-Pap1-GST, and detecting Sub1 with V5 antibody. (B) Two-hybrid analysis between the indicated proteins and a C-terminal Nab6 fragment. (C) Western blot of pull-downs using Nab6-V5-His6 (the last 210 amino acids) and Pap1-GST or ΔN-Pap1-GST and detecting Nab6 with V5 antibody.
We extensively screened a yeast two-hybrid library with Pap1 as bait and only identified two proteins, the known Pap1-interactor Fip1, and a fragment containing the last 210 amino acids of Nab6, a protein that crosslinks to nuclear poly(A)+ mRNA [21]. The Nab6 interaction was lost in the absence of the Pap1 N-terminus (Fig. 4B). Pull-downs using Pap1-GST and the C-terminal region of Nab6 confirmed that the interaction is direct and requires the Pap1 N-terminus (Fig. 4C).
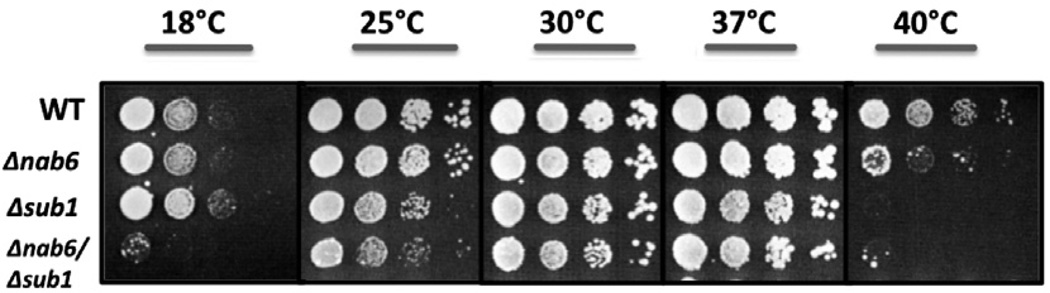
To further probe the role of Nab6 and Sub1, strains containing single and double deletions of these two genes were analyzed for effects on growth. At 40 °C, the Δnab6 strain is thermosensitive and the Δsub1 strain is not viable (Fig. 5A). Deletion of both genes caused cold-sensitivity, suggesting that Nab6 and Sub1 play a redundant role in yeast survival at 18 °C. Polyadenylation assays using extracts from the wild-type and deletion strains showed no changes in the processing efficiency or the length of poly(A) tails (data not shown), indicating that Nab6 and Sub1 do not have strong activating or inhibitory effects on poly(A) addition under optimal growth conditions.
Fig. 5.
Effects of deleting the NAB6 and SUB1 genes on cell growth. Serial dilutions of cultures of wild-type (WT) cells or strains with the indicated deletions were spotted on YPD medium and incubated at different temperatures for up to 120 h.
4. Discussion
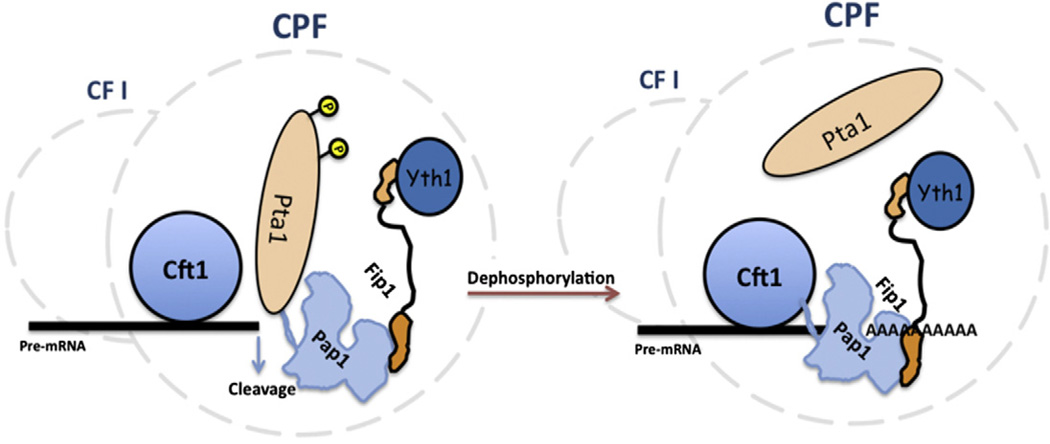
We have discovered novel interactions with a region at the N-terminus of the yeast poly(A) polymerase that is critical for this enzyme’s function in mRNA 3′ end maturation. Two of these contacts are made through Pta1 and Cft1, essential proteins that are conserved in the processing machinery of other eukaryotes as symplekin and CPSF-160, respectively. As part of CPF, Pta1 and Cft1 are needed for pre-mRNA cleavage and poly(A) addition, and most likely function as scaffolds to organize the processing complex and appropriately position the nuclease and poly(A) polymerase to act on the RNA [1,16,19]. Based on published work and on the new findings presented here, we propose a model in which Pta1 and Cft1 exert opposing regulatory effects on Pap1 while it is tethered to the processing complex through Fip1 interacting at its C-terminus (Fig. 6). As might be expected for regulatory functions, these interactions are normally not sufficient to hold Pap1 in the CPF complex, as disruption of the Fip1/Yth1 interaction results in loss of Pap1 from CPF [12].
Fig. 6.
Model of Pap1 regulation through interactions at the Pap1 N-terminus. Only the Cft1, Fip1, Pap1, Pta1, and Yth1 subunits of CPF are depicted to emphasize how interactions with Pap1 might shift during a cycle of processing. We propose that phosphorylation of Pta1 before and during the cleavage step strengthens its interaction with Pap1 and inhibits poly(A) addition. Dephosphorylation following cleavage relieves the inhibition of Pap1, allowing Pap1 to bind Cft1, which helps position Pap1 at the new 3′ end to initiate poly(A) addition. Fip1, acting as a flexible tether, facilitates these transitions [10]. Rephosphorylation of Pta1 may help terminate poly(A) addition. Dashed lines indicate the entire CPF and CF I complexes.
We show that addition of a fragment containing the essential 1–300 amino acids of Pta1 suppresses polyadenylation in wild type extract. This effect could be due to the Pta1 fragment out-competing a positive interaction provided by full-length Pta1. However, our earlier work [22] showed that phosphorylation of Pta1 inhibited poly(A) addition. Furthermore, phosphorylation of CPF releases Fip1 without affecting Pap1 association [22], implying the formation of stable interactions with other subunits that hold Pap1 in the complex. We propose that Pta1 phosphorylation strengthens its interaction with Pap1 and inhibits the enzyme.
Because deletion of the Pap1 N-terminus prevents specific poly(A) addition and is lethal in yeast cells [14], interactions with this part of Pap1 may also stimulate polyadenylation. Contact with Cft1 could provide this activity. However, an inhibitory role for Cft1 cannot be ruled out, as the mammalian homologue, CPSF-160, also interacts with mammalian poly(A) polymerase (PAP), and inhibits both specific and non-specific polyadenylation [6]. This interaction may not be comparable to the one observed in yeast, as it involves the C-terminal domain of PAP [5,6] and not its N-terminus, as is the case with Pap1 and Cft1.
We also report an interaction between the Pap1 N-terminus and Sub1 and Nab6. Sub1 is a transcriptional co-activator and modulates 3′ end processing and termination through its interaction with Rna15, a subunit of the CF I cleavage/polyadenylation factor [20], and references therein). Sub1 also interacts with Pta1, and its over-expression suppresses the processing defects of a pta1 mutant [18].
Nab6 is a poly(A)+ RNA-binding protein that preferentially associates with mRNAs encoding cell wall proteins [23]. Its over-expression suppresses growth defects caused by loss of Rrp6 [21], an exonuclease involved in the degradation of improperly processed RNAs, non-coding RNAs, and byproducts of RNA processing in the nucleus [24]. Deletion of RRP6 also relieves the inhibition of polyadenylation caused by defects in mRNP assembly [25], promotes the polyadenylation of GAL1 RNA [26], and causes longer poly(A) tails in vivo and in vitro [27]. Over-expression of Nab6 in the rrp6Δ strain might suppress the growth defect by inhibiting Pap1 activity, thereby promoting normal poly(A) tail synthesis. A role for Nab6 in down-regulating polyadenylation is further supported by the observation that NAB6 deletion partially rescues growth defects due to mutations in CF I subunits [21].
We show that deletions of NAB6 and SUB1 causes growth defects at extreme temperatures, suggesting a role for these proteins in Pap1 regulation in suboptimal growth conditions. Involvement of cleavage/polyadenylation factors in coping with stress is not unprecedented; the orthologue of the CPF subunit Yth1 plays a role in the plant response to oxidative stress [28]. A redundant role for Nab6 and Sub1 in controlling poly(A) activity under certain conditions is consistent with the observation that both proteins interact genetically with components of the osmotic stress response pathway [29,30].
In summary, we have discovered four proteins that interact with the Pap1 N-terminus. Two belong to the cleavage/polyadenylation complex, and these interactions probably explain why the Pap1 N-terminus is dispensable for catalytic activity but essential for the enzyme to function as part of the processing complex. The most likely role of these contacts would be to ensure proper timing of polyadenylation during a cycle of mRNA 3′ end processing. We have also identified two proteins whose interactions with the Pap1 N-terminus may modulate polyadenylation during unfavorable growth conditions. These findings expand the ways in which the cell can control the quality and functionality of the mRNA that it produces. Given the conserved nature of the Pap1 N-terminus, it will be interesting in future work to see if the mammalian enzyme is regulated by similar interactions.
Structured summary of protein interactions:
PAP1 binds to Fip1 by anti bait coimmunoprecipitation (View interaction)
PAP1 binds to Fip1 by pull down (View interaction)
PAP1 physically interacts with PTA1 by two hybrid (View interaction)
PAP1 binds to Sub1 by pull down (View interaction)
PAP1 physically interacts with Fip1 by two hybrid (View Interaction: 1, 2)
PAP1 binds to Nab6 by pull down (View interaction)
Nab6 physically interacts with PAP1 by two hybrid (View interaction)
Cft1 binds to PAP1 by pull down (View interaction)
PTA1 binds to PAP1 by pull down (View interaction)
Acknowledgements
This work was supported by NIH GM041752 to C.L.M. We thank the Moore and Bohm labs for helpful discussions, and M. Bucheli for providing nuclear extract and advice on peptide columns.
Abbreviations
- CF I
cleavage factor I
- CPF
cleavage/polyadenylation factor
- CPSF
cleavage/polyadenylation specificity factor
- DTT
dithiothreitol
- GAD
gal activation domain
- GBD
gal binding domain
- GST
glutathione S-transferase
- PAP
poly(A) polymerase
- PMSF
phenylmethylsulfonyl fluoride
- SDS
sodium dodecyl sulfate
References
- 1.Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev. RNA. 2011;2:321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Virtanen A, Kleiman FE. To polyadenylate or to deadenylate: that is the question. Cell Cycle. 2010;9:4437–4449. doi: 10.4161/cc.9.22.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizrahi N, Moore C. Posttranslational phosphorylation and ubiquitination of the Saccharomyces cerevisiae poly(A) polymerase at the S/G(2) stage of the cell cycle. Mol. Cell. Biol. 2000;20:2794–2802. doi: 10.1128/mcb.20.8.2794-2802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 7.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 8.Ko B, Gunderson SI. Identification of new poly(A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J. Mol. Biol. 2002;318:1189–1206. doi: 10.1016/s0022-2836(02)00240-1. [DOI] [PubMed] [Google Scholar]
- 9.Millevoi S, Decorsiere A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. A physical and functional link between splicing factors promotes pre-mRNA 3′ end processing. Nucleic Acids Res. 2009;37:4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezeokonkwo C, Zhelkovsky A, Lee R, Bohm A, Moore C. A flexible linker region in Fip1 is needed for efficient mRNA polyadenylation. RNA. 2011;17:652–664. doi: 10.1261/rna.2273111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmling S, Zhelkovsky A, Moore CL. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 2001;21:2026–2037. doi: 10.1128/MCB.21.6.2026-2037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barabino SM, Ohnacker M, Keller W. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 2000;19:3778–3787. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meinke G, Ezeokonkwo C, Balbo P, Stafford W, Moore C, Bohm A. Structure of yeast poly(A) polymerase in complex with a peptide from Fip1, an intrinsically disordered protein. Biochemistry. 2008;47:6859–6869. doi: 10.1021/bi800204k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhelkovsky AM, Kessler MM, Moore CL. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s) J. Biol. Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]
- 15.Zhelkovsky A, Helmling S, Moore C. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazy MA, He X, Singh BN, Hampsey M, Moore C. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol. Cell. Biol. 2009;29:2296–2307. doi: 10.1128/MCB.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikorski TW, Ficarro SB, Holik J, Kim T, Rando OJ, Marto JA, Buratowski S. Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol. Cell. 2011;44:397–409. doi: 10.1016/j.molcel.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abruzzi K, Denome S, Olsen JR, Assenholt J, Haaning LL, Jensen TH, Rosbash M. A novel plasmid-based microarray screen identifies suppressors of rrp6Delta in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:1044–1055. doi: 10.1128/MCB.01299-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X, Moore C. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol. Cell. 2005;19:619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler JS, Mitchell P. Rrp6, Rrp47 and cofactors of the nuclear exosome. Adv. Exp. Med. Biol. 2010;702:91–104. [PubMed] [Google Scholar]
- 25.Saguez C, Schmid M, Olesen JR, Ghazy MA, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 26.Vodala S, Abruzzi KC, Rosbash M. The nuclear exosome and adenylation regulate posttranscriptional tethering of yeast GAL genes to the nuclear periphery. Mol. Cell. 2008;31:104–113. doi: 10.1016/j.molcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid M, Poulsen M, Olszewski P, Saguez C, Moore C, Jensen T. Rrp6p controls mRNA polyA tail length and its decoration with polyA binding proteins. Mol. Cell. 2012 doi: 10.1016/j.molcel.2012.05.005. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE. 2008;3:e2410. doi: 10.1371/journal.pone.0002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosonina E, Willis IM, Manley JL. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol. Cell. Biol. 2009;29:2308–2321. doi: 10.1128/MCB.01841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]