Abstract
Translocations involving the T cell receptor alpha/delta (TCRα/δ) chain locus, which bring oncogenes in the proximity of the TCRα enhancer, are one of the hallmark features of human T cell malignancies from ataxia telangiectasia (AT) and non-AT patients. These lesions are frequently generated by the fusion of DNA breaks at the TCRα/δ locus to a disperse region centromeric of the immunoglobulin heavy chain (IgH) locus. Aberrant VDJ joining accounts for TCRα/δ associated DNA cleavage, but the molecular mechanism that leads to generation of the "oncogene partner" DNA break is unclear. Here we show that in ATM deficient primary mouse T cells, IgH/TCRα/δ fusions arise at a remarkably similar frequency as in human AT lymphocytes. Recombinase-activating gene (RAG) is responsible for both TCRα/δ as well as IgH associated breaks on chromosome 12 (Chr12), which are subject to varying degrees of chromosomal degradation. We suggest a new model for how oncogenic translocations can arise from two non-concerted physiological DSBs.
Keywords: translocations, T cell leukemia, V(D)J recombination, ATM
Introduction
One of the most frequent cytogenetic abnormalities found in prolymphocytic leukemia, adult T cell leukemia and in leukemias from AT patients are translocations and inversions between the TCRa locus (14q11) and a disperse region near band 14q32.1 The breakpoints at 14q32 span a region of 450 kb that is centromeric of the telomere-proximal IgH locus and frequently includes the TCL1 oncogene.1,2 In primary lymphocytes from normal individuals, non-clonal t(14;14)(q11;q32) translocations appear at a frequency of 0.002%.3 AT patients exhibit a 250-fold increase in the probability of developing leukemia or lymphoma, and the frequency of non-clonal t(14;14)(q11;q32) translocations is also increased 250-fold (0.5%) in primary AT lymphocytes.4
Atm−/− mice provide a model for studying lymphoid tumor development in AT patients. Atm−/− mice are predisposed to thymic lymphomas,5 all of which harbor chromosome 14 (Chr14) rearrangements involving the TCRa locus.6 TCRa/d translocations are also common in primary Atm−/− T cells,6 and these rearrangements are dependent on expression of the RAG endonuclease which introduces DSBs at Ig and TCR loci during V(D)J recombination.7 Mouse Chr12 is homologous to human chromosome 14, and is a translocation partner for TCRa/d in approximately 50% of Atm−/− thymic lymphomas.6 Analogous to human T cell tumors, the breakpoints on chromosome 12 in Atm−/− mouse lymphomas map to a disperse region centromeric of the IgH locus, and there is a frequent loss of the IgH locus on the translocated chromosome.6 Since there is a wide dispersion of breakpoints in a region outside of the IgH locus for both human t(14:14)(q11;q32) and mouse T(12;14) translocations, this suggests that unlike the TCRa/d locus, the “oncogenic” partner might not be targeted by RAG.
Recently, it was shown that ATM functions in two complementary ways to maintain chromosomal integrity during V(D)J recombination.7-11 In the absence of ATM, the ability to repair antigen receptor coding intermediates at Ig and TCR loci is significantly compromised.9,11 In addition to its role in DNA double strand break (DSB) repair, ATM prevents the transmission of DNA double strand breaks (DSBs) to daughter cells,7 potentially by activation of a p53 dependent apoptotic checkpoint.12 Impairment in both of these repair and checkpoint functions likely accounts for the high level of antigen receptor-associated DSBs in mature lymphocytes, some of which are generated earlier in development as a result of failed V(D)J recombination.7,12 Although broken antigen receptor loci with telomere-free ends persist through several rounds of cell division, they are characterized by varying degrees of chromosomal erosion.7 Prior to loss of essential genetic material, terminally deleted chromosomes have the -potential to fuse with chromosomes broken in subsequent generations, as suggested by the increase in IgH associated translocations after irradiation.7 Here we show that RAG endonuclease dependent IgH breaks in ATM−/− T cells can persist with varying degree of chromosomal loss and fuse at a high frequency to RAG-dependent TCRa/d breaks. RAG dependent DSBs on both chromosomes might also be the source of the highly selected translocation in prolymphocytic leukemia and adult T cell leukemias.
Results and Discussion
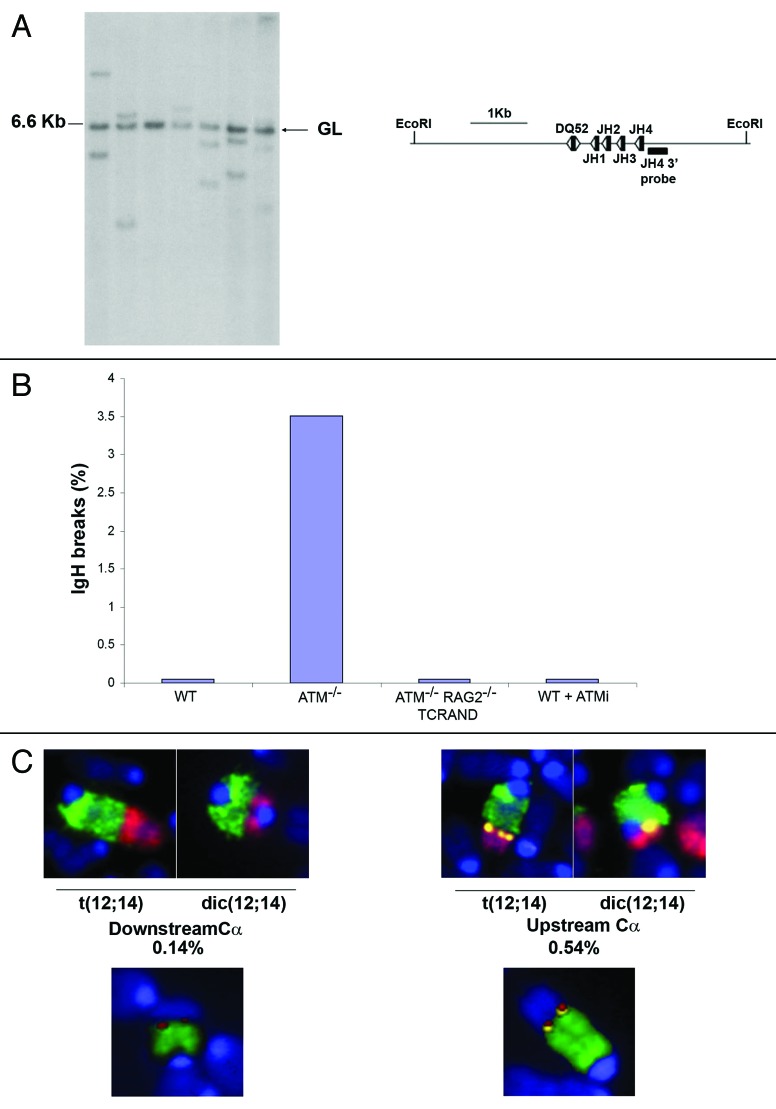
Recombination between immunoglobulin Dh and Jh segments occurs at a high frequency not only in developing B cells but also in early T lineage progenitors in the thymus.13-17 Nevertheless, productive assembly of Ig genes is restricted to B cells, since Ig variable Vh segments are not recombined in T cells. To determine the frequency of Dh-Jh rearrangements in ATM deficient cells, we performed Southern blot analysis on mature T cell hybridomas generated from mature ab T cells from two Atm−/− mice. All of the 257 hybridomas analyzed gave a 6.6 kb band indicative of an un-rearranged IgH allele, as would be expected given that both IgH alleles of the BW5147 fusion partner are un-rearranged. However, 126 of the hybridoma clones had one or two hybridizing fragments of diverse sizes indicative of Dh-Jh rearrangements (Fig. 1A). Among these 126 clones, 59 had two diverse fragments indicating that in these cells both IgH alleles in the ab T cells had undergone rearrangements. Thus, at least 49% of ATM deficient T cells have undergone Dh-Jh rearrangements on one or both alleles.
Figure 1. IgH rearrangements, breaks and translocations in ATM deficient T cells. (A) Southern blot analysis of Ig Dh-Jh rearrangements in mature ab T cell hybridomas. Rearrangements were identified by hybridizing EcoRI digested genomic DNA with a probe downstream of Jh4, as indicated on the schematic. Germline band at 6.6 kb is indicated. (B) Percentage of metaphases with abnormalities specifically associated with the IgH locus. Metaphases from Atm+/+ (WT), Atm−/−, Atm−/−RAG2-/-TCRAND and WT T cells treated with the ATM inhibitor (ATMi) were prepared 48 h after lymph node T cell stimulation and analyzed using the IgH/telomere FISH assay as described.18 At least two independent mice were analyzed for each genotype. (C) Examples of translocations between Chr12 and Chr14. Metaphases were hybridized with a Chr12 painting probe (green), a Chr14 painting probe (red) and a IgH Ca BAC probe (yellow), and counterstained with DAPI (blue) (top) or a Chr12 painting probe (green), a TCRa probe (red) and a IgH Ca BAC probe (yellow) (lower). Among 2012 metaphase spreads analyzed by FISH, 0.14% showed Chr12/Chr14 fusions in which the breakpoints on Chr12 were centromeric of Ca (left), and in 0.54% of cases, Ca was precisely at the breakpoint (right). M-FISH analysis estimated Chr12/Chr14 fusions at a somewhat higher frequency (1.11%, see Table 1). FISH-labeled metaphases were imaged using a Zeiss AxioImager M1 wide-field upright epifluorescence microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY), equipped with a motorized scanning stage and monochrome progressive scan video CCD camera (JAI America Inc., Woburn, MA), controlled by Metafer image acquisition and metaphase pattern recognition software (Metasystems Group Inc., Watertown, MA). Automated identification of metaphases used the pattern recognition algorithm in the MSearch module of the Metafer software (version 4.0). For M-FISH, metaphases were hybridized with the 21x mouse 21-color probe cocktail from Metasystems GmbH (Watertown, MA), and analyzed using Isis software.
Since failed V(D)J recombination intermediates are transmitted to downstream progenitors in the absence of ATM,7 we speculated that aberrant Ig Dh-Jh rearrangements generated in immature Atm−/− T cells might accumulate as IgH-associated breaks in mature T cells. To test this, we analyzed metaphase spreads from day two Atm−/− lymph node T cells by fluorescence in situ hybridization (FISH) with probes specific for chromosome 12 (Chr12), IgH Ca, and telomere repeats.18,19 Among 199 metaphases analyzed from two independent Atm−/− mice, seven of them (3.5%) showed IgH associated chromosome breaks evidenced by loss of telomeric sequences (Fig. 1B). In two of the aberrant metaphases, there was also a failure to hybridize with the IgH Ca probe, which is centromeric of the VDJ cluster. Cytogenetic analysis using DAPI-banding revealed that both of these breaks localized near band F1, which is the same Chr12 region observed in some of the t(12;14) translocations in Atm−/− tumors.6 In contrast to Atm−/− cells, the IgH locus was intact in all of the T cells derived from WT littermate controls (n = 200 metaphases) (Fig. 1B).
To determine whether Ig heavy chain specific chromosome breaks in mature T cells are products of V(D)J recombination, we performed the IgH FISH assay on metaphase spreads from Atm−/−Rag2−/−TCRAND T cells.7 TCRAND is a transgenic T cell receptor that reconstitutes T cell development and generates mature clonotypic T cells in the absence of RAG.20 In contrast to lymph node T cells from Atm−/− mice, no IgH chromosome breaks were found in T cells from Atm−/−Rag2−/−TCRAND mice (n = 70 metaphases) (Fig. 1B). Thus, IgH-associated DNA damage in mature Atm−/− T cells is dependent on RAG mediated cleavage. To control for the possibility that IgH breaks in ATM deficient cells are generated by RAG re-expression in mature T cells, we treated WT lymph node T cells with KU55933 (ATMi), a small molecule inhibitor of ATM kinase activity. This treatment increased the total number of aberrant metaphases to levels similar to those found in Atm−/− T cells (data not shown). However, unlike WT B cells stimulated to undergo class switch recombination,7 treatment of T cells with ATMi did not produce IgH-specific DNA damage (n = 106 metaphases) (Fig. 1B). Thus, IgH associated breaks, characterized by varying degrees of chromosomal loss, are not caused by de novo cleavage in mature ATM deficient cells, but more likely, they are produced earlier as a result of failed Dh-Jh recombination during thymocyte development. Nevertheless, these RAG-dependent chromosome breaks initiated in the thymus persist in the peripheral T cell compartment.
RAG-dependent IgH breaks in ATM deficient T cells occur at a frequency of 3.5% (Fig. 1B), while RAG-dependent TCRa/d breaks are found in 5.8% of metaphases (n = 1212 metaphases analyzed, not shown). Given the high level of unresolved/persistent breaks, we wished to determine the frequency of translocation between these loci. Metaphase spreads from Atm−/− T cells lymph node cells were identified using an automated image acquisition system and image analysis software (for details see Materials and Methods). Among 2012 metaphase spreads analyzed by FISH with probes specific for Chr12, Chr14 and IgH Ca, 14 of them (0.68%) harbored t(12;14) translocations or dic(12;14) dicentric chromosomes (Fig. 1C). This is remarkably similar to the incidence of t(14;14)(q11;q32) translocations (0.5%) or inv(14) inversions (0.65%) found in human AT lymphocytes.4 In three of 14 translocations, the breakpoints on Chr12 were located centromeric of Ca (Fig. 1C), consistent with chromosomal degradation. Using probes specific for TCRa, Chr12 and IgH Ca, we detected fusions in which TCRa/IgHCa were at the breakpoint and fusions between TCRa and Chr12, in which Ca hybridization was lost (Fig. 1C).
The frequency of the Chr14-Chr12 translocation in primary ATM−/− T cells produced by RAG mediated cleavage of both chromosomes (6.9 x 10−3) is 2–3 orders of magnitude higher than c-myc/IgH translocations induced in primary Atm−/− B cells (2 x 10−6) by AID dependent DSBs,18 or by I-SCE1 or RAG endonuclease translocation reporters21,22 (3 x 10−5). To determine whether Chr12 and Chr14 were indeed favored partners for translocation, we used multi-color FISH (M-FISH) to karyotype metaphases from three independent ATM−/− mice (Table 1). Among 450 metaphases examined, 61 (13.5%) exhibited translocations. Among these translocations, 25 (41%) involved Chr14 and 19 (31%) involved Chr12; therefore these two chromosomes are involved in 78.5% of all translocations. Fusions between Chr12 and Chr14 were highly favored, as 17% of all Chr14 translocations involved Chr12, and 26% of all Chr12 partners were Chr14. Strikingly, in addition to Chr12/Chr14 translocations, another favored fusion partner for Chr14 was Chr13 (3.5% of all metaphases). However, unlike the Chr12/Chr14 translocation, which occurs in 50% of ATM−/− thymic lymphomas,6 translocations involving Chr13 were not found among eight thymic lymphomas previously analyzed by spectral karyotyping.6 Although we do not currently know the etiology of the Chr13 translocation, our data provide evidence that some high frequency translocation events in primary cells are highly selected for while others are not.
Table 1. Frequencies of translocations involving chromosomes 12 and 14 in ATM−/− T cells (n = 450 metaphases scored, from three experiments).
| Type | Translocation | % Frequency |
|---|---|---|
| T(14;12) or T(12;14) |
|
1.11% |
| T(12;x) or T(x;12) |
Dic(6;12) |
2.22% |
| |
T(12;13) |
|
| |
T(12;13) |
|
| |
T(12;15) |
|
| |
T(10;12) |
|
| |
T(19;12) |
|
| |
T(12;13) |
|
| |
T(10;12) |
|
| |
T(10;12) |
|
| |
T(10;12) |
|
| T(14;13) or T(13;14) |
|
3.56% |
| T(14;x) or T(x;14) |
T(11;14) |
1.78% |
| |
T(14;6) |
|
| |
T(8;14) |
|
| |
T(14;3) |
|
| |
T(14;9) |
|
| |
Dic(14:14) |
|
| |
T(6;14) |
|
| Dic(14:14) |
T(12;x) indicates partners for chromosome 12 translocations, excluding chromosome 14. T(14;x) indicates partners for chromosome 14 translocations, excluding chromosome 12.
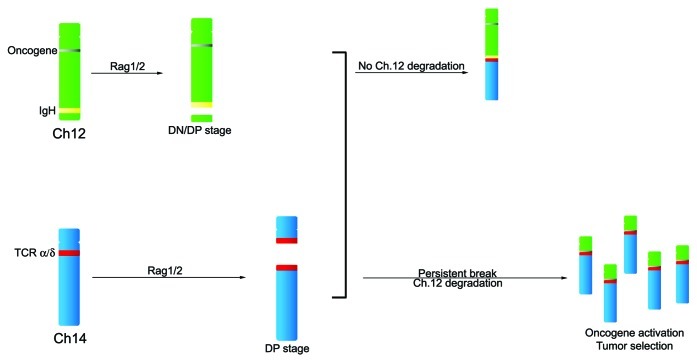
Abnormal V(D)J joining likely accounts for the TCRa/d translocations found in primary lymphocytes and lymphomas from ATM-deficient patients and mice. In human T cell tumors, the TCRa joining (J) region is frequently fused with a region 10 Mb centromeric of IgH that includes the TCL1 oncogene locus. The molecular mechanism that leads to generation of DNA breaks downstream of IgH has remained unclear. Although our data do not rule out that stochastic processes or fragile DNA segments can contribute to DNA breakage, we suggest that even breakpoints that are centromeric to the IgH locus might arise from failed V(D)J recombination. In Atm−/− thymocytes, unresolved IgH breaks generated by RAG mediated cleavage persist throughout T cell development and are subject to exo-nuclease digestion.7,8 These IgH associated breaks with varying degrees of chromosome loss may fuse to the TCRa/d locus during Va-Ja rearrangement. If chromosomal degradation occurs juxtaposing an oncogene on the eroded chromosome to the TCRa enhancer (or other transcription promoting element), deregulation of the transforming gene can ensue, resulting in clonal T cell expansion (Fig. 2). In this way, chromosomal breakpoints centromeric of the IgH locus may be selected for during lymphomagenesis. IgH-TCRa/d fusions occur at a remarkably high frequency in ATM deficient lymphocytes, but they might also occasionally arise in normal cells because of the high level of DNA damage at these loci and the imperfect nature of the DNA damage -checkpoint.3,23
Figure 2. Model for translocations in ATM deficient T cells. Immunoglobulin Ig Dh-Jh recombination associated DSB in immature (DN/DP CD4−/−CD8−/−/CD4+CD8+) thymoctyes may persist until they translocate to the TCRa/d locus during failed Va-Ja or Vd-DdJd recombination. Oncogenes localized centromeric of IgH might be brought into the vicinity of the TCRa enhancer (or other element) if the unprotected telemore-free Chr12 end is subject to degradation.
Acknowledgments
We thank Joseph Cheng for help with the image analysis system and for comments on the manuscript. This work is supported in part by the Intramural research Program of the NIH, National Cancer Institute, Center for Cancer Research. B.S. is supported by NIH grant AI074953. The authors declare no financial conflict of interest.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/9085
References
- 1.Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87:423–38. [PubMed] [Google Scholar]
- 2.Virgilio L, Isobe M, Narducci MG, Carotenuto P, Camerini B, Kurosawa N, et al. Chromosome walking on the TCL1 locus involved in T-cell neoplasia. Proc Natl Acad Sci U S A. 1993;90:9275–9. doi: 10.1073/pnas.90.20.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht F, Hecht BK, Kirsch IR. Fragile sites limited to lymphocytes: molecular recombination and malignancy. Cancer Genet Cytogenet. 1987;26:95–104. doi: 10.1016/0165-4608(87)90137-3. [DOI] [PubMed] [Google Scholar]
- 4.Aurias A, Dutrillaux B, Buriot D, Lejeune J. High frequencies of inversions and translocations of chromosomes 7 and 14 in ataxia telangiectasia. Mutat Res. 1980;69:369–74. doi: 10.1016/0027-5107(80)90101-3. [DOI] [PubMed] [Google Scholar]
- 5.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–71. doi: 10.1016/S0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 6.Liyanage M, Weaver Z, Barlow C, Coleman A, Pankratz DG, Anderson S, et al. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–6. [PubMed] [Google Scholar]
- 7.Callén E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–70. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 9.Huang CY, Sharma GG, Walker LM, Bassing CH, Pandita TK, Sleckman BP. Defects in coding joint formation in vivo in developing ATM-deficient B and T lymphocytes. J Exp Med. 2007;204:1371–81. doi: 10.1084/jem.20061460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matei IR, Guidos CJ, Danska JS. ATM-dependent DNA damage surveillance in T-cell development and leukemogenesis: the DSB connection. Immunol Rev. 2006;209:142–58. doi: 10.1111/j.0105-2896.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 11.Vacchio MS, Olaru A, Livak F, Hodes RJ. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor alpha locus coding end breaks. Proc Natl Acad Sci U S A. 2007;104:6323–8. doi: 10.1073/pnas.0611222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Difilippantonio S, Gapud E, Wong N, Huang C-Y, Mahowald G, Chen H-T, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–33. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurosawa Y, von Boehmer H, Haas W, Sakano H, Trauneker A, Tonegawa S. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981;290:565–70. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- 14.Forster A, Hobart M, Hengartner H, Rabbitts TH. An immunoglobulin heavy-chain gene is altered in two T-cell clones. Nature. 1980;286:897–9. doi: 10.1038/286897a0. [DOI] [PubMed] [Google Scholar]
- 15.Cory S, Adams JM, Kemp DJ. Somatic rearrangements forming active immunoglobulin mu genes in B and T lymphoid cell lines. Proc Natl Acad Sci U S A. 1980;77:4943–7. doi: 10.1073/pnas.77.8.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Born W, White J, Kappler J, Marrack P. Rearrangement of IgH genes in normal thymocyte development. J Immunol. 1988;140:3228–32. [PubMed] [Google Scholar]
- 17.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–74. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 18.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–9. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–14. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Kaye J, Hsu ML, Sauron ME, Jameson SC, Gascoigne NR, Hedrick SM. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–9. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 21.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–80. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–9. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]