Abstract
Heterochromatin protein 1 paralogs (HP1α, β and γ in mammals) are not only central in heterochromatin organization, but have also been linked to transcriptional activation at euchromatic regions, maintenance of telomere stability and, most recently, to the DNA damage response (DDR). However, how HP1 proteins contribute to the DDR at a molecular level, and whether HP1 paralogs within the same organism, as well as their respective orthologs, have overlapping or unique roles in the DDR, remain to be elucidated. Herein, we have combined the analysis of the efficiency and kinetics of recruitment of key repair proteins to sites of DNA damage with specific DNA repair assays to demonstrate that human HP1 paralogs differentially modulate homology-directed repair (HDR) pathways, including homologous recombination (HR) and single-strand annealing (SSA). We find that while HP1α and β stimulate HR and SSA, HP1γ has an inhibitory role. In addition, we show that the stimulatory role of HP1α and β in HDR is linked to the DNA-end resection step of DNA breaks, through the promotion of RPA loading and phosphorylation at damage sites. Altogether, our findings provide mechanistic insight into how human HP1 proteins participate in the recombination process, emerging as important chromatin regulators during HDR.
Keywords: DNA damage response, DNA end resection, DNA repair, HP1, chromatin, epigenetics, homologous recombination, non-histone chromatin proteins
Introduction
Both the normal cellular metabolism and external genotoxic agents constantly threaten genome integrity by inducing DNA damage.1 The elaborate network known as the DNA damage response (DDR) is thus continuously working to preserve genome stability.1 Notably, DDR machineries function within chromatin, a nucleoprotein complex containing histone and non-histone proteins.2 While histones are central in nucleosome organization,3 non-histone proteins promote higher levels of organization and compaction, as in the typical case of heterochromatin.4,5 The heterochromatin protein 1 (HP1) family, which comprises three closely related paralogs in mammals (HP1α, β and γ),6 represents a prominent group of non-histone proteins with essential roles in heterochromatin formation/maintenance and heterochromatin-related gene silencing.7 A current model for HP1 function in pericentric heterochromatin involves SUMOylation of HP1 and non-coding RNA for its targeting.8 Maintenance pathways exploit the interaction of HP1 N-terminal chromodomain (CD) with histone H3 tri-methylated on Lysine 9 (H3K9me3), a modification that is highly enriched in heterochromatin,7 as well as its capacity to homo- and hetero-dimerize via its C-terminal chromoshadow domain (CSD). In addition, several HP1 partners9,10 bind to the interface between two CSDs, which promote the activity of HP1 paralogs in various nuclear processes, such as transcriptional activation and telomere stability.11 Interestingly, even though mammalian HP1 paralogs and their orthologs in other metazoans share high levels of homology,6 they can differ substantially in their properties and functions,6,11,12 as illustrated with HP1γ that is mostly enriched in euchromatin.13
Several recent reports have linked mammalian HP1 paralogs, as well as HP1 orthologs from flies and worms, to key steps of DNA damage signaling and DNA repair, thus suggesting an active role for these proteins in the DDR.2 Following UV irradiation, exposure to α particles, soft X-rays and laser micro-irradiation, mammalian HP1 paralogs show reproducible accumulation at sites of damage.14-17 While the mechanism of recruitment to sites of DNA damage for all HP1 paralogs involves their direct interaction with the largest subunit of the chromatin assembly factor 1 (CAF-1),17 several lines of evidence indicate that their roles might not completely overlap. In mammalian cells, HP1α knockdown impairs the recruitment of the homologous recombination protein RAD51 to sites of local damage as well as cell survival after ionizing radiation.17 In contrast, knockdown of the HP1 Drosophila ortholog HP1a does not impair RAD51 recruitment.18 Intriguingly as well, knocking-out of C. elegans HP1 orthologs (hpl-1 and hpl-2) leads to opposite outcomes on cell survival after ionizing radiation (IR): hpl-1 KO shows increased resistance to IR, while hpl-2 KO is highly sensitive.14 Therefore, it is necessary to sort out the particular contribution of each HP1 paralog in the DDR and the molecular mechanisms at stake.
Herein, using specific DNA repair assays and exploring the recruitment of key repair proteins to sites of DNA damage in mammalian cells, we show that HP1 paralogs impact differentially on the DDR, leading both to inhibition and stimulation of the repair of double-stand breaks (DSBs) by homologous recombination. In addition, we show that the HR-promoting role of HP1 is linked to the stimulation of DNA end resection. Our results also demonstrate that other resection-dependent processes, such as DNA repair by single-strand annealing (SSA), are similarly affected by HP1 knockdown. Together, these findings reveal that HP1 proteins are important players during the DDR. In addition, given their unexpected contrasting impact on DNA repair efficiency, our data put forward the HP1 family as proteins with regulatory potential during initial steps of the DDR.
Results
HP1 paralogs differentially modulate homologous recombination repair
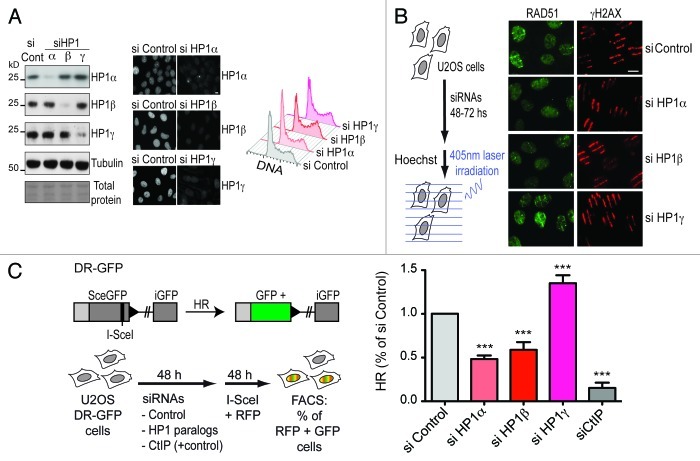
We recently showed that HP1α knockdown in human cells impairs the recruitment of the recombinase RAD51 to sites of DNA damage and the efficiency of DNA repair by homologous recombination (HR).17 Here, we decided to study HP1α and the other two mammalian paralogs of HP1 (β and γ) under equivalent experimental settings. Initially, using human U2OS cells we set up optimal knockdown conditions for each HP1 paralog (Fig. 1A). Importantly, HP1 downregulation did not impair cell cycle progression (Fig. 1A), which is critical for a proper evaluation of DNA repair-associated processes such as HR that is restricted to S and G2 phases of the cell cycle. We evaluated the impact of HP1 downregulation on RAD51 accumulation at sites of DNA damage induced by laser micro-irradiation (Fig. 1B). As expected, HP1α knockdown inhibited RAD51 recruitment to sites of DNA damage. In a similar manner, HP1β knockdown also impaired RAD51 accumulation substantially. HP1γ knockdown, however, did not lead to any apparent defect in RAD51 accumulation, while the amount of DNA damage estimated by the levels of H2AX phosphorylation was comparable in all cases (Fig. 1B). We also evaluated the contribution of HP1 paralogs to the efficiency of HR using a previously characterized and widely used system to monitor homology-directed repair.19,20 This system assesses the repair of a single DSB induced by the rare-cutting I-SceI endonuclease within a mutant GFP gene stably integrated in human cells, which, when repaired by homologous recombination, generates a wt-GFP. After 24–48 h, GFP expression can be detected by flow cytometry (Fig. 1C). To monitor and normalize our data, considering transfection efficiencies for each sample, we co-transfected I-SceI and RFP encoding plasmids (Fig. 1C). As a positive control we performed knockdown of CtIP, a key player in the DNA end resection step of homologous recombination.21 We observed that both HP1α and β knockdown led to a significant decrease in the efficiency of homologous recombination (Fig. 1C). This observation is in line with our initial finding showing impaired accumulation of the recombination protein RAD51 (Fig. 1B). Surprisingly, however, upon HP1γ knockdown, we observed a significant stimulation of HR efficiency (Fig. 1C). A similar stimulatory effect was reported after knocking down the NHEJ protein KU80.22 This was also observed under our experimental settings (Fig. S1D), thus validating the sensitivity of our method. We are confident that these cannot reflect off-target effects, given that we obtained similar findings with two different sets of siRNAs for each HP1 paralog, which lead to equivalent levels of knockdown without affecting cell cycle progression (Fig. S1). Together, these results demonstrate that HP1 paralogs contribute differentially to the efficiency of HR.
Figure 1. HP1 paralogs differentially modulate RAD51 recruitment and homology-directed repair. (A) Control of knockdown efficiency and cell cycle progression after siRNA treatment against the three mammalian HP1 paralogs. We transfected U2OS cells with siRNAs against HP1α, HP1β and HP1γ, and 48 h after, we monitored the knockdown efficiency by WB (left panel) and by immunofluorescence (middle panel). Bar: 10 μm. Parallel samples were also collected for flow cytometry analysis of the cell cycle progression (right panel). (B) RAD51 recruitment to DNA damage sites analyzed by immunofluorescence 10 min after laser micro-irradiation (as shown in the experimental scheme) in U2OS cells treated with the indicated siRNAs. γH2AX is used as a marker to monitor the efficiency of DNA damage induction. (C) Homology-directed repair efficiency after transient knockdown of HP1 paralogs normalized to control siRNA. CtIP knockdown is used as a positive control. Error bars: s.d. from three independent experiments. Further statistical analysis was performed using one-way ANOVA (p ≤ 0.0001), followed by Tukey-Kramer post-test. Knockdown of all HP1 paralogs presented highly significant differences in their mean compared with siRNA control (*** = p ≤ 0.001).
HP1α and β but not HP1γ promote DNA end resection
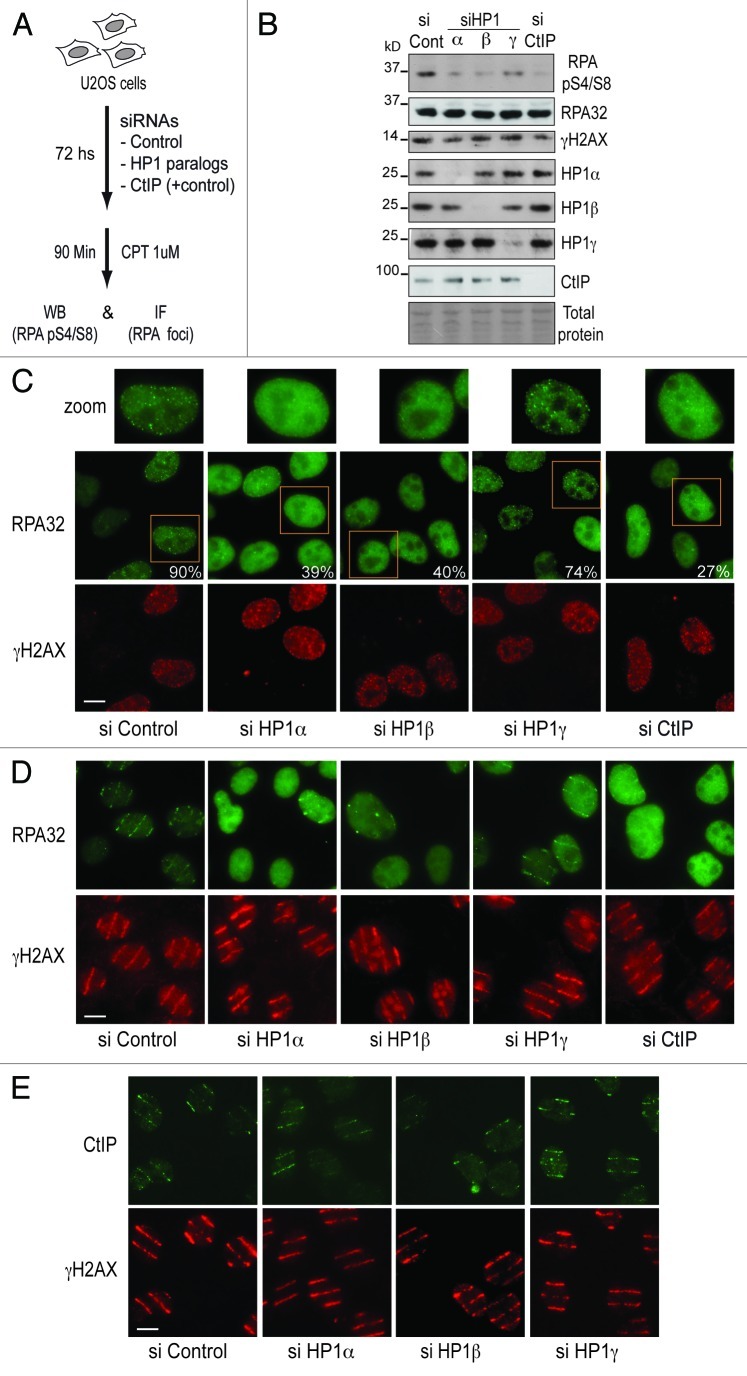
To explore how HP1 proteins impact on HR at a molecular level, we considered whether RAD51 loading could be affected due to impaired DNA end resection at the sites of DNA breaks. Such hypothesis is supported by the fact that HP1α knockdown decreases the levels of RPA32 phosphorylation after DNA damage induction with camptothecin,17 thus suggesting that the processing of single-stranded DNA might be affected. We thus examined the effect of knocking down the different HP1 paralogs on camptothecin-induced RPA32 phosphorylation and foci formation (Fig. 2A). As a positive control, we depleted the resection protein CtIP.21 We observed that HP1α and β knockdown impaired both the phosphorylation of RPA (Fig. 2B) and the efficiency of RPA foci formation (Fig. 2C), without noticeably affecting the levels of H2AX phosphorylation (Fig. 2B and C). Remarkably, these effects of HP1α and β knockdown were comparable to the ones observed after CtIP knockdown (Fig. 2B and C), thus arguing for an important role of HP1α and β in promoting DNA end resection. HP1γ knockdown however only slightly affected RPA phosphorylation and foci formation (Fig. 2B and C). To further support a distinct contribution of HP1 paralogs to DNA-end resection, and to avoid misinterpretations that may arise from the processing of single-stranded DNA during normal DNA replication, we used laser micro-irradiation as an alternative to camptothecin to induce DNA breaks. In this context, while both HP1α and β proved critical for efficient RPA recruitment to DNA damage stripes, HP1γ knockdown showed only a marginal defect (Fig. 2D). These differential effects of HP1 paralogs in DNA-end resection are in agreement with the effects observed on the recruitment of the downstream HR protein RAD51 (Fig. 1B). We then evaluated the recruitment of the upstream resection protein CtIP to sites of DNA damage induced by laser micro-irradiation after HP1 depletion, and found that accumulation of CtIP occurred without substantial alteration (Fig. 2E). Thus, these experiments suggest that HP1α and β roles during DNA end resection takes place either downstream or independently of CtIP.
Figure 2. HP1α and HP1β -but not HP1γ- promote RPA loading and phosphorylation at sites of DNA damage independently of CtIP. (A) Experimental scheme used to induce DNA damage by treating U2OS cell with 1 μM camptothecin (CPT). After siRNA-mediated knockdown of HP1α, HP1β, HP1γ and CtIP (used as a positive control of the experiment) we examined RPA32 phosphorylation by western blot (WB) and the efficiency of RPA foci formation by immunofluorescence (IF). (B) WB of U2OS cells depleted of HP1 paralogs to analyze the efficiency of RPA phosphorylation after 90 min of CPT treatment. We monitored the total levels RPA32 and the extent of DNA damage induced using antibodies against RPA32 and γH2AX respectively. (C) We show a representative field for the efficiency of RPA foci formation after HP1 knockdown using γH2AX as marker of DNA damage (a zoomed nucleus for each siRNA is shown on the top). The numbers depicted of the right corner of the RPA32 panels represent the mean of cells with distinguishable RPA foci in the γH2AX positive population. 150 γH2AX-positive cells were analyzed in each siRNA-treated sample. Bar: 10 μm. (D) We generated local DNA damage as described in Figure 1B and analyzed by microscopy the accumulation of RPA32 to sites of laser micro-irradiation after 10 min, using γH2AX as control of efficient DNA damage induction. Bar: 10 μm. (E) Using laser micro-irradiation we assessed the efficiency of recruitment of CtIP to sites of DNA damage 10 min post-irradiation using γH2AX as control of efficient DNA damage induction. Bar: 10 μm.
HP1 paralogs modulate resection-dependent double-strand break repair
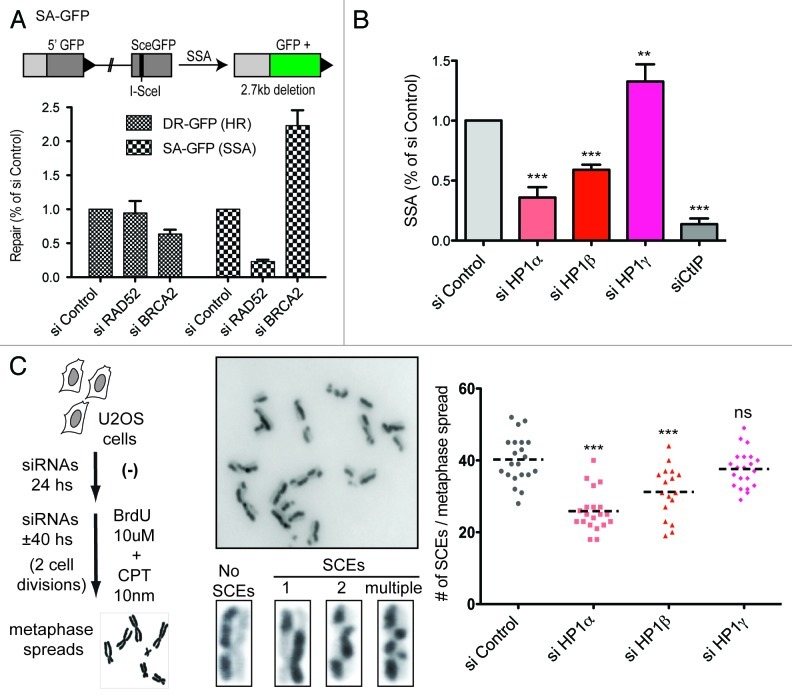
Given the impact of HP1 paralogs on homologous recombination repair (Fig. 1) and DNA-end resection (Fig. 2), we decided to explore if HP1 proteins could also influence other resection-dependent processes. To this end, we initially exploited a system enabling us to assess the efficiency of homology directed repair by single-strand annealing (SSA).22 In a similar fashion to the DR-GFP system, this assay relies on the restoration of a GFP gene that also presents an I-SceI cutting site, which is truncated by the insertion of 2.7 Kb DNA fragment. When the DSB is induced by I-SceI transfection, the 2.7 kb fragment is removed and the GFP sequence restored by a SSA event (Fig. 3A). To verify that this method specifically measures SSA, we compared the effect of the knockdown of RAD52 (which is critical for SSA) and of BRCA2 (which is critical for HR).23 We observed that BRCA2 knockdown specifically impaired HR, whereas RAD52 knockdown specifically impaired SSA (Fig. 3A). Thus, each assay proved specific for each HDR mechanism. Strikingly, we found that in agreement with our results for HR, HP1α and β both promoted SSA, while HP1γ impaired it (Fig. 3B).
Figure 3. HP1 paralogs differentially modulate resection-dependent double-strand break repair. (A) On the top panel we show a scheme of the SA-GFP system that was used to estimate the efficiency of homology-directed repair by single-strand annealing. On the bottom panel we show a comparison of the repair efficiency by homologous recombination (HR) and single-strand annealing (SSA) using U2OS carrying either the DR-GFP or the SA-GFP system. (B) We show the results of three independent experiments, depicting the mean and standard deviation of the repair by single-strand annealing (SSA) normalized to the control siRNA. Statistical analysis was performed using one-way ANOVA (p ≤ 0.0001), followed by Tukey-Kramer post-test. Knockdown of all HP1 paralogs presented highly significant differences in their mean compared with siRNA control (*** = p ≤ 0.001; ** = p ≤ 0.01). (C) Sister chromatid exchange (SCE) efficiency after transient knockdown of HP1 paralogs. On the left panel we show a brief summary of the experimental protocol used to quantify SCEs. We also show a field at high magnification showing part of metaphase and enhanced images of individual chromosomes to exemplify the range of the SCEs phenotypes observed. On the right panel we show the result of a representative experiment, plotting the absolute number of SCEs in 20 independents metaphases for each treatment. The dotted line represents the mean. Further statistical analysis was performed using one-way ANOVA (p ≤ 0.0001), followed by Tukey-Kramer post-test. Knockdown of HP1α and HP1β presented highly significant differences in their mean compared with siRNA control (*** = p ≤ 0.001). The knockdown of HP1γ leads to not significant (ns) differences in the average amount of SCEs.
To further delineate the role of HP1 paralogs in HDR, we decided to evaluate their potential contribution to sister chromatid exchanges (SCEs), a form of recombination that allows repair of potentially lethal DNA breaks during DNA replication.24 SCEs can be monitored by the incorporation of bromodeoxyuridine (BrdU) for two DNA duplication cycles, which leads to a staining gap in one chromatid and a gain of staining in the corresponding sister chromatid (Fig. 3C). Upon knockdown of each HP1 paralog, we observed that HP1α and β promoted SCEs formation (Fig. 3C). Intriguingly, HP1γ did not significantly affect this process (Fig. 3C), thus suggesting that the inhibitory role of HP1γ does not apply to S-phase related HR.
Discussion
Our work provides a first characterization of the distinct specific contribution of HP1 proteins (HP1α, β and γ) as regulators of the DDR in mammals. Supporting our findings, knockouts of HP1 orthologs in C. elegans (hpl-1 and hpl-2) display contrasting phenotypes in the survival after ionizing irradiation; hpl-1 KO is protective, while hpl-2 KO is detrimental.14 Although HP1 proteins diverged substantially during evolution, hpl-1 displays increased homology with HP1γ and hpl-2 with HP1β.6 Thus, it is conceivable that the opposite roles of HP1 proteins in the regulation of HDR are ancestral and well-conserved among eukaryotes, HP1α and β being stimulatory and HP1γ being inhibitory. Future work on the evolutionary properties extended to other organisms could help to deepen our understanding of the mechanistic basis of such an opposite behavior of HP1 paralogs on HDR efficiency. In particular, it will be important to consider how the ability of HP1 paralogs to heterodimerize7 might impact on their contribution to DNA repair. Furthermore, evaluating if compensatory mechanisms can be activated due to the deficiency of a given paralog that could lead to the mislocalization and malfunction of other paralogs should also be explored. Beyond the evolutionary aspect, the context of cancer cells will be particularly interesting to consider given that frequent unbalance between HP1 paralogs is observed.25 In addition, we should also consider the important role that HP1 proteins play in transcriptional regulation12 and examine whether the different phenotypes observed herein could also be related, at least in part, to indirect transcriptional modulation of relevant DDR genes. Thus, transcriptome analysis under conditions of specific knockdown of the different HP1 paralogs would provide an entry point into this question that would then deserve a comprehensive study.
A second set of important findings in this work establishes that HP1 role in HDR is linked to DNA-end resection. Although it is unlikely that HP1 proteins represent previously unnoticed core components of the resection machinery, the fact that the phenotypes observed for RPA accumulation and phosphorylation in response to DNA damage after HP1α and β knockdown are similar to the ones observed after CtIP knockdown underline their importance during the resection process. Given that CtIP recruitment is not affected after HP1 knockdown (Fig. 2E), to investigate potential links with other relevant proteins that work downstream CtIP, like the Bloom helicase (BLM) and the exonuclease EXO1,26,27 will be therefore critical to clarify the role of HP1 proteins during DNA-end resection.
Finally, an important question that remains open is how HP1γ exerts such an opposite effect compared to HP1α and β during HDR. Our data suggest that such a role is not linked to an opposite contribution to DNA-end resection or RAD51 recruitment. In fact, while HP1γ knockdown does not affect RAD51 accumulation at damage sites (as HP1α and β), we do not observe an increase in its accumulation that could explain the stimulation of HR (Fig. 1B and C). A similar conclusion could be made from the resection data. While HP1γ clearly impacts differently both RPA phosphorylation and loading at damage sites, these effects are not simply opposite to the ones observed for HP1α and β, but rather show a similar trend (Fig. 2). In conclusion, while the stimulatory role of HP1α and β could certainly be linked to the promotion of DNA-end resection, the inhibitory role is most likely linked to a different process. More insights into the molecular mechanisms can be anticipated soon, since several new interactors have been recently identified for HP1 proteins,9,10 including proteins involved in sister chromatid cohesion such as the cohesins loader NIPBL. In addition, HP1γ specifically interacts with TIN2, a central component of the shelterin complex, an interaction important to promote telomeric cohesion.28 These partnerships open up intriguing possibilities to explain the role of HP1 paralogs during HDR. Indeed, a recent report shows that NIPBL recruitment to DNA double-strand breaks requires HP1γ in human cells.29 An exciting hypothesis that arises from these findings is the existence of an HP1-related mechanism to ensure that resection only takes place in the presence of a sister chromatid. In this scenario, the HP1 proteins would act to permit the resection when HR has a chance to be productive. Another, yet not mutually exclusive, possibility is that HP1 regulates the accumulation of important mediators of the DDR that ultimately impact on HDR efficiency. In line with this hypothesis, HP1α knockdown impairs both 53BP1 and BRCA1 recruitment to sites of damage.17 Given that 53BP1 and BRCA1 have opposite effects on DNA-end resection,30-33 further research on this particular issue would certainly be critical to shed light on the molecular details of HP1 role during this process.
Materials and Methods
Cell lines, culture and transfection
We used human U2OS cells (provided by J. Bartek, Institute of Cancer Biology and Centre for Genotoxic Stress Research), U2OS DR-GFP (provided by M. Jasin, Sloan-Kettering Institute) and U2OS SA-GFP cells (described in ref. 34). We cultured all cells lines in DMEM (Invitrogen) supplemented with 10% (vol/vol) FCS (Eurobio), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) in a humidified atmosphere with 5% CO2 at 37°C. We maintained U2OS DR-GFP and U2OS SA-GFP cells in medium supplemented with 1 μg/ml puromycin (Invitrogen).
We transfected siRNAs (50–100 nM) into cells at 20–40% confluence depending on the length of the experiment, using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer instructions. We performed plasmids transfections using Lipofectamine 2000 (Invitrogen).
siRNA sequences
The sequences of the siRNA duplexes used (MWG eurofins) were as follows: siLuciferase (control), CGUACGCGGAAUACUUCGA;35 siHP1α, #1-CCTGAGAAAAACTTGGATT used in the majority of the experiments36 and #2-GGAATGAACATGAGACTTA ; siHP1β, #1-AGGAATATGTGGTGGAAAA used in the majority of the experiments36 and #2-AGGTAAGACAGTAGGGAAA; siHP1γ, #1-AGGTCTTGATCCTGAAAGA used in the majority of the experiments36 and #2-GGTGAACAGTGGTTAATAA; siCtIP, GCUAAAACAGGAACGAAUC21; siKU80 (Dharmacon Smartpool); siBRCA2, AACTGAGCAAGCCTCAGTCAA37; siRAD52, ACACATTAGCCTTGAACAA.
DNA damage induction
Camptothecin
We treated cells with 1 μM camptothecin (Sigma-Aldrich) and harvested them at the indicated time points. For the generation of sister chromatid exchanges, we used 10 nM for the complete length of the experiment.
Laser-induced damage
To generate localized DNA lesions, we used as the main technique in this work a method described previously,17 with particular modifications. We pre-sensitized the cells with 10 μg/ml viable Hoechst dye 33258 (Sigma-Aldrich) for 5 min at 37°C. We performed laser microirradiation using an inverted confocal microscope (LSM 510 Meta; Carl Zeiss, Inc.) equipped with a 37°C heating chamber and a 25-mW 405 nm diode laser focused through a 63× Plan-Apochromat/1.4 NA oil objective. We performed one iteration at a laser output of 50%, which in our microscope corresponds to a power of ≤ 320 μW, as monitored with a Coherent Fieldmate detector model OP2-vis. To target a large number of nuclei, we scanned 25 adjacent fields in a pattern of evenly spaced parallel lines using the tile scan tool coupled to a motorized stage.
Cell cycle analysis by flow cytometry
We performed cell cycle analysis as described previously.38 In brief, we collected siRNA-treated cells by trypsinization using the same culture media to avoid loosing of detached cells. Then, we pelleted the cells by centrifugation at 500 g, and we immediately fixed them in ice-cold ethanol with gentle vortexing. After a minimum of 2 h at -20°C, we pelleted the cells and resuspended them in PBS containing 50 μg/ml propidium iodide and 50 μg/ml RNase. We collected the samples using a C6 flow cytometer (Accuri) and analyzed them using FlowJo software (TreeStar Inc.). We analyzed a minimum of 10,000 cells/sample.
HR and SSA assays
We used an HR assay generated previously in U2OS cells containing an integrated HR reporter substrate DR-GFP,19 with some modifications. In brief, 48 h after siRNA transfection in 35-mm dishes, we cotransfected U2OS+DR-GFP cells with pCBASce, a plasmid expressing the I-SceI endonuclease and pCS2-mRFP, a plasmid expressing mRFP to follow the transfection efficiency (both plasmids provided by S. Jackson, Wellcome Trust/Cancer Research UK Gurdon Institute, University of Cambridge). Two days after I-SceI transfection, we harvested cells and performed flow cytometry analysis on C6 flow cytometer (Accuri) to determine HR-mediated DNA repair events. We analyzed the mRFP-positive cell population to avoid the bias caused by differences in transfection efficiencies. We analyzed FACS data by using CFlow software to evaluate the percentage of GFP-positive cells relative to the number of mRFP-positive cells (HR efficiency). We showed the results normalized to control siRNA as a percentage of sicontrol. We followed exactly the same protocol to estimate single-strand annealing using U2OS SA-GFP cells that were established previously.34
Antibodies
Primary antibodies used during immunoblot (IB) or immunofluorescence (IF) experiments are listed below. Mouse anti-CtIP (IB, 1:50, IF, 1:5; provided by R. Baer21); mouse anti-γH2AX (IB, 1:1,000, IF, 1:2,000; 05-636; Millipore); rabbit anti-γH2AX (IF 1:500; 07-164; Millipore); rabbit anti-γH2AX (IF 1:500; 2,577; Cell signaling) mouse anti-HP1α (IF, 1:1,000; 2HP-1H5-AS; Euromedex); rabbit anti-HP1α (IB, 1:1,000; H-2164, Sigma-Aldrich); mouse anti-HP1β (IB, 1:1,000; IF, 1:2,000; 1MOD-1A9; Euromedex); mouse anti-HP1γ (IB, 1:1,000; IF, 1:4,000; 2MOD-1G6); rabbit anti-human RAD51 (IB, 1:1,000; IF, 1:500; PC130; EMD); rabbit anti-RPA32 (IB, 1:500; 691-P1ABX; Neomarkers); mouse anti-RPA32 (IF, 1:500; 2,175; ABCAM); rabbit anti-RPA32 phospho-Ser4/Ser8 (IB, 1:1,000; A300-245A; Bethyl Laboratories Inc.); mouse anti-tubulin (IB, 1:10,000; T 9026; Sigma-Aldrich. Secondary antibodies used were: for IB, HRP-conjugated affinity-purified sheep anti-mouse or donkey anti-rabbit (1:5000; Jackson ImmunoResearch Laboratories, Inc.); and for IF, goat anti-mouse or anti-rabbit coupled to Alexa Fluor 488 or 594 (1:1,000; Invitrogen).
Immunofluorescence
At indicated times after DSB induction, we performed immunostaining as described previously.39 For in situ cell fractionation, we washed the coverslips twice in PBS, rinsed them with cytoskeleton buffer (CSK) containing 10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2 and a cocktail of protease inhibitor (Complete, EDTA-free tablets; Roche). We subsequently performed a Triton X-100 extraction by incubating the coverslips in CSK containing 0.5% Triton X-100 for 5 min. We fixed cells with 2% wt/vol paraformaldehyde for 20 min at RT. For standard immunostaining, we incubated the coverslips for 10 min with PBS/0.1%T (PBS containing 0.1% vol/vol Triton X-100) to permeabilize the cells. After blocking for 30 min with BSA 2.5%, we incubated the coverslips with the appropriate primary and secondary antibodies for 1 h. We fixed cells with 2% wt/vol paraformaldehyde for 20 min at RT. We mounted samples onto slides in Vectashield mounting medium with DAPI (Vector Laboratories). We used an epifluorescence microscope (Axio Imager Z1; Carl Zeiss, Inc.) piloted with MetaMorph software (Molecular Devices), a 63 × PA/1.4 NA oil objective lens and a chilled charge-coupled device camera (CoolSnap HQ2; Photometrics) for image acquisition.
Sister chromatid exchanges
We incubated siRNA-treated cells for approximately 40 h (to allow cells to replicate twice) with 10 uM BrdU and 10 nM CPT, and we obtained metaphases as previously described.40 We stain the metaphases with Hoechst for 15 min in SSC buffer 2X and then we irradiated the slides slightly submerged in SSC buffer 2X for 15 min. After, we washed the slides for 10 min at 50°C in SSC buffer 2X, we dried them, and we performed a final staining with DAPI for 10 min. We mounted samples onto slides in Vectashield mounting medium (Vector Laboratories).
Supplementary Material
Acknowledgments
We thank Sophie Polo, Vanesa Gottifredi and Jose Luis Bocco for critical reading of the manuscript. We also thank Jeremy Stark for helpful suggestions and for providing us the U2OS SA-GFP cells. We are grateful to the PICT-iBiSA Imaging facility of Centre de Recherche de l’Institut Curie. G.A.’s laboratory is supported by la Ligue Nationale contre le Cancer (Equipe labelisee Ligue 2010), the European Commission Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082), ERC Advanced Grant 2009-AdG_20090506 “Eccentric” and the ANR. G.S. was funded by a fellowship of the Association pour la Recherche sur le Cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23215
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–34. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 4.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 5.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–86. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng W, Ball AR, Jr., Yokomori K. HP1: heterochromatin binding proteins working the genome. Epigenetics. 2010;5:287–92. doi: 10.4161/epi.5.4.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 8.Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, et al. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet. 2011;43:220–7. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 9.Nozawa R-S, Nagao K, Masuda H-T, Iwasaki O, Hirota T, Nozaki N, et al. Human POGZ modulates dissociation of HP1α from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12:719–27. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- 10.Rosnoblet C, Vandamme J, Völkel P, Angrand P-O. Analysis of the human HP1 interactome reveals novel binding partners. Biochem Biophys Res Commun. 2011;413:1–6. doi: 10.1016/j.bbrc.2011.08.059. [DOI] [PubMed] [Google Scholar]
- 11.Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169–74. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Kwon SH, Workman JL. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays. 2011;33:280–9. doi: 10.1002/bies.201000138. [DOI] [PubMed] [Google Scholar]
- 13.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–86. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. Paving the way for H2AX phosphorylation: chromatin changes in the DNA damage response. Cell Cycle. 2009;8:1494–500. doi: 10.4161/cc.8.10.8501. [DOI] [PubMed] [Google Scholar]
- 16.Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A. 2009;75:619–25. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- 17.Baldeyron C, Soria G, Roche D, Cook AJL, Almouzni G. HP1α recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–44. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–42. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:1–12. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–16. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–9. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dialynas GK, Terjung S, Brown JP, Aucott RL, Baron-Luhr B, Singh PB, et al. Plasticity of HP1 proteins in mammalian cells. J Cell Sci. 2007;120:3415–24. doi: 10.1242/jcs.012914. [DOI] [PubMed] [Google Scholar]
- 26.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–72. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011;10:344–8. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, et al. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 2011;25:1807–19. doi: 10.1101/gad.17325211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka Y, Suzuki K, Yamauchi M, Mitsutake N, Yamashita S. Recruitment of the cohesin loading factor NIPBL to DNA double-strand breaks depends on MDC1, RNF168 and HP1γ in human cells. Biochem Biophys Res Commun. 2011;411:762–7. doi: 10.1016/j.bbrc.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowndes NF. The interplay between BRCA1 and 53BP1 influences death, aging, senescence and cancer. DNA Repair (Amst) 2010;9:1112–6. doi: 10.1016/j.dnarep.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) 2012;11:441–8. doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunn A, Bennardo N, Cheng A, Stark JM. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem. 2011;286:42470–82. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45:505–16. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Koning L, Savignoni A, Boumendil C, Rehman H, Asselain B, Sastre-Garau X, et al. Heterochromatin protein 1α: a hallmark of cell proliferation relevant to clinical oncology. EMBO Mol Med. 2009;1:178–91. doi: 10.1002/emmm.200900022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan S, Meng Q, Auborn K, Carter T, Rosen EM. BRCA1 and BRCA2 as molecular targets for phytochemicals indole-3-carbinol and genistein in breast and prostate cancer cells. Br J Cancer. 2006;94:407–26. doi: 10.1038/sj.bjc.6602935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soria G, Belluscio L, van Cappellen WA, Kanaar R, Essers J, Gottifredi V. DNA damage induced Pol eta recruitment takes place independently of the cell cycle phase. Cell Cycle. 2009;8:3340–8. doi: 10.4161/cc.8.20.9836. [DOI] [PubMed] [Google Scholar]
- 39.Soria G, Speroni J, Podhajcer OL, Prives C, Gottifredi V. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. J Cell Sci. 2008;121:3271–82. doi: 10.1242/jcs.027730. [DOI] [PubMed] [Google Scholar]
- 40.Bayani J, Squire JA. Sister chromatid exchange. Curr Protoc Cell Biol 2005; Chapter 22:Unit 22.7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.