Abstract
Helicobacter pylori (H. pylori) is a highly successful human-specific gastric pathogen, infecting over half the world’s population. Virulent H. pylori isolates harbour the cytotoxin-associated genes pathogenicity island (cag-PAI), the majority of which have no known function. In this study, we used cell infection assay and reverse transcriptase PCR, identified that CagL recombinant protein, one of the cag-PAI proteins, induced GES-1 cells to express cytokine IL-8. Then we performed western blot and translocation assay. Our result showed CagL polyclonal antibody counteracted translocation of CagA. This will provide a foundation for the further studies on its biological function.
Keywords: Helicobacter pylori, T4SS, CagA, CagL
Introduction
Helicobacter pylori (H. pylori) is a gram-negative, spiral-shaped, microaerophilic bacterium that colonizes the human gastric mucosa and is recognized as the causative agent of chronic active gastritis, gastric and duodenal ulcers or lymphoma of the MALT (mucosa-associated lymphoid tissue) system, and is a risk factor for the development of adenocarcinoma [1–3]. This microorganism has been categorized as class I carcinogen by the World Health Organization [1]. The pathogenesis of H. pylori mainly depends on the exposure of several bacterial factors, including cytotoxin-associated gene A (CagA), the type IV secretion system (T4SS), vacuolating cytotoxin A, outer inflammatory protein A and several adherence factors, to the host [4–6]. Due to their pivotal role in H. pylori pathogenesis, these factors are currently being intensively studied to elucidate how they induce specific host responses.
Cag-PAI, a 40 kb stretch of DNA, encodes components of a T4SS. This T4SS forms a pilus for the injection of virulence factors into host target cells such as the CagA oncoprotein [7, 8]. This is accomplished by a specialized adhesin of the pilus surface, the CagL, which binds to and activates integrin α5β1 receptor on gastric epithelial cells through an arginine–glycine–aspartate motif. This interaction triggers CagA delivery into target cells as well as activation of focal adhesion kinase and Src [9]. Furthermore CagL is a potential antigen candidate for H. pylori vaccine.
In this study, the cagL from H. pylori has been cloned and expressed, moreover, we have found that the CagL recombinant protein could induce GES-1 cells to express cytokine IL-8 mRNA and participate in translocation of cagA protein. The results will provide a foundation for the further studies on H. pylori pathogenesis.
Materials and Methods
Bacterial Strains and Growth Conditions
Helicobacter pylori strain NCTC 11637 was cultured on agar plates containing 5 % fetal bovine serum (FBS) (BioMerieux) and incubated for 48 h at 37 °C under a microaerophilic atmosphere. Bacteria were harvested in PBS, pH 7.4 H. pylori strains were grown in Brucella broth with 5 % FBS for 24 h, with shaking under a microaerophilic atmosphere. Then they were harvested by centrifugation.
Preparation of DNA Template
Genomic DNA of H. pylori was prepared by the routine phenol–chloroform method and DNase-free Rnase treatment. The obtained DNA was dissolved in TE buffer, and its concentration and purity were determined by ultraviolet spectrophotometry.
Polymerase Chain Reaction
Primers were designed to amplify the sequence of cagL from strain H. pylori NCTC 11637 based on the published sequences of strain H. pylori 26695. The sequences of cagL sense primer with an endonuclease site of BamHI and antisense primer with an endonuclease site of XhoI were 5′-GAT GGATCC GAA GATA TA AC AA G C G GTTT-3′ and 5′-GCCCTCGAGTTTAACAATGATCTTACTTGA-3′ respectively. The parameters for PCR were 94 °C for 5 min, ×1; 94 °C for 30 s, 55 °C for 30 s, 72 °C for 60 s, ×30; and then 72 °C for 10 min, ×1. The results of PCR were observed under UV light after electrophoresis in 10 g/l agarose, pre-stained with ethidium bromide.
Cloning and Sequencing
The amplified target DNA fragment from cagL was cloned into pGEM-T vector (pGEM-T -cagL) by using the T-A Cloning kit according to the manufacturer’s instruction. A professional company (BBST) was responsible for nucleotide sequence analysis of the inserted fragment. The cagL target fragment and PET28a(+) were recovered for ligation. The recombinant expression vector PET28a(+)-cagL was transformed into E. coli BL21DE3, which was named as PET28a(+)-cagL-BL21 (DE3). The target cagL fragment inserted in PET28a(+) plasmid was sequenced again.
Expression and Identification of the Fusion Protein
PET28a (+)-cagL-BL21 was cultured in LB medium at 37 °C and induced by IPTG at concentrations of 1.0 mmol/l. The bacterial were harvested at different time of 2, 3, 4, 5 h. The supernatant and precipitate were separated by centrifugation after the bacterial pallet was ultrasonically broken (300 V, 5 s × 3). The molecular mass and output of CagL were measured by SDS-PAGE. The expressed CagL was collected by Ni–NTA affinity chromatography. The mouse anti-His and HRP-labeling sheep anti-mouse IgG were used as the first and second antibodies, respectively, to identify the CagL fusion protein by Western blot.
Preparation and Identification of the Polyclonal Antibody
Rabbit antiserum was prepared against the CagL purified protein in a 6 weeks old New Zealand White rabbit. The rabbit was immunized with 1 ml of purified protein (0.5 mg/ml) emulsified in 1 ml of Freund’s complete adjuvant and followed immunization with 1 ml of purified protein (0.5 mg/ml) emulsified in 1 ml of Freund’s incomplete adjuvant at an interval of 14 days. After four times of injection, 1 ml of the protein solution was then given intravenously at weekly intervals for a further 4 weeks. The rabbit was bled 1 week after the last immunisation and the serum was collected. ELISA assay was used to detect the titer of the antibody. Antibody was also identified by Western blot.
Cell culture and Infection Experiment
GES-1 were grown in DMEM medium containing 20 % heat-inactivated FBS in a humidified 5 % CO2 atmosphere. Cells were seeded in tissue culture plates before infection. Eighty percent confluent monolayers were washed in PBS and were incubated in serum and antibiotic-free medium. Before infection, H. pylori was neutralized by CagL polyclonal antibody serum at different concentrations of 5, 10, 20 %V/V) and incubated for 4 h at 37 °C under a microaerophilic atmosphere. The bacteria were added to the host cells at a multiplicity of infection of 300 for 6 h. Cultures were maintained at 37 °C under a 5 % CO2 atmosphere. As a control, cells were infected with PBS and non-neutralization H. pylori for the indicated time periods. All experiments were repeated 3–4 times.
Preparation of Cell Lysates and Western Blotting
The infected cells were harvested in lysis buffer(20 mmol/l Tris-hydrochloride (pH, 7.5), 150 mmol/l sodium chloride, 1 % Triton X-100, 1 % NP-40, 3 mmol/l sodium vanadate, 20 mmol/l sodium fluoride, 1 mmol/l phenylmethylsulphonyl fluoride, 10 mg/ml aprotinin, and 10 mg/ml leupeptin) and separated by SDS-PAGE, transferred onto PVDF membranes. To analyze translocation of CagA, a monoclonal CagA antibody (Aalto Bio Reagents Ltd., Dublin, Ireland) and Horseradish peroxidase-conjugated anti-mouse (Amersham, Germany) antibody were used as the first and second antibodies by Western blot and detected with the kit system for ECL (Amersham, Germany). As a control, the anti-β-actin antibody was used.
RNA Isolation and Reverse Transcriptase PCR
After stimulation, total RNA was isolated using the RNAeasy Midi Kit (Qiagen, Germany) as recommended by the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed into single-stranded cDNA with Superscript II RT (Invitrogen, Germany) and oligo(dT)primers. Amplification of IL-8 (forward: 5′-TACTCCAAACCTT TCCACCC-3′, reverse: 5′-CCTACAACAGACCCACACA AT-3′) and GAPDH (forward: 5′-CCACCCATGGCAAATCCATGGC-3′, reverse: 5′-TCTAG ACGGCAGCGGCAGGTCAGGTCCACC-3′) was described before. PCR products were visualized by ethidium bromide staining after agarose gel electrophoresis.
Statistical Analysis
Data were analyzed using Student’s t test and were expressed as mean values of at least 3 independent experiments ± standard deviations. Differences were considered to be statistically significant at P < 0.05.
Results
Construction of Recombinant Plasmid and Restriction Enzyme Confirmation
The fragment of cagL was cloned into pGEM-T vector and expression vector PET28a(+), resulting in recombinant plasmids named pGEM-T-cagL and PET28a(+)-cagL. The recombinant plasmids were all digested by BamHI and XhoI simultaneously. Then digestive products were visualized on 1 % agarose gel electrophoresis.
The pGEM-T-cagL and PET28a(+)-cagL plasmids were identified by PCR, 2 restriction enzymes digestion (BamHI and XhoI), and single restriction enzyme digestion with BamHI, respectively. The results showed that an approximately 650 bp band was amplified by PCR, band approximately 650 bp and 3000, 650 and 5,400 bp were observed following 2 restriction enzymes digestion for pGEM-T-cagL and PET28a(+)-cagL plasmids, respectively, and about 4000, 6000 bp bands were observed by single restriction enzyme digestion for pGEM-T-cagL and PET28a(+)-cagL plasmids, respectively. The sequencing results of both cloning plasmids showed that the band was specific, which was the same as the designed results.
Sequence Analysis
The homologies of nucleotide and putative amino acid sequences of the cloned cagL gene compared with the published cagL sequences were from 96 to 98 % and from 97 to 99 % respectively. The potential antigenic epitopes of the CagL was analyzed by DNASTAR7.0 software. The results showed that this protein has a plurality of structural domain of antigenic activity.
Expression and Identification of Target Fusion Proteins
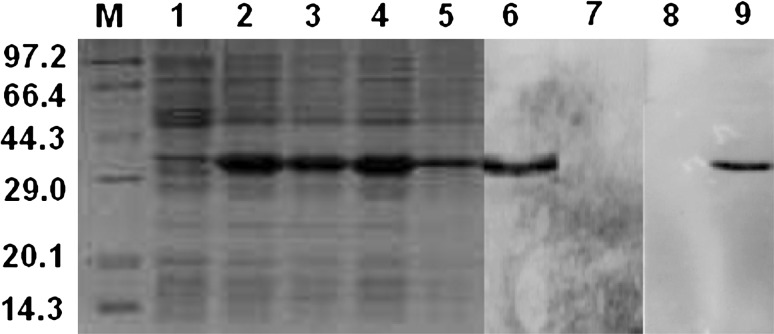
Gel automatic scan analysis showed that the final concentration of IPTG was 1 mmol/l and the expression of CagL rose remarkably after 4 h of induction, which accounted for 48 % of total bacterial proteins. The result of Western blot showed the clearly identifiable band, which was similar to that predicted (Fig. 1).
Fig. 1.
The effect of inducing time on the expression amount of recombinant CagL and analysis of specificity of polyclonal antibody against CagL Expression of CagL was induced by 1 mmol/l IPTG. M protein mark, 1 negative control (PET28a(+) vector), 2–5E. coli BL21(DE3) containing PET28a(+)/cagL recombinant plasmid induced with IPTG after 2–5 h respectively, 6 Western blot analysis of CagL recombinant protein, 7 negative control, 8 Western blot analysis of serum of negative control, 9 Western blot analysis of polyclonal antibody against CagL
Polyclonal Antibody Specifically Recognizes CagL Protein
Six immunizations with the CagL were given to rabbit. Antisera containing CagL antibody was harvested from rabbits 59 days after the primary immunization day. ELISA results showed that the titer of the antisera was 1:320000. Western blot results showed that these antisera were able to recognize recombinant CagL (Fig. 1).
Polyclonal Antibody Counteracts Translocation of CagA
We evaluated whether there was any difference in the levels of CagA translocation into H. pylori-infected GES-1 cells before and after neutralization of polyclonal antibody. Whole-cell lysates were prepared and analyzed by Western blotting to quantify the amount of CagA protein delivered into cells. When H. pylori were pretreated with different concentrations of polyclonal antibody, the level of CagA translocation decreased in a dose-dependent manner (Fig. 2). Compared with nontreated group, there was a significantly smaller amount of CagA translocation. Together, these results suggest that the function activity of CagL is required to mediate the translocation of CagA by the T4SS efficiently.
Fig. 2.
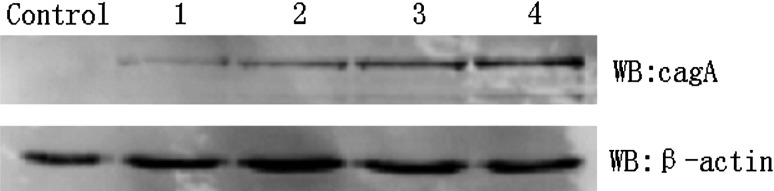
The result of CagA’s translocation after neutralization GES-1 cells were infected with H. pylori strain and H. pylori neutralized by CagL polyclonal antibody for 6 h, respectively. Then, the translocation of CagA was compared between them. Control GES-1 cells without H. pylori treated, 1–3 GES-1 cells co-cultured with H. pylori of CagL polyclonal antibody neutralization (CagL polyclonal antibody density degrade gradually), 4 GES-1 cells co-cultured with H. pylori without neutralization
CagL Induces Expression of IL-8
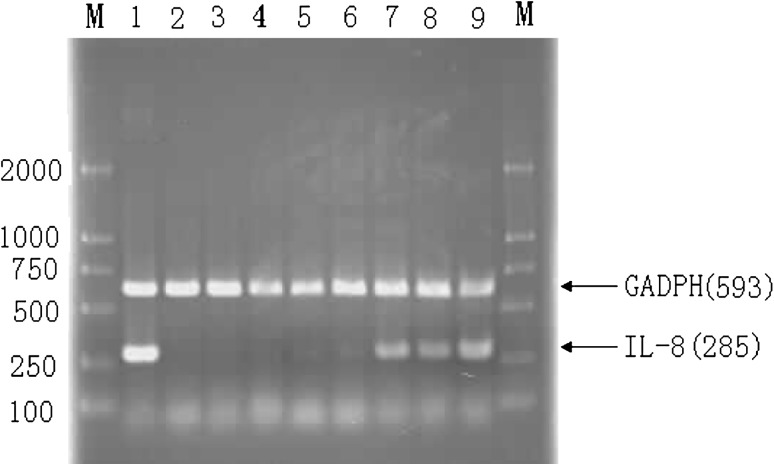
It was previously reported that H. pylori leads to IL-8 induction via the NF-kB signaling pathway in cells. Furthermore, the CagL protein is a specialized adhesin of the pilus surface, which binds to and activates integrin α5β1 receptor on gastric epithelial cells. Thus, we sought to assess whether CagL affects IL-8 expression, we analyzed IL-8 mRNA in non-stimulated and CagL stimulated GES-1 cells. A significantly higher level of IL-8 mRNA was detected in cells stimulated with CagL in a dose-dependent manner (Fig. 3), indicating that CagL can mediate immune response of cells, which may contribute to translocation of CagA.
Fig. 3.
IL-8 mRNA expression in GES-1 cells stimulated with PBS, LPS and recombinant CagL protein. GES-1 and LPS, PBS, different concentrations of recombinant CagL were cultured for 4 h, respectively. M DNA Marker DL2000, 1 Positive control (GES-1 cells co-cultured with LPS), 2 GES-1 cells co-cultured with PBS, 3 GES-1 cells co-cultured with the extractive of bacterium transformed with PET28a(+), 4–9 GES-1 cells co-cultured with different concentrations of recombinant CagL (1, 5, 10, 15, 20 and 30 μg/ml)
Discussion
T4SS are highly versatile secretion machineries that transfer effectors from bacteria to either bacteria of the same or different species, or from bacteria to fungi, plants or mammalian target cells. The assembly of T4SS transporters in the inner and outer membranes of the bacteria [10–15] and the transport of substrates through the assembled T4SS is well documented [16]. However, almost nothing is known about how T4SS substrates are delivered across the cell membrane in their eukaryotic host. It is also unclear whether components of the T4SS specifically recognize their target cell before injecting effector molecules. Indeed, Most T4SS of different pathogens do not translocate their effectors into the culture supernatant [4, 12]. This suggests that functional activation of these T4SS requires a signal from the host cell, for example, the interaction with a specific receptor. Host cell integrins were recently shown to directly interact with the H. pylori CagL and are so far the only T4SS receptor known. Integrins are cell adhesion receptors that mediate cell–cell, cell–extra cellular matrix and cell–pathogen interactions by binding to Arg-Gly-Asp (RGD) motifs [9]. Integrin function is dynamically regulated by processes termed outside-in and inside-out signalling [17]. Similarly, CagL, a protein highly conserved among pathogenic H. pylori strains, is the only cagPAI-encoded gene product that contains an RGD motif which mediates binding of the pilus to integrin α5β1 and is required for injection of CagA [9]. Binding of CagL to integrins induces local membrane ruffling, indicative of a general effect on membrane dynamics which is the first T4SS effect on host cells. After established the T4SS injection needle, H. pylori also affects the actin cytoskeleton, transcriptional responses and cell-to-cell junctions. Therefore, whether the CagL-mediated signalling might facilitate type-IV secretion by altering membrane dynamics is an intriguing proposition to be investigated further.
The CagL-integrin-mediated CagA injection is crucial for deregulation of focal adhesions. However, rearrangement of the actin cytoskeleton leading to cell migration appears to be mainly independent of CagA, but requires a functional T4SS [18–21]. In particular, H. pylori mutants that do not express the CagL are unable to stimulate cell migration, while H. pylori strains that are deficient for CagA still activate motility to a certain extent [18]. This led to the hypothesis that CagL-mediated stimulation of the β1 integrin/FAK/Src pathway is involved in the cytoskeletal rearrangement. The interaction between CagL and integrin may lead to dissociation of the E-cadherin/p120ctn/β-catenin complex from the actin cytoskeleton occurred rapidly by interrupting its binding to α-catenin, which may aberrantly activate p120 and regulates expression of the carcinogenic effector, mmp-7.
In this study, we have cloned and sequenced cagL from H. pylori. The polyclonal antibody against cagL was obtained. We have demonstrated that CagL recombinant protein can induce host cells to express cytokine IL-8 mRNA and CagL polyclonal antibody can inhibit translocation of CagA protein, the date indicate that CagL can mediate immune response of cells and the function activity of CagL is required to mediate the translocation of CagA by the T4SS efficiently. Therefore, CagL might be a novel drug target for combating the severe gastric diseases caused by cagPAI-positive H. pylori strains. In a word, our study provided a foundation for the further studies on cagL biological function and H. pylori pathogenesis.
Acknowledgments
We thank Prof. Seung-chul Baik (Gyeongsang National University College of Medicine, Republic of Korea) for kindly providing us the suicide plasmid pBlue-KM40 and technical assistant for mutant construct of H. pylori. This work was supported by grants from the National Natural Science Funds (81271795) and Research Foundation for Advanced Talents in Jiangsu University (12JDG064).
Footnotes
Hua Wang and Shiteng Huang contributed equally to this work.
References
- 1.Backert S, Schwarz T, Miehlke S, Kirsch C, et al. Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. Infect Immun. 2004;72:1043–1056. doi: 10.1128/IAI.72.2.1043-1056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. Helicobacter pylori and gastroduodenal disease. Annu Rev Med. 1992;43:135–145. doi: 10.1146/annurev.me.43.020192.001031. [DOI] [PubMed] [Google Scholar]
- 3.Sanders MK, Peura DA. Helicobacter pylori-associated diseases. Curr Gastroenterol Rep. 2002;4:448–454. doi: 10.1007/s11894-002-0019-x. [DOI] [PubMed] [Google Scholar]
- 4.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Clyne M, Dolan B, Reeves EP. Bacterial factors that mediate colonization of the stomach and virulence of Helicobacter pylori. FEMS Microbiol Lett. 2007;268:135–143. doi: 10.1111/j.1574-6968.2007.00648.x. [DOI] [PubMed] [Google Scholar]
- 6.Rieder G, Fischer W, Haas R. Interaction of Helicobacter pylori with host cells: function of secreted and translocated molecules. Curr Opin Microbiol. 2005;8:67–73. doi: 10.1016/j.mib.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Odenbreit S, Püls J, Sedlmaier B, Gerland E, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 8.Rohde M, Püls J, Buhrdorf R, Fischer W. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 9.Kwok T, Zabler D, Urman S, Rohde M, et al. Helicobacter pylori exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 10.Backert S, Churin Y, Meyer TF. Helicobacter pylori type IV secretion, host cell signalling and vaccine development. Keio J Med. 2002;51:6–14. doi: 10.2302/kjm.51.supplement2_6. [DOI] [PubMed] [Google Scholar]
- 11.Baron C. From bioremediation to biowarfare: on the impact and mechanism of type IV secretion systems. FEMS Microbiol Lett. 2005;253:163–170. doi: 10.1016/j.femsle.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci A, Rappuoli R. Tyrosine-phosphorylated bacterial proteins: Trojan horses for the host cell. J Exp Med. 2000;191:587–592. doi: 10.1084/jem.191.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schröder G, Lanka E. The mating pair formation systems of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Yeo HJ, Waksman G. Unveiling molecular scaffolds of the type IV secretion system. J Bacteriol. 2004;186:1919–1926. doi: 10.1128/JB.186.7.1919-1926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Al-Ghoul L, Wessler S, Hundertmark T, Kruger S, et al. Analysis of the type IV secretion system-dependent cell motility of Helicobacter pylori-infected epithelial cells. Biochem Biophys Res Commun. 2004;322:860–866. doi: 10.1016/j.bbrc.2004.07.199. [DOI] [PubMed] [Google Scholar]
- 19.Churin Y, Kardalinou E, Meyer TF, Naumann M. Pathogenicity island-dependent activation of Rho GTPases Rac1 and Cdc42 in Helicobacter pylori infection. Mol Microbiol. 2001;40:815–823. doi: 10.1046/j.1365-2958.2001.02443.x. [DOI] [PubMed] [Google Scholar]
- 20.Moese S, Selbach M, Kwok T, Brinkmann V, et al. Helicobacter pylori induces AGS cell motility and elongation via independent signaling pathways. Infect Immun. 2004;72:3646–3649. doi: 10.1128/IAI.72.6.3646-3649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weydig C, Starzinski-Powitz A, Carra G, Löwer J, et al. CagA-independent disruption of adherence junction complexes involves E-cadherin shedding and implies multiple steps in Helicobacter pylori pathogenicity. Exp Cell Res. 2007;313:3459–3471. doi: 10.1016/j.yexcr.2007.07.015. [DOI] [PubMed] [Google Scholar]