Abstract
Objective
Transcriptional coactivator p300 has been shown to play a variety of roles in the transcription process and mutation of p300 has been found in certain types of human cancers. However, the expression dynamics of p300 in breast cancer (BC) and its effect on BC patients’ prognosis are poorly understood.
Methods
In the present study, the methods of tissue microarray and immunohistochemistry (IHC) were used to investigate the protein expression of p300 in BCs. Receiver operating characteristic (ROC) curve analysis, Spearman’s rank correlation, Kaplan-Meier plots and Cox proportional hazards regression model were utilized to analyze the data.
Results
Based on the ROC curve analysis, the cutoff value for p300 high expression was defined when the H score for p300 was more than 105. High expression of p300 could be observed in 105/193 (54.4%) of BCs, in 6/25 (24.0%) of non-malignant breast tissues, respectively (P=0.004). Further correlation analysis showed that high expression of p300 was positively correlated with higher histological grade, advanced clinical stage and tumor recurrence (P<0.05). In univariate survival analysis, a significant association between high expression of p300 and shortened patients’ survival and poor progression-free survival was found (P<0.05). Importantly, p300 expression was evaluated as an independent prognostic factor in multivariate analysis (P<0.05).
Conclusion
Our findings provide a basis for the concept that high expression of p300 in BC may be important in the acquisition of a recurrence phenotype, suggesting that p300 high expression, as examined by IHC, is an independent biomarker for poor prognosis of patients with BC.
Key words: Breast cancer, p300, Tumor recurrence, Prognosis
INTRODUCTION
Breast cancer (BC) is the most common malignancy among women, and accounted for approximately 1.15 million new cases and 411,000 deaths worldwide in 2002[1]. Particularly in the last two decades, incidence and mortality rate of BC have climbed sharply in China[2]. The survival rate for BC patients has increased dramatically due to earlier detection and new treatment protocols in the last decade[3]. Presently, various clinicopathologic factors, such as lymph node status, histological grade, tumor size, vascular invasion, hormone receptor status and HER2 expression are utilized to predict BC prognosis and provide accurate treatment[4]. However, these factors are insufficient and approximately 20% to 30% of BC patients will die from BC within five years of primary diagnosis[5].
p300, a member of the histone acetyltransferase family of transcriptional coactivator, has been found to play a critical role in the transcription process and catalyzes histone acetylation through its histone acetyltransferase activity[6, 7]. Transcriptional coactivator p300 has been indicated to regulate different cellular processes including differentiation, cell-cycle regulation, proliferation, apoptosis and DNA damage response[8]. A role for p300 in tumor suppression has been proposed by the evidence that disturbance of p300 function by viral oncoproteins is essential for the transformation of rodent primary cells[9,10]. However, several studies revealed that transcriptional coactivator p300 is a positive regulator of cancer progression and related to tumorigenesis of various human cancers[11-13]. It has been suggested that the cooperation between CtBP1 and p300 appears to be central in discriminating nuclear receptor repression versus stimulation of genes at early times after hormone exposure in breast cancer cells[14]. Stronger p300 expression was observed in malignant epithelia compared to normal mammary glands in animal models and human breast carcinoma[15]. In addition, Marije et al suggested that p300 is a cofactor highly correlated with p53 accumulation and HIF-1а levels in invasive breast cancer[16].
Up to date, the clinicopathologic/prognostic significance of p300 in BCs is poorly understood. In this study, immunohistochemistry (IHC) was utilized to examine the distribution and frequency of p300 expression in a well-characterized breast cancers prepared by tissue microarray. In order to avoid predetermined cutpoint, receiver operating characteristic (ROC) curve analysis was applied to define the cutoff score for separating p300 highly expressed tumors from p300 low expressed tumors. Subsequently, the clinicopathologic/prognostic significance of p300 expression in BCs was analyzed.
MATERIALS AND METHODS
Patients and Tissue Specimens
In the current study, the paraffin-embedded pathologic specimens from 193 patients with BC were collected from the archives of Department of Pathology, Sun Yat-Sen University Cancer Center, Guangzhou, China, between September 1997 and October 2004. The cases selected were based on distinctive pathologic diagnosis of BC, undergoing primary and curative resection for tumor without preoperative anticancer treatment, availability of resection tissue and follow-up data. The mean age of these patients was 48.1 years. Average follow-up time was 50.57 months (median, 47.97 months; range, 3.73 to 94.77 months). Additional 25 non-malignant breast specimens were obtained from reduction mammoplasties.
Patients whose cause of death remained unknown were excluded from our study. Clinicopathologic characteristics for these patients including age, tumor size, histological grade, clinical stage and relapse were detailed in Table 1. Histological grade was based on the criteria proposed by Elston and Ellis[17]. Tumor stage was defined according to American Joint Committee on Cancer/International Union Against Cancer tumor-node-metastasis (TNM) classification system[18]. Institute Research Medical Ethics Committee of Sun Yat-Sen University granted approval for this study.
Table 1. Correlation between p300 expression and clinicopathologic features in breast cancer patients.
| Indices | All cases | p300 protein |
||
|---|---|---|---|---|
| Low expression | High expression | Pa | ||
| Age at diagnosis (years) | 0.971 | |||
| ≤48.1b | 105 | 48 (45.7%) | 57 (54.3%) | |
| >48.1 | 88 | 40 (45.5%) | 48 (54.5%) | |
| Tumor size (cm) | 0.281 | |||
| ≤3.2c | 126 | 61 (48.4%) | 65 (51.6%) | |
| >3.2 | 67 | 27 (40.3%) | 40 (59.7%) | |
| Histological grade | 0.000 | |||
| Grade I | 30 | 29 (96.7%) | 1 (3.3%) | |
| Grade II | 116 | 48 (41.4%) | 68 (58.6%) | |
| Grade III | 47 | 11 (23.4%) | 36 (76.6%) | |
| Lymph node metastasis | 0.152 | |||
| N0 | 93 | 48 (51.6%) | 45 (48.4%) | |
| N1 | 54 | 25 (46.3%) | 29 (53.7%) | |
| N2 | 23 | 9 (39.1%) | 14 (60.9%) | |
| N3 | 23 | 6 (26.1%) | 17 (73.9%) | |
| Clinical stage | 0.038 | |||
| I | 20 | 13 (65.0%) | 7 (35.0%) | |
| II | 124 | 59 (47.6%) | 65 (52.4%) | |
| III | 49 | 16 (32.7%) | 33 (67.3%) | |
| Recurrence | 0.007 | |||
| Absent | 160 | 80 (50.0%) | 80 (50.0%) | |
| Present | 33 | 8 (24.2%) | 25 (75.8%) | |
aChi-square test; bMean age; cMean size.
Tissue Microarray (TMA) Construction
Tissue microarray was constructed as the method described in our previous study[19]. Briefly, formalin-fixed, paraffin-embedded tissue blocks and the corresponding H&E-stained slides were overlaid for TMA sampling. The slides were reviewed by a senior pathologist (M-Y. C.) to determine and mark out representative tumor areas. Triplicates of 1.0 mm diameter cylinders were punched from representative tumor areas of individual donor tissue block and re-embedded into a recipient paraffin block at defined position, using a tissue arraying instrument (Beecher Instruments, Silver Spring, MD, USA).
Immunohistochemistry (IHC)
The TMA slides were dried overnight at 37°C, deparaffinized in xylene, rehydrated through graded alcohol, immersed in 3% hydrogen peroxide for 20 minutes to block endogenous peroxidase activity, and antigen-retrieved by pressure cooking for 3 minutes in ethylenediamine tetraacetic acid (EDTA) buffer (pH=8.0). Then the slides were preincubated with 10% normal goat serum at room temperature for 30 minutes to reduce nonspecific reaction. Subsequently, the slides were incubated with mouse monoclonal anti-p300 (Abcam, Cambridge, MA, USA) at a concentration of 3ng/ml for 2 hours at room temperature. The slides were sequentially incubated with a secondary antibody (Envision; Dako, Glostrup, Denmark) for 1 hour at room temperature, and stained with 3,3-diaminobenzidine (DAB). Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. A negative control was obtained by replacing the primary antibody with a normal murine IgG. Known immunostaining positive slides were used as positive controls.
IHC Evaluation
Immunoreactivity for p300 protein was evaluated in semi-quantitative method as described previously[20]. Each TMA spot was assigned an intensity score from 0–3 (I0, I1–3) and proportion of tumor cells for that intensity over the total number of tumor cells was recorded as 5% increments from a range of 0–100 (P0, P1–3). A final H score (range 0–300) was achieved by adding the sum of scores obtained for each intensity and proportion of area stained (H score = I1XP1+ I2XP2+I3XP3).
Selection of Cutoff Score
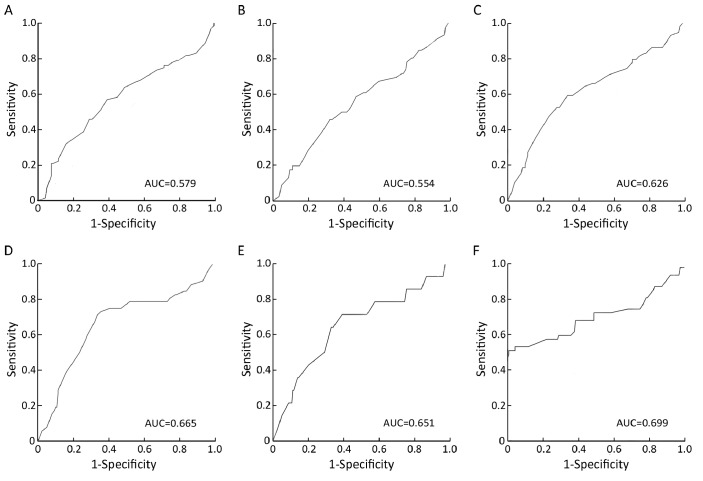
ROC curve analysis was utilized to determine cutoff value for separating tumors with p300 high expression from tumors with p300 low expression by using the 0,1-criterion[21]. At the p300 H score, the sensitivity and specificity for each outcome under study was plotted, thus generating various ROC curves (Figure 1). The score was selected as the cutoff value, which was closest to the point with both maximum sensitivity and specificity. Tumors designated as “low expression” for p300 were those with scores below or equal to the cutoff value, while “high expression” tumors were those with scores above the value. In order to perform ROC curve analysis, the clinicopathologic features were dichotomized: tumor size (≤3.2 cm, or >3.2 cm), histological grade (Grade I+Grade II or Grade III), lymph node metastasis (N0+N1 or N3), clinical stage (I+II or III), relapse (absence or presence) and survival status (death due to BC or censored).
Figure 1.
ROC curve analysis was used to determine the cutoff value for high expression of p300 protein. The sensitivity and specificity for each outcome were plotted: tumor size (P=0.067, A), lymph node status (P=0.270, B), clinical stage (P=0.006, C), tumor recurrence (P<0.001, D), survival status (P=0.060, E), and histological grade (P<0.001, F).
Statistical Analysis
Statistical analysis was performed by using the SPSS statistical software package (standard version 13.0; SPSS, Chicago, IL, USA). ROC curve analysis was applied to determine the cutoff score for high expression of p300. The correlation between p300 expression and clinicopathological features of BC patients was evaluated by χ2-test. Univariate and multivariate survival analyses were performed using the Cox proportional hazards regression model. Survival curves were obtained with the Kaplan-Meier method. Predictive accuracy was quantified using the Harrell concordance index. Differences were considered significant if the P-value from a two-tailed test was <0.05.
RESULTS
Expression of p300 in BC and Non-malignant Breast Tissues by IHC
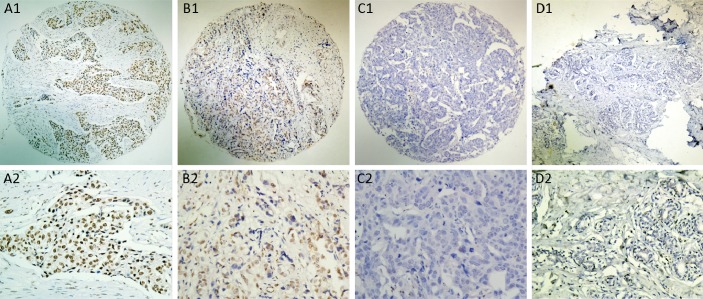
For p300 IHC staining in BCs and non-malignant breast tissues, immunoreactivity was predominantly observed in the nuclei within tumor cells, though occasionally yellowish brown granules could also be seen in the cytoplasm (Figure 2). p300 expression could be evaluated informatively in 193 BCs by the TMA constructed previously. The non-informative TMA samples included samples with too few tumor cells (<300 cells per case) and lost samples. H scores for p300 in BC ranged from 0 to 300 (Figure 2A-2C). According to ROC curve analysis, H score for p300 above the cutoff value 105 was defined as high expression, while below or equal to the cutoff value was considered as low expression. In this study, high expression of p300 could be detected in 105/193 (54.4%) of BCs, in 6/25 (24.0%) of non-malignant breast tissues, respectively (P=0.004, Fisher’s exact test).
Figure 2.
The protein expression patterns of p300 in BC and non-malignant breast tissues. A: High expression of p300 was observed in a BC (case 86), in which more than 80% tumor cells revealed high immunostaining of p300 in nuclei (A1: ×40, A2: ×200). B: A BC case (case 54) demonstrated low expression of p300, in which less than 50% of tumor cells showed moderate immunoreactivity of p300 protein in nuclei (B1: ×40, B2: ×200). C: Nearly negative expression of p300 protein was demonstrated in a BC case (case 35, C1: ×40, C2: ×200). D: The non-malignant breast tissue (case 12) showed nearly negative expression of p300 protein in nuclei (D1: ×40, D2: ×200).
Selection of Cutoff Value for p300 Expression
The ROC curves for each clinicopathological parameter (Figure 1) clearly show the point on the curve closest to (0.0, 1.0) which maximizes both sensitivity and specificity for the outcome as described in our previous study[19]. Tumors with scores above the obtained cutoff value were considered as p300 high expression leading to the greatest number of tumors classified based on clinical outcome presence or absence. The corresponding area under the curve (AUC, 95% CI) were collected and listed in Figure 1. The H score for p300 high expression was determined to be more than 105.
Association of p300 Expression with BC Patients’ Clinicopathologic Features
The high or low expression rates of p300 in BCs with respect to several standard clinicopathologic features are listed in Table 1. The p300 expression rate was higher in patients with higher histological grade (P<0.0001), advanced stage (P=0.038) and tumor recurrence (P=0.007, Table 1). There was no significant correlation between p300 expression and other clinicopathologic features, such as patient age (≤48.1 years vs >48.1 years), tumor size and lymph node status (P>0.05, Table 1).
Relationship between Clinicopathologic Features, p300 Expression, and BC Patients’ Survival: Univariate Survival Analysis
In order to confirm the representativeness of the BC cohort in our study, we analyzed well-established prognostic features of patients’ survival. Kaplan-Meier analysis demonstrated a significant impact of well-known clinicopathologic prognostic parameters, such as tumor size, lymph node status, clinical stage, and tumor relapse (P<0.05) on patients’ overall survival and progression-free survival (Table 2). Assessment of survival in total BCs showed that high expression of p300 was correlated with adverse overall survival of BC patients (P=0.016, Table 2, Figure 3A). In addition, high expression of p300 in BCs was evaluated to correlate closely with poor progression-free survival (P =0.011, Table 2, Figure 3B).
Table 2. Clinicopathologic features and p300 expression for prognosis of 193 patients with breast cancer by univariate survival analysis (log-rank test).
| Indices | All cases | OS (months) |
PFS (months) |
||
|---|---|---|---|---|---|
| Median | P | Median | P | ||
| Age at diagnosis (years) | 0.439 | 0.386 | |||
| ≤48.1a | 105 | 44.77 | 40.40 | ||
| >48.1 | 88 | 50.86 | 49.22 | ||
| Tumor size (cm) | 0.020 | 0.023 | |||
| ≤3.2b | 126 | 51.92 | 49.07 | ||
| >3.2 | 67 | 36.43 | 32.37 | ||
| Histological grade | 0.747 | 0.445 | |||
| Grade I | 30 | 64.67 | 54.85 | ||
| Grade II | 116 | 47.42 | 44.27 | ||
| Grade III | 47 | 45.10 | 41.70 | ||
| Lymph node metastasis | 0.000 | 0.000 | |||
| N0 | 93 | 54.75 | 53.37 | ||
| N1 | 54 | 53.70 | 49.47 | ||
| N2 | 23 | 47.30 | 46.37 | ||
| N3 | 23 | 33.47 | 24.20 | ||
| Clinical stage | 0.004 | 0.000 | |||
| I | 20 | 52.34 | 49.40 | ||
| II | 124 | 45.22 | 45.22 | ||
| III | 49 | 45.10 | 33.53 | ||
| Relapse | 0.000 | ||||
| Absent | 160 | 51.05 | |||
| Present | 33 | 32.07 | |||
| p300 expression | 0.016 | 0.011 | |||
| Low | 88 | 47.80 | 47.42 | ||
| High | 105 | 45.10 | 44.77 | ||
aMean age; bMean size; OS indicates overall survival; PFS indicates progression-free survival.
Figure 3.
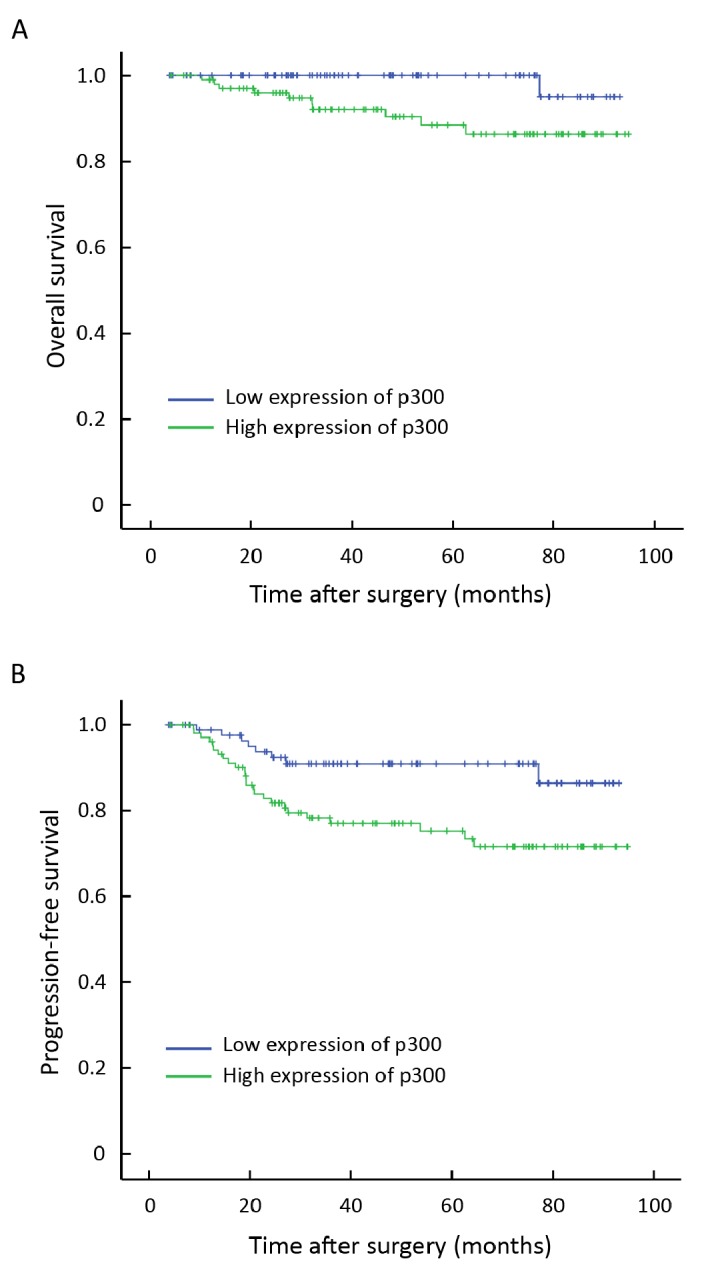
Survival curves for 193 BC patients according to p300 expression status (log-rank test). A: Overall survival, probability of survival of all patients with BC: low expression, n=88; high expression, n=105. B: Progression-free survival, probability of survival of all patients with BC: low expression, n=88; high expression, n=105.
Independent Prognostic Factors of BC: Multivariate Cox Regression Analysis
Since features observed to have a prognostic influence by univariate analysis may covariate, p300 expression and those clinicopathologic variables that were significant in univariate analysis (i.e. tumor size, lymph node status, clinical stage, and tumor recurrence) were further examined in multivariate analysis. Results showed that high expression of p300 was an independent prognostic factor for poor patient overall survival (hazard ratio, 3.369; 95%CI, 1.593-7.762, P=0.021; Table 3) and progression-free survival (hazard ratio, 2.046; 95%CI, 1.312-4.585, P=0.017; Table 3). Of the other features, lymph node status was evaluated as well an independent prognostic factor for patients’ overall survival and progression-free survival (P<0.05, Table 3).
Table 3. Multivariate Cox regression analysis for patient survival.
| Indices | Overall survival |
Progression-free survival |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Tumor sizea | 0.566 | 0.161-1.985 | 0.374 | 1.541 | 0.752-3.158 | 0.237 |
| Lymph node metastasisb | 3.657 | 1.644-8.136 | 0.001 | 2.194 | 1.438-3.348 | 0.000 |
| Clinical stagec | 0.578 | 0.096-3.490 | 0.550 | 0.965 | 0.402-2.317 | 0.936 |
| Relapsed | 4.117 | 1.873-9.049 | 0.001 | |||
| p300 expressione | 3.369 | 1.593-7.762 | 0.021 | 2.046 | 1.312-4.585 | 0.017 |
a≤3.2 cm vs >3.2 cm; bN0 vs N1 vs N2 vs N3; cStage I vs Stage II vs Stage III; dAbsent vs Present; eLow expression vs high expression; HR: Hazard ratio; CI: confidence interval.
DISCUSSION
Transcriptional coactivator p300 has the potential to participate in a variety of cellular functions, such as cell proliferation and differentiation, senescence and apoptosis[8]. Recently, several studies have documented an involvement of p300 in oncogenic processes, such as lung, colon, and prostate cancers and leukemia[22-26]. However, the status of p300 protein and its potential prognostic impact on BC patients have not been explored. In the present study, we examined the expression levels of p300 protein in BC tissues and non-malignant breast tissues by high-through TMA and IHC. Our results demonstrated that high expression of p300 was more frequently observed in BC tissues when compared to that in the non-malignant breast tissues. The expression of p300 in non-malignant breast tissues was either absent or at low levels. In contrast, high expression of p300 was frequently observed in large number of our BC tissues. Previous studies also described that mutation in p300 gene, accompanied by loss of the other allele, was observed in certain types of tumors, including colorectal, gastric and breast cancers[9, 10]. In addition, higher expression of p300 was observed in malignant epithelia compared to normal mammary glands in breast carcinoma tissues[15]. These findings provide evidence that the up-regulation of p300 expression may play an important role in tumorigenic process of BC.
To assess the significance of p300 protein in BC and avoid predetermined arbitrary cutpoint, ROC curve analysis was employed to determine cutoff value for p300 expression as described in our previous study[19]. Further correlation analysis revealed that high expression of p300 in BCs was correlated with higher histological grade, advanced clinical stage and tumor recurrence. Importantly, high expression of p300 was a strong and independent predictor of shortened overall survival as evidenced by univariate and multivariate analysis. In addition, high expression of p300 in BCs was evaluated to correlate significantly with poor progression- free survival by univariate and multivariate analysis. Our findings in this study suggest that high expression of p300 in BC may facilitate an increased tumor recurrence and/or worse prognosis of this tumor. Previous study also suggested that the cooperation between CtBP1 and p300 was central in discriminating nuclear receptor repression versus stimulation of genes at early times in BC cells after hormone exposure[14]. Thus, the examination of p300 expression by IHC could be used as an additional tool in identifying those patients at risk of BC progression; p300 expression analysis may also be useful in optimizing individual BC therapy management: favoring a more aggressive regimen in tumors with a high expression of p300.
Although several characteristics of p300 suggested that the protein might serve as a tumor suppressor, other studies reported an important role of p300 protein in oncogenic processes[8,15]. In prostate cancer, p300 expression was shown to be linked to cell proliferation and identified as a predictor of progression of this cancer[23]. In colon carcinoma, overexpression of p300 was an indicator of poor prognosis[22]. Moreover, p300 mRNA levels were observed to correlate with lymph node status in BC[26]. However, p300 protein levels did not show significant correlations with tumor grade or nodal positivity in other studies[27, 28]. In the present study, we did observe that high expression of p300 was associated with an aggressive feature of BC and was a strong and independent predictor of shorter cancer-specific survival. Considering that the mechanism by which coactivator p300 promotes gene transcription may vary among gene targets, it is not very difficult for us to understand that the function of p300 and its underling mechanism(s) to impact cancer progression may lead to this discrepancy.
Since advanced pTNM stage and histological grade are the best-established risk factors for the prognosis of patients with BC, these two parameters, based on specific clinicopathologic features and extent of disease, may have reached their limits in providing critical information influencing patient prognosis and treatment strategies. Furthermore, outcome of patients with same stage following surgery is substantially different and such large discrepancy has not been explored. Thus, there is a need for new objective strategies that can effectively distinguish between patients with favorable and unfavorable prognosis. In this study, our results support the ideas that p300 expression, as examined by IHC, can identify patients with BC that may show aggressive clinical course and poor outcome.
Our findings provide a basis for the concept that high expression of p300 may play an important role in the acquisition of a recurrence phenotype in BC, suggesting that the expression of p300, as examined by IHC, will be an important independent biomarker for shortened survival time of BC patients.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55:74-108 [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Ji R, Li J, et al. Correlation effect of EGFR and CXCR4 and CCR7 chemokine receptors in predicting breast cancer metastasis and prognosis. J Exp Clin Cancer Res 2010; 29:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry DA, Cronin KA, Plevritis SK, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med 2005; 353:1784-92 [DOI] [PubMed] [Google Scholar]
- 4.Goldhirsch A, Wood WC, Gelber RD, et al. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol 2007; 18:1133-44 [DOI] [PubMed] [Google Scholar]
- 5.Coleman MP, Quaresma M, Berrino F, et al. : Cancer survival in five continents: a worldwide population-based study (CONCORD). Lancet Oncol 2008; 9:730-56 [DOI] [PubMed] [Google Scholar]
- 6.Kundu TK, Palhan VB, Wang Z, et al. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol Cell 2000; 6:551-61 [DOI] [PubMed] [Google Scholar]
- 7.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem 2001; 276:13505-8 [DOI] [PubMed] [Google Scholar]
- 8.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev 2000; 14:1553-77 [PubMed] [Google Scholar]
- 9.Muraoka M, Konishi M, Kikuchi-Yanoshita R, et al. p300 gene alterations in colorectal and gastric carcinomas. Oncogene 1996; 12:1565-9 [PubMed] [Google Scholar]
- 10.Gayther SA, Batley SJ, Linger L, et al. : Mutations truncating the EP300 acetylase in human cancers. Nat Genet 2000; 24:300-3 [DOI] [PubMed] [Google Scholar]
- 11.Fan S, Ma YX, Wang C, et al. : p300 Modulates the BRCA1 inhibition of estrogen receptor activity. Cancer Res 2002; 62:141-51 [PubMed] [Google Scholar]
- 12.Bandyopadhyay D, Okan NA, Bales E, et al. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res 2002; 62:6231-9 [PubMed] [Google Scholar]
- 13.Li M, Luo RZ, Chen JW, et al. High expression of transcriptional coactivator p300 correlates with aggressive features and poor prognosis of hepatocellular carcinoma. J Transl Med 2011; 9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol Cell Biol 2009; 29:1749-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fermento ME, Gandini NA, Lang CA, et al. Intracellular distribution of p300 and its differential recruitment to aggresomes in breast cancer. Exp Mol Pathol 2010; 88:256-64 [DOI] [PubMed] [Google Scholar]
- 16.Vleugel MM, Shvarts D, van der Wall E, et al. p300 and p53 levels determine activation of HIF-1 downstream targets in invasive breast cancer. Hum Pathol 2006; 37:1085-92 [DOI] [PubMed] [Google Scholar]
- 17.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991; 19:403-10 [DOI] [PubMed] [Google Scholar]
- 18.Wittekind C. 2010 TNM system: on the 7th edition of TNM classification of malignant tumors. Pathologe 2010; 31:331-2. [DOI] [PubMed] [Google Scholar]
- 19.Cai MY, Zhang B, He WP, et al. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci 2010; 101:1543-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abubaker J, Bavi P, Al-Haqawi W, et al. PIK3CA alterations in Middle Eastern ovarian cancers. Mol Cancer 2009; 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlobec I, Steele R, Terracciano L, et al. Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol 2007; 60:1112-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishihama K, Yamakawa M, Semba S, et al. Expression of HDAC1 and CBP/p300 in human colorectal carcinomas. J Clin Pathol 2007; 60:1205-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debes JD, Sebo TJ, Lohse CM, et al. p300 in prostate cancer proliferation and progression. Cancer Res 2003; 63:7638-40 [PubMed] [Google Scholar]
- 24.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. Roles of CREB-binding protein (CBP)/p300 in respiratory epithelium tumorigenesis. Cell Res 2007; 17:324-32 [DOI] [PubMed] [Google Scholar]
- 25.Borrow J, Stanton VP, Jr, Andresen JM, et al. : The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 1996; 14:33-41 [DOI] [PubMed] [Google Scholar]
- 26.Kurebayashi J, Otsuki T, Kunisue H, et al. Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res 2000; 6:512-518 [PubMed] [Google Scholar]
- 27.De-Carvalho MC, Chimelli LM, Quirico-Santos T. Modulation of fibronectin expression in the central nervous system of Lewis rats with experimental autoimmune encephalomyelitis. Braz J Med Biol Res 1999; 32:583-92 [DOI] [PubMed] [Google Scholar]
- 28.Hudelist G, Czerwenka K, Kubista E, et al. Expression of sex steroid receptors and their co-factors in normal and malignant breast tissue: AIB1 is a carcinoma-specific co-activator. Breast Cancer Res Treat 2003; 78:193-204 [DOI] [PubMed] [Google Scholar]