Abstract
Modern clinical treatments of childhood acute lymphoblastic leukemia (ALL) employ enzyme-based methods for depletion of blood asparagine in combination with standard chemotherapeutic agents. Significant side effects can arise in these protocols and, in many cases, patients develop drug-resistant forms of the disease that may be correlated with up-regulation of the enzyme glutamine-dependent asparagine synthetase (ASNS). Though the precise molecular mechanisms that result in the appearance of drug resistance are the subject of active study, potent ASNS inhibitors may have clinical utility in treating asparaginase-resistant forms of childhood ALL. This review provides an overview of recent developments in our understanding of (a) the structure and catalytic mechanism of ASNS, and (b) the role that ASNS may play in the onset of drug-resistant childhood ALL. In addition, the first successful, mechanism-based efforts to prepare and characterize nanomolar ASNS inhibitors are discussed, together with the implications of these studies for future efforts to develop useful drugs.
Keywords: amino acids, asparaginase resistance, drug discovery, enzyme inhibitors, leukemia
INTRODUCTION
The remarkable inverse correlation between the susceptibility of leukemia cells to drug therapy and their capacity for intracellular asparagine biosynthesis was first described almost forty years ago (1). This observation provides a rationale for the current widespread use of L-asparaginase (ASNase) in chemotherapeutic protocols for treating childhood acute lymphoblastic leukemia (ALL) and some forms of acute myeloblastic leukemia (AML) (2–7). The molecular basis for the therapeutic utility of ASNase remains ill defined (8), although it is believed that normal and malignant lymphocytes depend on the uptake of asparagine from circulating plasma for growth (9). ASNase therefore likely exerts its effects indirectly by depleting asparagine in the blood, which is followed by an efflux of cytoplasmic asparagine from leukemic blasts (1). The use of Escherichia coli ASNase as a single agent leads to nearly complete remission in 40% to 60% of cases of ALL (10, 11), and, in combination with vincristine and prednisone, increases the remission rate up to 95% in cases of childhood ALL. Unfortunately, three factors limit the clinical utility of ASNase in cancer therapy (8, 12). First, the treatment produces a wide variety of side effects, including immunosuppression and pancreatitis (13, 14). Second, 10% to 12% of patients who achieve remission suffer a relapse with tumors that are resistant to further ASNase therapy (5, 14–16). Finally, ASNase administration may enhance the growth of resistant tumors and increase their metastatic activity (10, 17). The molecular basis of ASNase resistance, which is a major clinical problem, remains poorly understood despite a significant amount of ongoing research (8, 18). Because ASNase sensitivity in tumors cannot yet be predicted reliably, the major use of this enzyme remains confined to the treatment of childhood ALL, despite estimates that 5–10% of all solid tumors may be sensitive to therapies based upon the depletion of blood asparagine (16).
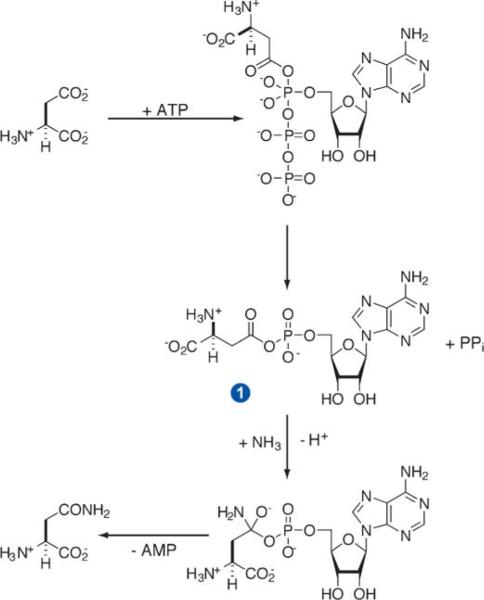
Human asparagine synthetase (ASNS) catalyzes the biosynthesis of l-asparagine from l-aspartate in an ATP-dependent reaction for which l-glutamine is the nitrogen source under physiological conditions (Scheme 1) (19). Recent work has demonstrated the importance of ASNS overexpression in conferring ASNase resistance in cell lines (20), and several lines of evidence suggest that inhibiting ASNS activity represents a viable strategy for treating ASNase-resistant leukemias in the clinic (1, 8, 21). Early large-scale screening studies employing a range of substrate and product analogs failed, however, to identify potent and selective ASNS inhibitors (22, 23). In part, the failure of these efforts reflected a lack of detailed knowledge concerning the structure of human ASNS and its functional role in cellular metabolism. Considerable progress has been made in all of these areas over the past few years, and several recent advances have set the stage for the identification and characterization of the first nanomolar inhibitors of human ASNS (24). This review provides an overview of recent developments in understanding (a) the structure and catalytic mechanism of ASNS, and (b) the role that ASNS plays in the onset of drug-resistant ALL. In addition, the molecular principles that have been employed to discover and characterize two potent classes of ASNS inhibitors are outlined, together with the implications of these studies for future efforts to develop clinically useful drugs against ASNase-resistant leukemia. Additional information, especially pertaining to the catalytic mechanism of ASNS, can be found in a previous review (19).
Scheme 1.

Overall transformation catalyzed by glutamine-dependent ASNS, showing the β-aspartyl-AMP intermediate. Ammonia may replace glutamine as a nitrogen source in vitro.
STRUCTURE AND MECHANISM OF ASPARAGINE SYNTHETASE
The use of rational, structure-based methods for obtaining small molecule enzyme inhibitors requires a detailed understanding of critical transition states and reaction interme diates that are bound tightly by the enzyme during catalysis. In addition, the identification of active site residues that have functional roles in the catalytic mechanism provides a set of “contact points” that can be exploited in inhibitor design because mutation of these residues will generally lead to loss of function in the protein. We therefore begin this review with an overview of ASNS structure and catalytic mechanism.
Structure of Asparagine Synthetase
The detailed kinetic and structural characterization of ASNS from mammalian sources historically proved to be difficult because of both reported low abundance and instability of the native enzyme during purification (25–29). In addition, only small amounts of recombinant, wild-type human ASNS could be obtained in a variety of early expression systems, the enzyme being purified to homogeneity using an affinity chromatography protocol (30–32). Access to recombinant human enzyme, albeit in small quantities, in these experiments did permit limited insights into properties such as substrate specificity (31, 32). As a result, until the very recent development of an efficient expression system for human ASNS (vide infra) (33), procedures for obtaining large amounts of the glutamine-dependent ASNS (AS-B) encoded by the asnB gene in Escherichia coli (34) were essential to detailed investigations of the structure and mechanism of the enzyme.
The C1A mutant of AS-B, in which the N-terminal cysteine residue is substituted by alanine, exhibits no glutamine-dependent activity (19) but retains substantial affinity for l-glutamine (KD of approximately 6 μM). Therefore this AS-B mutant could be crystallized as its ternary complex with glutamine and AMP, and the crystal structure of the complex determined to a resolution of 2.0Å (Figure 1a) (35). As expected on the basis of sequence alignment (36), and experimental studies of bovine ASNS using monoclonal antibodies (37), this structure showed that glutamine-dependent ASNS is composed of two distinct domains, each containing a separate catalytic site. The N-terminal domain of the enzyme has a tertiary structure that is observed in members of the N-terminal amidohydrolase (Ntn) enzyme superfamily (38, 39), which mediates a number of biologically important hydrolytic reactions (40–42). The N-terminal active site therefore catalyzes glutamine hydrolysis to yield glutamate and ammonia, and glutamine-dependent ASNS is classified as a Class II, or Ntn, amidotransferase (43). Other Ntn amidotransferases possessing structurally homologous glutamine-hydrolyzing domains include glutamine 5′-phosphoribosyl-1-pyrophosphate amidotransferase (GPATase) (43, 44) and glutamine fructose-6-phosphate amidotransferase (GFAT) (45, 46), which both play important functions in cellular metabolism. The mechanism of ASNS-catalyzed glutamine hydrolysis is discussed in detail elsewhere (19, 43). The complex three-dimensional fold of the C-terminal domain of AS-B is similar to that observed in ATP pyrophosphatases (47), an enzyme superfamily that includes guanosine-5′-monophosphate synthetase (GMPS) (48), argininosuccinate synthetase (49), ATP sulfurylase (50), carbapenam synthetase (51), β-lactam synthetase (BLS) (52, 53), and ThiI (54). Because all ATP pyrophosphatases convert ATP to AMP, the C-terminal active site of ASNS likely catalyzes activation of the side-chain carboxylate of aspartate to form an electrophilic intermediate, β-aspartyl-AMP (βAspAMP) 1, and inorganic pyrophosphate (PPi) (Scheme 2) (28, 55). As observed in other glutamine-dependent amidotransferases (44, 46, 56–61), the two active sites of AS-B are linked by a solvent-inaccessible, intramolecular “tunnel” that is sufficiently wide to allow passage of an ammonia molecule (Figure 1b) (62). Glutamine-dependent asparagine production is therefore accomplished using ammonia as a common intermediate to couple the two “half-reactions” carried out in the independent active sites of the enzyme. Thus, after being released in, and channeled from, the glutaminase site, a molecule of ammonia attacks bound βAspAMP 1 to give asparagine and AMP via a tetrahedral intermediate (Scheme 2).
Figure 1.
(a) Cartoon representation of the structure of the Cys-1-Ala mutant of Escherichia coli AS-B complexed with glutamine (blue space-filling model) and AMP (green space-filling model) showing the domain organization of the enzyme (35). Helices and β-strands are shown in yellow and red, respectively. The final 40 C-terminal residues are not observed in the crystal structure, presumably due to their disordered conformation in the absence of bound aspartate. (b) Cartoon showing the putative pathway by which ammonia (light blue spheres) travels between the glutaminase (top) and the synthetase (bottom) active sites in AS-B. The side chains of residues defining the ammonia tunnel that are variable and conserved throughout the family of known asparagine synthetases are colored green and red, respectively. Bound glutamine (top) and AMP (bottom) are rendered as gray-white “ball-and-stick” models. Reprinted from (63), Copyright 2003, with permission from Elsevier.
Scheme 2.
Hypothetical mechanism for the ASNS-catalyzed formation of βAspAMP 1 and its subsequent reaction with ammonia to form asparagine and AMP.
Kinetic Mechanism of Asparagine Synthetase
Given that all Ntn amidotransferases share a common N-terminal glutaminase domain, the development of selective ASNS inhibitors requires that they be targeted against the C-terminal, synthetase active site. As a result, recent work in this area has been concerned with obtaining a detailed understanding of (a) the steady-state kinetic mechanism for glutamine-dependent asparagine production (55, 63, 64), and (b) critical intermolecular interactions that play a role in mediating the synthesis of βAspAMP and its subsequent reaction with ammonia (Scheme 2). Although the general properties of ASNS from a variety of sources appear very similar, many different kinetic mechanisms have been proposed for their glutamine-dependent synthetase activity (65–67). As a result, it is only recently that a consensus has begun to emerge on the general details of substrate binding order and product release, particularly because kinetic analysis of ASNS-catalyzed asparagine formation is complicated by the high glutaminase activity of the enzyme. Steady-state experiments on AS-B (55), in combination with isotope partitioning techniques (68) and measurements of product stoichiometry, gave rise to an initial hypothesis that βAspAMP formation commits the enzyme to asparagine synthesis. Such a proposal is analogous to the kinetic mechanism of certain aminoacyl tRNA synthetases in which an acyl-AMP intermediate is synthesized prior to interaction of the enzyme with uncharged tRNA (69, 70). The question of when glutamine or ammonia binds to AS-B was experimentally unresolved in these studies, however (55), and it was proposed that glutamine (and therefore ammonia) binds to the E.βAspAMP complex after PPi release. If correct, this order of catalytic events implies that βAspAMP must be stabilized within the synthetase site so as to prevent futile ATP hydrolysis prior to binding of the nitrogen source for asparagine production. On the other hand, numerical simulations (71) showed that the original kinetic model for AS-B (55) was inconsistent with the dependence of the glutamate/asparagine ratio upon glutamine concentration at saturating levels of ATP and aspartate. In addition, and as observed for other glutamine-dependent amidotransferases (36, 72), ASNS does not catalyze ATP/PPi exchange (55). A new kinetic model has been recently developed that appears consistent with all known experimental data for the bacterial enzyme (63). Hence, in addition to suggesting that βAspAMP formation commits the enzyme to asparagine synthesis, implying that ammonia transfer from the N-terminal domain through the intramolecular tunnel is totally efficient, the model provides evidence for the hypothesis that glutamine can bind to the E.Asp.ATP ternary complex to yield a quaternary complex from which glutamate and ammonia can be released. Thus, coordination of catalytic activity in the two active sites of AS-B during glutamine-dependent asparagine synthesis appears to be remarkably small prior to βAspAMP formation, which is in sharp contrast to the coupling of glutaminase and synthetase activities seen for other Ntn glutamine-dependent amidotransferases (73–75). The lack of ATP/PPi exchange has been rationalized by assuming that PPi is released as the final product from the enzyme. Although this conclusion is supported by crystal structures of the BLS/CMA-AMP/PPi and BLS/DGPC/PPi complexes (53), and structural models for the AS-B/βAspAMP/PPi complex (vide infra) in which PPi is located deep in the active site cleft covered by the other ligands, PPi release is reported to occur prior to glutamine binding for Vibrio cholerae ASNS (64). Whatever the timing of product release, the latest kinetic model (63) supports the hypothesis that ASNS must bind βAspAMP with high affinity, raising the possibility that stable analogs of this intermediate might be potent ASNS inhibitors. In addition, it seems likely that the enzyme also stabilizes the transition state for addition of ammonia to βAspAMP 1 (Scheme 2). As a consequence, compounds that mimic this transition state may also have significant potential as clinically useful drugs (76, 77).
ASPARAGINE SYNTHETASE AND DRUG-RESISTANT LEUKEMIA
Children with acute lymphoblastic leukemia (ALL) are treated with a multidrug regimen that includes the enzyme L-asparaginase (ASNase). Although modern therapeutic protocols lead to remission rates of greater than 80%, relapse and drug resistance remain a problem. Consequently, the relationship between the expression of the ASNS and development of ASNase resistance is of interest from the viewpoint of both metabolic regulatory mechanisms and development of new therapeutic strategies.
Asparagine Synthetase Expression and the Cell Cycle
Basilico and colleagues determined that ASNS could complement temperature-sensitive hamster BHK ts11 cells, which are specifically blocked in progression through the G1 phase of the cell cycle when grown at the nonpermissive temperature (78, 79). Those authors showed that because of a point mutation in the ASNS gene, at the nonpermissive temperature, the BHK ts11 cells produce an inactive enzyme (79). This loss of ASNS activity leads to cell cycle arrest, as a consequence of a depletion of cellular asparagine, and a corresponding increase in ASNS mRNA due to regulatory mechanisms described below. Both of these effects could be reversed by the addition of exogenous asparagine to the cells maintained at the nonpermissive temperature. A link between asparagine content and the cell cycle is illustrated by several additional observations. For example, ASNS mRNA expression was substantially increased during the G1 phase of the cell cycle by refeeding serum-deprived Balb/c 3T3 cells. After serum repletion, ASNS mRNA was induced, but the addition of asparagine to the culture medium could prevent the induction (80). Hongo et al. (81) showed that ASNS activity is induced during lymphocyte activation by phytohemagglutinin, and the increase in activity coincides with the rate of DNA synthesis. Likewise, Cirafici (82) showed that thyroid-stimulating hormone treatment of quiescent rat thyroid cells causes entry into the S phase and there was a concurrent increase in ASNS mRNA content. Collectively, these observations suggest that ASNS expression is linked to cell growth and ASNS mRNA content is controlled in accordance with changes in the cell cycle.
Amino Acid Control of ASNS Gene Transcription
Amino acids are known to modulate a number of fundamental processes in mammalian cells, especially with regard to the “central dogma” of DNA to RNA to protein (83). Although circulating amino acids and intracellular protein turnover both act to buffer cells from variations in dietary protein/amino acid intake, fluctuations in the intracellular levels of individual amino acids do occur in response to diet, disease, and metabolic status. In this context, amino acids serve as signal transduction messengers to transmit the nutritional status of the entire organism to individual cells. One of the mechanisms by which amino acids mediate this signaling is altered transcription for specific genes via a signal transduction process referred to as the amino acid response (AAR) pathway. Detection of a limiting amount of any single amino acid has been linked to a ribosome-associated kinase, GCN2, that binds and, therefore, monitors the level of uncharged tRNAs (84–86). Starvation-activated GCN2 phosphorylates the eukaryotic initiation factor eIF-2α and, as a consequence, global translation initiation is suppressed. However, certain mRNAs that contain short upstream open reading frames exhibit increased translation under amino acid limiting conditions. An example in yeast is the transcription factor GCN4 (84), which has been reported to alter the transcription rate of several hundred genes in response to amino acid limitation (87). The mammalian counterpart to yeast GCN4 appears to be the basic leucine zipper (bZIP) transcription factor ATF4. The translation of pre-existing ATF4 mRNA is rapidly increased following amino acid deprivation (88–90), and ATF4 protein has been shown to mediate the increased transcription of AAR pathway target genes, including ASNS (91, 92).
Gong et al. (93) were the first to determine that ASNS mRNA content increased in cells deprived of amino acids, and subsequently, Hutson & Kilberg (94) also demonstrated increased ASNS mRNA content following total amino acid deprivation or depletion of a single essential amino acid. In this circumstance, the definition of “essential amino acid” must be considered in the context of individual cell types rather than the entire organism. Guerrini et al. (95) analyzed the ASNS gene and determined that an amino acid response element (AARE) was present in the promoter. In vivo footprinting by Barbosa-Tessmann et al. (96) identified five protein binding sites within the ASNS proximal promoter region that contribute to nutrient control of the human ASNS gene, three GC-rich sequences (GC-I, GC-II, and GC-III) and two sequences, originally labeled sites V and VI, and later renamed Nutrient Sensing Response Elements (NSRE-1, NRSE-2). The GC-rich sites are necessary to maintain the basal transcription rate and to permit maximal activation of the ASNS gene by the AAR pathway (97). Interestingly, expression of either Sp1 or Sp3 in Drosophila SL2 cells supported basal ASNS promoter activity, but only Sp3 expression permitted the starvation-induced ASNS-driven transcription. The location of the NSRE-1 footprint overlapped with the sequence identified by Guerrini et al. (95). Mutagenesis confirmed the presence of a second AARE, NSRE-2 (5′-GTTACA-3′, nt −48 to −43), positioned eleven nucleotides downstream of NSRE-1. Mutagenesis studies have documented that these two elements must be aligned on the same side of the DNA helix and only one turn away from each other (98), presumably to permit protein-protein interactions that occur between the transcription factors that bind to these two sites. Barbosa-Tessmann et al. (96, 99) demonstrated that both NSRE-1 and NSRE-2 were required for activation of the human ASNS gene following activation by either the AAR pathway or an ER stress pathway referred to as the unfolded protein response (UPR). Experimentally, the UPR pathway is often activated by conditions that perturb ER calcium levels (thapsigargin treatment) or conditions that cause accumulation of misfolded glycoproteins (glucose starvation or tunicamycin treatment). Electrophoresis mobility shift analysis (EMSA) experiments revealed increased nuclear protein binding to the ASNS NSRE-1 binding site (5′-TGATGAAAC-3′, nt −68 to −60) when extracts from either amino acid (AAR pathway) or glucose-deprived (UPR pathway) cells were tested (96). This broader nutrient detecting capability is the reason that the ASNS sites are referred to as nutrient-sensing response elements (NSRE), rather than solely as an AARE.
The binding proteins for the NSRE-2 site have not yet been identified, whereas progress has been made in identifying those for NSRE-1. In vitro binding analysis by EMSA revealed ATF4 binding to the NSRE-1 sequence that was increased when nuclear extracts from either histidine-deprived (AAR pathway) or glucose-deprived (UPR pathway) cells were tested (91). Consistent with translational control of ATF4 production following activation of the AAR pathway, the inhibition of protein synthesis blocked the starvation-dependent enhancement of ATF4/NSRE-1 complex formation. ATF4-dependent regulation of ASNS expression in vivo was suggested by the observation that ASNS promoter-driven transcription was induced by ATF4 overexpression (91, 92). Subsequently, chromatin immunoprecipitation (ChIP) analysis documented that there is increased ATF4 binding to the ASNS promoter in vivo following amino acid deprivation (92).
A survey of bZIP transcription factors by both EMSA experiments that ATF3 (91, 100) and C/EBPβ (101) also exhibited affinity for the ASNS NSRE-1 site and transient expression showed that both can modulate transcription driven by the ASNS promoter. Transient expression studies using combinations of ATF4, ATF3, and C/EBPβ then implied that ATF3 serves as the primary antagonist to ATF4 function (92, 102). Consistent with this hypothesis, Fawcett et al. (103) used nuclear extracts from arsenite-treated cells in EMSA studies to show a transient increase in ATF4 binding activity with affinity for a cis-element in the human C/EBP homology protein (CHOP) promoter referred to as a C/EBP-ATF composite site. ATF4 binding activity peaked at 2 h after arsenite exposure, but that activity was subsequently replaced at about 6 h by elevated ATF3 binding activity, a time at which the elevated transcription rate from the gene was declining back toward the basal rate. Those authors also used overexpression studies to document that elevated ATF4 activates the CHOP gene through the C/EBP-ATF composite site and that increased ATF3 production antagonizes the ATF4 function. The CHOP sequence (5′-TGATGCAAT-3′) that Fawcett et al. (103) identified is only two nucleotides different than the ASNS NSRE-1 site, which is also a C/EBP-ATF composite site. Chen et al. (92) extended the observations of Fawcett et al. (103) by showing through ChIP analysis that a similar sequence of events, i.e., activation via ATF4 and subsequent antagonism by ATF3, can be observed in vivo at the ASNS NSRE-1 site.
Based on the in vivo ChIP analysis, Chen et al. (92) have proposed a working model for ASNS transcription that describes two distinct phases in response to amino acid limitation (Figure 2). Phase I encompasses the first 4 h after amino acid withdrawal and phase II covers the time from 4–24 h. Within 30 min of amino acid depletion, translational control of ATF4 mRNA results in increased de novo synthesis and subsequent binding of ATF4 to the NSRE-1 site (92). In parallel with ATF4 binding, acetylation of histones H3 and H4 is increased and the general transcription factors TBP and TFIIB, as well as RNA polymerase II, are recruited to the promoter. It is assumed that the ATF4 complex requires a coactivator and/or other bridging proteins to the general transcription machinery, which are shown as gray modules in the model (Figure 2). During phase I of amino acid deprivation, a low level of C/EBPβ is constitutively bound to the ASNS promoter, whereas in phase II, C/EBPβ de novo synthesis and subsequent binding increases at a time when the transcription rate has peaked. Likewise, the synthesis and action of ATF3 also increases during this period (92). It is proposed that ATF3 and C/EBPβ act in concert to suppress, but not to completely reverse, the increased ASNS transcription during phase II, such that even out to 24 h of amino acid deprivation, transcription remains elevated relative to the status of the gene in amino acid-complete medium (92).
Figure 2.
A working model for control of the asparagine synthetase (ASNS) gene by the AAR or UPR pathways (92). Transcription factors shown in color have been localized to the ASNS promoter by chromatin immunoprecipitation analysis. Unidentified or putative components are shown in gray. Transcription from the ASNS gene reaches its highest rate at 1–4 h (phase I) following nutrient stress. ASNS transcription is still elevated relative to the “fed” state between 4–24 h (phase II) following nutrient stress, but the rate is reduced.
Asparagine Synthetase Expression and Asparaginase Therapy of Childhood ALL
Cancer cells exhibit rapid growth and cell division, and therefore have an increased nutritional need. Consequently, therapies have been developed to take advantage of this dependency on circulating nutrients in those cases where critical enzymes are either not expressed at sufficient levels or can be selectively inhibited. As mentioned above, childhood ALL is treated using a combination of chemotherapeutic drugs that include the enzyme ASNase (8, 13, 104, 105). The logic of ASNase therapy is that by delivering ASNase to the bloodstream plasma asparagine is quickly depleted causing a rapid efflux of cellular asparagine, which is also destroyed, and thus, the cells of the entire body are depleted of asparagine. Most cells express sufficient ASNS to counteract this asparagine starvation and survive, but in general, childhood ALL cells express ASNS at a low level, and therefore, treatment with ASNase is extremely effective in blocking growth of this particular form of leukemia. The bacterially derived ASNase enzymes currently used clinically also have an inherent glutaminase activity that is about 2% to 3% of the ASNase activity (19). As a result, because l-glutamine is the nitrogen donor for the ASNS-catalyzed reaction (see above), the depletion of this amino acid may also play a role in ASNase action (92).
Hutson et al. (108) showed that exposure to ASNase of a cultured human ALL-derived cell line, MOLT-4, can result in selection for a population of ALL cells that are drug resistant (107). Whether or not these resistant cells exist within the parental population or if they are induced by ASNase treatment is not yet known. Aslanian & Kilberg (109) documented that many adaptive changes occur in the ASNase-resistant cells to alter amino acid transport and metabolism (Figure 3). Collectively, these changes appear to support increased asparagine biosynthesis by increasing the intracellular levels of the ASNS substrates aspartate and glutamine, and by enhancing uptake of asparagine and glutamine via secondary active Na+-dependent transporters (109). For example, the enzymatic activity of glutamine synthetase is increased, presumably by a post-transcriptional mechanism, as glutamine synthetase mRNA is not changed. The ASNase-resistant cells also exhibit reduced efflux of asparagine via Na+-independent transporters. These drug-resistant MOLT-4 cells express elevated levels of ASNS mRNA and protein (108, 109), which Aslanian et al. (20) demonstrated remain elevated, even after removal of the ASNase from the culture medium for 6 weeks or more. In a result consistent with these observations it was also established that ASNase resistance, analyzed by cell growth or apoptosis following a drug challenge, was not completely reversible following long-term culture in the absence of ASNase (20). The relationship between ASNase resistance and increased ASNS expression had been noted before in non-ALL cell types (15, 110, 111), leading to the assumption that resistance to this particular drug was the result of a compensatory increase in ASNS expression. However, it was possible that elevated ASNS was a consequence of ASNase resistance rather than the cause. To establish which of these hypotheses was true, Aslanian et al. (20) showed that over-expression of ASNS in the ASNase-sensitive parental MOLT-4 cells caused these cells to acquire the ASNase-resistant phenotype without drug selection, thus proving that elevated ASNS levels alone are sufficient to generate drug resistance.
Figure 3.

Selected metabolic changes that take place in ASNase-resistant MOLT-4 leukemia cells (123). Treatment with ASNase causes a rapid degradation of extracellular asparagine (Asn) and a subsequent depletion of intracellular Asn. Compensatory changes, shown with red arrows, include increases in: transcription from the asparagine synthetase gene, glutamine synthetase activity (post-transcriptional), and active glutamine transport. Conversely, there is a decrease, shown in yellow, in Asn efflux through Na+-independent exchange. There is little or no aspartate uptake by these cells, so synthesis via transamination may play a role in supplying this substrate for the ASNS-catalyzed reaction.
Despite these cell culture studies, microarray analyses and qPCR quantification of ASNS mRNA have illustrated that the relationship between AS expression and the manifestation of ASNase resistance in primary cells of childhood ALL patients is more complicated. For example, Holleman et al. (112) showed that in vitro resistance to ASNase was correlated with elevated ASNS expression, as assayed by array analysis of primary isolates of ALL cells. In contrast, Fine et al. (113) observed a similar correlation between high expression of ASNS and increased ASNase resistance in a collection of 16 different ALL-derived cultured cell lines, but that relationship did not hold true when primary ALL cells were tested in vitro for their ASNS mRNA content and sensitivity to ASNase. Among the ALL genetic subtypes, those cells with a t(12;21) chromosomal translocation, which leads to synthesis of a TEL/AML1 fusion protein (114), or those with hyper-diploidy, are more sensitive to ASNase (115, 116). Stams et al. (117) hypothesized that TEL/AML1(+) cells are more sensitive to ASNase because the fusion protein, which functions as a transcriptional repressor, may inhibit the expression of ASNS. Surprisingly, they observed that the TEL/AML1(+) cells contained a fivefold higher amount of ASNS mRNA compared to TEL/AML1(−) cells, a result consistent with the data of Krejci et al. (118). Furthermore, Stams et al. (117) discovered that in the TEL/AML1(+) cells there was no correlation between ASNS mRNA content and the in vitro sensitivity to ASNase. Interestingly, those authors went on to show that even though the TEL/AML1(−) ALL cells are less sensitive to ASNase, there is an inverse correlation between ASNS mRNA expression and drug sensitivity in vitro (119). Given that 78% of childhood ALL patients are TEL/AML1(−) (120), it is important to gain a further understanding of the relationship between ASNS expression and ASNase action. Collectively, the results obtained with patient samples show that we still do not fully understand the mechanisms of ASNase action and the development of drug resistance.
SYNTHESIS AND CHARACTERIZATION OF ASPARAGINE SYNTHETASE INHIBITORS
The need for cell-permeable, small molecules capable of inhibiting cellular ASNS in a potent and highly selective manner was recognized almost 40 years ago by the pioneers who developed L-ASNase-based protocols for the treatment of ALL (1). Even so, efforts to obtain such compounds were sporadic, perhaps because a lack of definitive evidence to support the hypothesis that ASNS overexpression really was a primary cause of drug-resistant ALL. As is evident from the previous section of this review, dissecting the cellular mechanisms that underpin the onset of refractory leukemia cells remains a complicated problem that has still not clarified this question. The increased knowledge of ASNS structure and mechanism has rekindled interest in the synthesis of transition state analogs, which might not only be clinically useful compounds but also tools for establishing the relevance of ASNS overexpression to drug resistance.
Evidence Supporting the Clinical Utility of Asparagine Synthetase Inhibitors
In the absence of small molecule inhibitors, preliminary evidence supporting the hypothesis that specific inhibitors of human ASNS have potential clinical utility in treating ASNase-resistant ALL and related leukemias was provided by electroporation studies involving delivery of anti-ASNS monoclonal antibodies to ASNase-resistant mouse L5178Y D10/R cells (21). Parental L5178 cells, derived from a murine leukemia cell line (121), have nearly undetectable ASNS activity and therefore their growth is dependent on the presence of asparagine in the external medium. In contrast, an ASNase-resistant subclone (D10/R) did not require extracellular asparagine because there was a high level of ASNS-catalyzed, intracellular asparagine production. Two mouse monoclonals (3F3 and 2B4) raised against native, bovine ASNS (122), were tested for their ability to inhibit the enzymatic activity of cellular ASNS using extracts from the L5178Y D10/R cells (21). Although both antibodies were able to recognize and bind to ASNS, only the 3F3 monoclonal could inhibit asparagine production in the cell extracts. This monoclonal antibody was therefore electroporated into the ASNase-resistant D10/R cells to evaluate whether suppressing ASNS activity could slow growth in the absence of extracellular asparagine. After establishing that there was no intracellular degradation of the heavy chains of the electroporated antibodies, cells that incorporated the antibody 3F3 were shown to require exogenous asparagine for growth. These experiments suggested that the effect of 3F3 was specifically caused by blocking the action of ASNS, its target enzyme (21). Subsequent controls demonstrated that both ASNase-sensitive and -resistant L5178Y cells contained immunoreactive material, and that the observations on the electroporated DR10/Y cells were not associated with genomic alterations in the form of translocations, gene amplification, or increased P-glycoprotein.
Using Structural Homology and Chemical Constraints to Model the Synthetase Active Site
Unfortunately for efforts to employ rational, structure-based strategies to discover potent, selective ASNS inhibitors that resemble βAspAMP or the transition state for ammonia addition, disorder in the C-terminal domain of the C1A/Gln/AMP complex does not permit observation of two loop regions (Ala-250 to Leu-267 and Cys-422 to Ala-426) and the final forty C-terminal residues of the enzyme by X-ray crystallography (35). Insight into the structure(s) of the ASNS synthetase site during catalytic turnover, and the potential role of residues involved in aspartate activation and ammonia addition, has been obtained, however, by recognizing the striking chemical similarity of the reactions employed to synthesize asparagine from aspartic acid and a functionalized β-lactam from the adenylated form of N2-(carboxyethyl)-l-arginine (CEA) 2 (Scheme 3) (123, 124). Thus, the enzyme BLS, which is found in the biosynthetic pathway leading to the important secondary metabolite clavulanic acid (125), activates its substrate by forming an AMP derivative in which the carbonyl group is attacked intramolecularly by a nitrogen nucleophile. In agreement with modern models for enzyme evolution (126, 127), the common ancestry of ASNS and BLS is evident from the degree of similarity in their three-dimensional structures (Figure 4), and the catalytic machinery for substrate activation by adenylation is highly conserved in both enzymes. For example, the contiguous segment SGGLDSS in AS-B, which binds to PPi during substrate adenylation, is also present in BLS. Using the crystal structure of a ternary complex between BLS, CEA and AMPCPP (an unreactive ATP analog), MgATP could be positioned within the synthetase site of AS-B. This initial model was then used to build a quaternary complex (C1A/Gln/Asp/MgATP) in which aspartate was placed in an analogous location within the protein to that observed for CEA in the BLS/CEA/AMPCPP crystal structure. This modeling exercise was facilitated by assuming two chemical constraints. First, by analogy to the chemistry of aminoacyl tRNA synthetases, attack of the side-chain carboxylate of bound aspartate on the α-phosphate of ATP likely proceeds via in-line attack with inversion at the phosphorus atom (Scheme 2) (128). Second, studies with non-natural, conformationally constrained aspartate analogs, prepared using the diastereoselective alkylation of l-aspartate diester derivatives (129, 130), demonstrated that the polar functional groups are located on one face in the bound conformation of aspartate (131). After refinement using constrained molecular dynamics (MD) simulations (132) in combination with simulated annealing algorithms (133), and subsequent energy minimization, the resulting model was then modified by connecting the side-chain carboxylate of aspartate to the α-phosphate of the ATP moiety to form the βAspAMP intermediate. In this model, PPi was positioned over the pyrophosphatase loop region of the enzyme so that (a) the hydrogen bonding interactions with the enzyme corresponded to those observed in the GMPS/AMP/PPi crystal structure (48), and (b) noncovalent interactions with the active site Mg2+ ion were maintained. Further MD refinement and energy minimization then gave a computational model for the AS-B/βAspAMP/PPi complex (Figure 5), which was checked for consistency with the results of mutagenesis and kinetic studies employing AS-B (19, 34, 131, 134–136). Importantly for inhibitor development, we were able to identify completely conserved residues having side chains positioned within 5 Å of either βAspAMP or PPi. It is therefore likely that these residues have key functional roles in aspartate binding, acyl-AMP formation, and catalyzing the attack of ammonia on βAspAMP, although experiments testing this hypothesis have not yet been published.
Scheme 3.

A comparison of the chemical reactions catalyzed by BLS and ASNS. (a) Aspartate is activated by adenylation to yield βAspAMP, which undergoes intermolecular attack by ammonia to yield asparagine. (b) CEA, the substrate for BLS, is activated as an acyl-AMP derivative, which then undergoes intramolecular nitrogen attack to give the β-lactam product.
Figure 4.
Cartoon representations of the X-ray crystal structures of (a) BLS complexed to its substrate (CPK-colored space-filling model) and AMPCPP (green space-filling model) in the C-terminal domain (52) and (b) Escherichia coli AS-B complexed with glutamine (not shown) and AMP (green space-filling model) (35). The striking structural conservation in both enzymes suggests either a common ancestor or recruitment of AS to provide BLS during the evolution of clavulanic acid biosynthesis. In both structures, helices and β-strands are shown in yellow and red, respectively, whereas water molecules are represented by red spheres.
Figure 5.
Views of the working molecular model for the AS-B/βAspAMP/PPi complex. (a) Interactions between PPi and residues defining the ATP pyrophosphatase loop motif. (b) Protein/βAspAMP interactions involving conserved residues Glu-352, Tyr-357, Lys-376, Asp-384, Arg-387, and Lys-449 that illustrate recognition of the α-amino and α-carboxylate groups present in the βAspAMP intermediate 1. (Color coding: C – gray; H – white; O – red; N - blue; P – purple. Light blue lines show locations of putative hydrogen bonds.)
Sulfonamide Derivatives as Inhibitors of Human Asparagine Synthetase
Early studies employing a variety of glutamine, ATP, and aspartic acid analogs failed to identify compounds exhibiting significant in vivo or in vitro activity against ASNS (22, 23). Aromatic sulfonylfluoride derivatives of glutamine and asparagine also failed to inhibit ASNS (137), and observations suggesting that cineoles, a group of monoterpenes isolated from the essential oils of aromatic plants (138), inhibit asparagine production in various plant tissues (139) have proven difficult to reproduce (140). Progress, however, (a) obtaining multi-milligram amounts of human ASNS from a baculovirus-based expression system (33), (b) determining the high-resolution, three-dimensional structure of the bacterial enzyme AS-B (35), and (c) recognizing that aminoacyl tRNA synthetases are targets for new antibacterial and antifungal drugs (141–143) stimulated renewed efforts by Richards and coworkers to obtain potent, small molecule ASNS inhibitors (24, 144). For example, the antibiotic mupirocin 3 (Figure 6) exerts its action by selectively inhibiting prokaryotic isoleucyl-tRNA synthetases (145, 146), and phosmidosine 4 (Figure 6) (147, 148) likely inhibits prolyltRNA synthetase by effectively mimicking the acyladenylate intermediate formed by this enzyme. These findings therefore resulted in the development of efficient synthetic routes for preparing chemically stable analogs of acyladenylates (148–150) in the expectation that such compounds will exhibit antibacterial activity coupled with low mammalian toxicity (151, 152). In light of these studies (145–150), β-asparaginyladenylate 5 (Figure 6) was synthesized (153) and proved to have micro-molar affinity for the bacterial enzyme ASB (S.K. Boehlein, J.-Q. Wang, Y. Ding, N.G.J. Richards & S.M. Schuster, unpublished results). The relatively lengthy synthetic route to obtain this compound and the presence of a charged phosphoamidate functional group argue against its likely clinical utility. In addition, the coupling reaction was insufficiently robust for use in constructing molecular libraries that could be assayed for ASNS inhibitory activity using modern high-throughput screening methods (154–157).
Figure 6.
Structures of compounds 3–11 (see text for details).
Given that N-acylsulfonamide derivatives are high-affinity, slow-binding inhibitors of isoleucyl-tRNA synthetases (158–160) and have “drug-like” physical properties (161, 162), a sulfonamide analog of the βAspAMP intermediate (6) (Figure 6) was also prepared and characterized for its ability to inhibit recombinant, wild-type human ASNS (24). These in vitro experiments clearly showed that sulfonamide derivative 6 is a slow-onset, tight-binding inhibitor that interacts with the free enzyme, and revealed the importance of carboxyl and amino groups in the recognition and binding of this class of βAspAMP analog. These observations are consistent with the computationally-derived model of the ASB/βAspAMP/PPi complex (Figure 3b). Detailed steady-state kinetic analysis using standard models (163) revealed the KI* value for 6 to be approximately 700 nM, making this compound the first submicromolar ASNS inhibitor identified in the literature.
A Nanomolar Inhibitor of Human Asparagine Synthetase
Perhaps the most important progress toward the discovery of potent inhibitors of glutamine-dependent ASNS has been obtained from recent work employing the adenylated sulfoximine 7 (Figure 6). The conceptual impetus for synthesizing and characterizing this compound was the observation that methionine sulfoximine 8 (Figure 6) is a potent inhibitor of glutamine synthetase (164). Thus, this natural product binds to the enzyme in the glutamate site (165, 166) where it undergoes phosphorylation to yield the sulfoximine phosphate derivative 9 (167), which was proposed to be an analog for the transition state formed during the attack of ammonia on γ-glutamylphosphate 10 (Figure 6) (168). This interesting example of a mechanism-based inhibitor therefore stimulated the synthesis of the analogous sulfoximine 11 (Figure 6) from homocysteine with the goal of obtaining a potent inhibitor of ammonia-dependent asparagine synthetases (169), a class of enzymes that, to date, have only been found in prokaryotes (170, 171). On this point, we note that there is no evolutionary relationship of glutamine- and ammonia-dependent asparagine synthetases, the latter family being related to aminoacyl tRNA synthetases (172, 173). Incubation of 11 with AS-A, the ammonia-dependent ASNS isolated from Escherichia coli (174, 175), however, showed that this compound was a poor (millimolar) inhibitor because it was not adenylated when bound within the synthetase active site. A synthetic route to the required adenylated sulfoximine was therefore developed by Hiratake and coworkers, yielding the target compound as a mixture of diastereoisomers (7a and 7b) (Figure 6) at the sulfur center (169). Not only did this mixture prove to exhibit potent inhibition of AS-A (169) but it was also shown to be a slow-onset inhibitor of the glutamine-dependent AS-B when the bacterial enzyme was undergoing catalytic turnover (144). Although, at first sight, it may be difficult to see why this compound should resemble the transition state for ammonia addition to the βAspAMP intermediate, semiempirical quantum mechanical (QM) calculations (N.G.J. Richards, unpublished results), using the PM3 parameter set (176), clearly show that the methyl group in the sulfoximine moiety is electron deficient, giving a significant positive electrostatic potential about the three hydrogen atoms. The methyl group therefore resembles the steric and electrostatic properties of ammonia in the transition state for CN bond formation (Figure 7). This analysis is supported by the observation that an aspartate side chain is positioned close to the methyl group of 7b in the X-ray crystal structure of its complex with AS-A (169).
Figure 7.
Computational visualization of transition state mimicry by the adenylated sulfoximine functional group. (a) Semiempirical (PM3) transition state for the attack of ammonia on a computational model of the acyladenylate intermediate formed in the ASNS synthetase site. (b) Graphical representation of the transition state structure showing the isodensity surface color-coded by the electrostatic potential. Note that the hydrogen atoms in ammonia gain substantial positive charge (red). (c) Optimized (PM3) structure for a model phosphorylated sulfoximine. (d) Graphical representation of the phosphorylated sulfoximine in (c) showing the isodensity surface color-coded by the electrostatic potential. Note that the hydrogen atoms of the methyl group have similar steric and electrostatic properties to those on ammonia in the transition state shown in (b). [Color coding in (a) and (c): C – gray; H – white; O – red; N – blue; P – purple; S – yellow. Dotted lines show noncovalent, electrostatic interactions.]
Incubation of 7, as the diastereoisomeric mixture 7a and 7b, with recombinant, wild-type human ASNS at various ATP concentrations, showed this compound to be a potent, slow-onset, tight-binding inhibitor of the enzyme in vitro. Use of standard mathematical models (163) gave values for the constants KI and KI* of 285 nM and 2.5 nM, respectively, making the adenylated sulfoximine the most potent inhibitor of the human enzyme reported to date (J.A. Gutierrez & N.G.J. Richards, unpublished results). In common with many other tight-binding inhibitors (177, 178), the adenylated sulfoximine 7 binds to the free form of the human enzyme, and reactivation and mass spectrometric experiments have demonstrated that it does not covalently modify ASNS. Efforts to cocrystallize the recombinant, human ASNS with 7 (albeit using the diastereoisomeric mixture) have failed, however, to yield crystals suitable for high-resolution X-ray structure determination. Because all kinetic experiments were performed using a 1:1 mixture of the sulfoximine diastereoisomers 7a and 7b (Figure 6), and given that residues defining the synthetase site are completely conserved throughout the glutamine-dependent ASNS family, computational docking methods (179, 180) were used to position each of diastereoisomeric adenylated sulfoximine derivatives into the working model of the AS-B/βAspAMP/PPi complex (N.G.J. Richards, unpublished results). After constrained MD-based refinement, as described above, only diastereoisomer 7b could be positioned such that its methyl substituent was positioned correctly relative to the C-terminal end of the ammonia channel (Figure 8). In addition, the side chain of Glu-348 (AS-B numbering), which is conserved in the primary structures of known asparagine synthetases, was positioned adjacent to the electron-deficient methyl group of the docked inhibitor in the model, suggesting that this residue might function as a general base to deprotonate ammonia in the transition state leading to the tetrahedral adduct (Scheme 2). In efforts to validate the computational model of the AS-B/7b active site complex, site-specific AS-B mutants were prepared in which this residue was replaced by aspartate (E348D) and alanine (E348A), and assayed for their ability to form asparagine and PPi. In contrast to wild-type AS-B, which forms these products in a 1:1 ratio (63, 89), the E348D AS-B mutant formed two molecules of PPi per asparagine. In addition, the E348A AS-B mutant loses the ability to catalyze asparagine synthesis when glutamine or ammonia is the nitrogen source, although it retains significant levels of ATP pyrophosphatase activity (J.A. Gutierrez & N.G.J. Richards, unpublished results). Both of these observations are consistent with the role deduced for Glu-348 on the basis of the AS-B/inhibitor model, suggesting that this in silico structure may prove a useful asset in the discovery of simpler, and more cell permeable, structures with activity as ASNS inhibitors using computational (181) and other structure-based strategies (182, 183).
Figure 8.
Computational model of the adenylated sulfoximine ASNS inhibitor 7b docked into the synthetase site of AS-B. For ease of comprehension, only selected protein residues are shown, which are all conserved within glutamine-dependent ASNS. Note the distance between the Glu-348 side-chain carboxylate (pink) and the methyl group (cyan) of the ASNS inhibitor. The methyl substituent mimics the location of ammonia in the transition state formed during attack on the βAspAMP intermediate 1. Residues shown in green (Leu-232 and Ser-346) define the C-terminal end of the channel through which ammonia enters the synthetase active site after being released in the N-terminal glutaminase domain. (Color coding: C – gray; H – white; O – red; N – blue; P – purple; S – yellow.)
PERSPECTIVES
A number of valuable initiatives can be envisaged, some of which are ongoing, to further our knowledge of the relationship between ASNS expression and ASNase sensitivity or resistance in childhood ALL. First, to complement the microarray analysis of patient samples at the time of diagnosis, similar analyses performed on individual patients during the course of therapy should be of considerable interest. However, this proposed approach has some technical limitations because over the course of therapy in most patients there will be a significant decline in the number of lymphoblastic cells, and consequently the background of ASNS content in other cell types within the sample will complicate any quantitative analysis and interpretation of the data. It must also be recognized that ALL therapy involves a multidrug regimen, so that the effect of ASNase treatment is difficult to analyze in isolation. Parallel proteomic approaches to (a) document the expression of ASNS protein and enzymatic activity, thereby yielding a more definitive analysis of the actual cellular rates of asparagine synthesis, and (b) screen the entire proteome of the cell for the changes in specific protein expression that take place in ASNase-sensitive versus resistant cells, or during ASNase therapy, will also be important (184, 185). Such proteomic analyses will be critical because it is becoming increasingly clear that fluctuations in cellular amino acid content, similar to those induced by ASNase treatment, can cause significant effects on global mRNA translation rates, as well as on the translation of specific mRNA species. For example, data from cell culture studies indicate that in response to long-term ASNase treatment, cells are irreversibly altered by unknown mechanisms such that ASNase resistance and increased transcription from the ASNS gene is retained long after drug exposure has been terminated. It will be important to determine if such an irreversible elevation of ASNS expression occurs in subpopulations of patients undergoing therapy, and, if so, the identification of the molecular mechanism(s) by which that enhanced expression occurs might lead to new therapeutic targets. Finally, extending the current studies on the relationship between ASNS expression, ASNase effectiveness, hyperdiploidy, and the expression of the t(12;21) product TEL-AML1 is likely to be important in designing rational ASNase therapy in ALL genetic subtypes.
The demonstration that adenylated sulfoximine 7b is a nanomolar ASNS inhibitor sets the stage for cell-based assays to determine the biological effects of inhibiting the enzyme in ASNase-resistant MOLT-4 cell lines. On the other hand, the ability of this compound to pass through cellular membranes is clearly limited by the presence of amine and carboxylate functional groups, which have been shown to be critical substituents for aspartate recognition and binding. Therefore obtaining functionalized sulfoximine derivatives that are “second-generation” ASNS inhibitors is dependent on identifying uncharged functional groups, such as hydroxyl groups or heteroaromatic rings, which can substitute for these ionizable groups. Accomplishing this goal will likely require the development of suitable, targeted molecular libraries (151, 186), for which access to (a) novel, and more efficient, synthetic methods for constructing the sulfoximine functionality, and (b) validated computational models of the AS-B/7b active site complex that can be employed in de novo design strategies (181) will be essential. Novel assays for detecting asparagine formation that are sufficiently sensitive for use in automated, high-throughput screening also remain to be developed. Finally, the use of profiling methods (187) should offer a rapid approach to delineating the specificity of adenylated sulfoximines when introduced into cells, particularly regarding their ability to bind to aspartyl tRNA synthetases.
SUMMARY POINTS
Crystal structures of the evolutionarily related enzymes AS-B and BLS have permitted the construction of a computational model for the complex between β-aspartyladenylate, a key reaction intermediate, and conserved residues in the ASNS synthetase active site.
Determination of a kinetic mechanism for ASNS has suggested that analogs of β-aspartyladenylate, and the transition state for nucleophilic attack of ammonia on this intermediate, will be potent inhibitors of the human enzyme.
ASNS transcription is enhanced by amino acid deprivation via a signal transduction pathway that senses amino acid availability, resulting in the enhanced synthesis and function of selected transcription factors belonging to the bZIP family.
When delivered to patients, ASNase degrades plasma asparagine and glutamine thereby depleting the cellular levels of these amino acids. ALL cells express a low level of ASNS activity and are therefore particularly sensitive to ASNase-based therapies.
Selection and characterization of ASNase-resistant ALL cell lines in culture suggests that ASNase resistance is correlated with the elevation of ASNS expression, although this relationship appears to be more complicated in primary ALL cells isolated from patients and probably depends on the genetic background of each individual.
The first slow-onset, tight-binding inhibitor for recombinant human ASNS, with nanomolar binding affinity, has been identified, and its interactions with the ASNS synthetase site characterized using molecular modeling methods.
FUTURE ISSUES TO BE RESOLVED
The effects of the N-acylsulfonamide and sulfoximine-derived ASNS inhibitors on ASNase-resistant cell lines need to be evaluated and assays developed to ensure their specificity of interaction with intracellular ASNS.
The validation, and refinement, of computational models describing the interaction of protein residues with the N-acylsulfonamide and sulfoximine-derived ASNS inhibitors must be undertaken prior to their subsequent use in designing molecular libraries fhat can be screened for compounds with “drug-like” properties and good cell permeability.
Further insight into the role that ASNS plays in the treatment of childhood ALL will require more intensive research into the regulation of its expression by amino acid deprivation and the consequences on cell growth of inhibiting its synthetase activity.
Also needed is the development of proteomics-based methods for (a) quantitating the expression of ASNS protein and enzymatic activity, thereby yielding a more definitive analysis of the actual cellular rates of asparagine synthesis, and (b) identifying changes in protein expression, which take place in ASNase-sensitive and -resistant cells.
ACKNOWLEDGMENTS
We thank the many coworkers and collaborators who have contributed to the studies discussed in this review, and apologize for omitting many details of their efforts because of space limitations. This work was supported by grants from the National Institutes of Health (CA09126, CA107437, and DK52064) and the Chiles Endowment Biomedical Research Program of the Florida Department of Health.
Glossary
- ASNS
asparagine synthetase
- AS-B
Escherichia coli glutamine-dependent ASNS
- Prednisone
an orally active, synthetic corticosteroid used to suppress the immune system
- L-asparaginase (ASNase)
a serine-dependent hydrolase that catalyzes the hydrolysis of L-asparagine to L-aspartic acid
- Acute lymphoblastic leukemia (ALL)
a disease in which patients produce primitive lymphoid cells instead of cells that would normally develop along the lymphoid lineage into mature B- or T-lymphocytes
- Acute myeloblastic leukemia (AML)
a disease in which patients produce cancerous primitive cells instead of cells that would normally develop along the myeloid lineage into myeloid white blood cells
- Vincristine
an alkaloid that binds to tubulin monomers thereby preventing the formation of spindle microtubules and stopping separation of duplicated chromosomes
- Ntn
N-terminal amidohydrolase
- GMPS
guanosine-5′ -monophosphate synthetase
- BLS
β-lactam synthetase
- βAspAMP
β-aspartyl-AMP
- PPi
inorganic pyrophosphate
- BHK ts11 cells
a hamster cell line that expresses a mutant form of ASNS and, consequently, is blocked at G1 in the cell cycle
- Amino acid response (AAR) pathway
the pathway by which mammalian cells sense and respond to a deficiency of protein/amino acid
- bZIP
basic leucine zipper transcription factor
- NSRE
nutrient-sensing response element
- UPR
unfolded protein response
- EMSA
electrophoresis mobility shift analysis
- ChIP
chromatin immunoprecipitation
- MOLT-4 cells
a T cell–derived leukemic cell line isolated from a patient with ALL
- Molecular dynamics (MD)
a computational method in which Newton's equations of motion are solved to yield a trajectory showing the dynamical motions of a protein structure
- Simulated annealing
a technique to optimize the structure of a protein by performing an MD simulation in which the temperature is systematically lowered to obtain a low-energy conformation
LITERATURE CITED
- 1.Cooney DA, Handschumacher RE. Annu. Rev. Pharmacol. 1970;10:421–40. doi: 10.1146/annurev.pa.10.040170.002225. [DOI] [PubMed] [Google Scholar]
- 2.Ertel IJ, Nesbit MG, Hammon D, Weiner J, Sather H. Cancer Res. 1979;39:3893–96. [PubMed] [Google Scholar]
- 3.Amylon MD, Shuster J, Pullen J, Berard C, Link MP, et al. Leukemia. 1999;13:335–42. doi: 10.1038/sj.leu.2401310. [DOI] [PubMed] [Google Scholar]
- 4.Sanz GF, Sanz MA, Rafecas FJ, Martinez JA, Martin-Aragonés G, et al. Cancer Treat Rep. 1986;70:1321–24. [PubMed] [Google Scholar]
- 5.Barr RD, DeVeber LL, Pai KM, Andrew M, Halton J, et al. Am. J. Pediatr. Hematol. Oncol. 1992;14:136–39. doi: 10.1097/00043426-199205000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Pui CH, Relling MV, Campana D, Evans WE. Rev. Clin. Exp. Hematol. 2002;6:161–80. doi: 10.1046/j.1468-0734.2002.00067.x. [DOI] [PubMed] [Google Scholar]
- 7.Ohnuma T, Holland JF, Meyer P. Cancer. 1972;30:376–81. doi: 10.1002/1097-0142(197208)30:2<376::aid-cncr2820300212>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarti R, Schuster SM. Int. J. Pediatr. Hemotol./Oncol. 1997;4:597–611. [Google Scholar]
- 9.Graham ML. Adv. Drug Deliv. Rev. 2003;55:1293–302. doi: 10.1016/s0169-409x(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 10.Tallal L, Tan C, Oettgen H, Wollner N, McCarthy M, et al. Cancer. 1970;25:306–20. doi: 10.1002/1097-0142(197002)25:2<306::aid-cncr2820250206>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 11.Sutow WW, Garcia F, Starling KA, Williams TE, Lane DM, et al. Cancer. 1971;28:819–24. doi: 10.1002/1097-0142(1971)28:4<819::aid-cncr2820280403>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Muller HJ, Boos J. Crit. Rev. Oncol. Hematol. 1998;28:97–113. doi: 10.1016/s1040-8428(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 13.Hersh EM. Transplantation. 1971;12:368–76. doi: 10.1097/00007890-197111000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Terebello HR, Anderson K, Wiernik PH, Cuttner J, Cooper RM, et al. Am. J. Clin. Oncol. 1986;9:411–15. doi: 10.1097/00000421-198610000-00010. [DOI] [PubMed] [Google Scholar]
- 15.Kiriyama Y, Kubota M, Takimoto T, Kitoh J, Tanizawa A, et al. Leukemia. 1989;3:294–97. [PubMed] [Google Scholar]
- 16.Lobel JS, O'Brien RT, McIntosh S, Aspnes GT, Capizzi RL. Cancer. 1979;43:1089–94. doi: 10.1002/1097-0142(197903)43:3<1089::aid-cncr2820430346>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Capizzi RL, Bertino JR, Skeel RT, Creasey WA, Zanes R, et al. Ann. Intern. Med. 1971;74:893–901. doi: 10.7326/0003-4819-74-6-893. [DOI] [PubMed] [Google Scholar]
- 18.Pieters R, Klumper E, Kaspers GJL, Veerman AJP. Crit. Rev. Oncol. Hematol. 1997;25:11–26. doi: 10.1016/s1040-8428(96)00223-5. [DOI] [PubMed] [Google Scholar]
- 19.Richards NGJ, Schuster SM. Adv. Enzymol. Relat. Areas Mol. Biol. 1998;72:145–98. doi: 10.1002/9780470123188.ch5. [DOI] [PubMed] [Google Scholar]
- 20.Aslanian AM, Fletcher BS, Kilberg MS. Biochem. J. 2001;357:321–28. doi: 10.1042/0264-6021:3570321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti R, Wylie DE, Schuster SM. J. Biol. Chem. 1989;264:15494–500. [PubMed] [Google Scholar]
- 22.Cooney DA, Driscoll JS, Milman HA, Jayaram HN, Davis RD. Cancer Treat. Rep. 1976;60:1493–557. [PubMed] [Google Scholar]
- 23.Cooney DA, Jones MT, Milman HA, Young DM, Jayaram HN. Int. J. Biochem. 1980;11:519–39. doi: 10.1016/0020-711x(80)90261-x. [DOI] [PubMed] [Google Scholar]
- 24.Koroniak L, Ciustea M, Gutierrez JA, Richards NGJ. Org. Lett. 2003;5:2033–36. doi: 10.1021/ol034212n. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y-Z, Knox EW. Enzyme. 1975;19:314–28. doi: 10.1159/000459006. [DOI] [PubMed] [Google Scholar]
- 26.Hongo S, Motosugu F, Shioda S, Nakai Y, Takeda M, et al. Arch. Biochem. Biophys. 1992;295:120–25. doi: 10.1016/0003-9861(92)90496-j. [DOI] [PubMed] [Google Scholar]
- 27.Mehlhaff P, Luehr CA, Schuster SM. Biochemistry. 1985;24:1104–10. doi: 10.1021/bi00326a006. [DOI] [PubMed] [Google Scholar]
- 28.Luehr CA, Schuster SM. Arch. Biochem. Biophys. 1985;237:335–46. doi: 10.1016/0003-9861(85)90285-1. [DOI] [PubMed] [Google Scholar]
- 29.Patterson MK, Orr GR. J. Biol. Chem. 1968;243:376–80. [PubMed] [Google Scholar]
- 30.Van Heeke G, Schuster SM. Protein Eng. 1990;3:739–44. doi: 10.1093/protein/3.8.739. [DOI] [PubMed] [Google Scholar]
- 31.Sheng S, Moraga-Amador DA, Van Heeke G, Schuster SM. Prot. Exp. Purif. 1992;3:337–46. doi: 10.1016/1046-5928(92)90010-t. [DOI] [PubMed] [Google Scholar]
- 32.Sheng S, Moraga-Amador DA, Van Heeke G, Allison RD, Richards NGJ, et al. J. Biol. Chem. 1993;268:16771–80. [PubMed] [Google Scholar]
- 33.Ciustea M, Gutierrez JA, Abbatiello SE, Eyler JR, Richards NGJ. Arch. Biochem. Biophys. 2005;440:18–27. doi: 10.1016/j.abb.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 34.Boehlein SK, Richards NGJ, Schuster SM. J. Biol. Chem. 1994;269:7450–57. [PubMed] [Google Scholar]
- 35.Larsen TM, Boehlein SK, Schuster SM, Richards NGJ, Thoden JB, et al. Biochemistry. 1999;38:16146–57. doi: 10.1021/bi9915768. [DOI] [PubMed] [Google Scholar]
- 36.Zalkin H. Adv. Enzymol. Relat. Areas Mol. Biol. 1993;66:203–309. doi: 10.1002/9780470123126.ch5. [DOI] [PubMed] [Google Scholar]
- 37.Pfeiffer NE, Mehlhaff PM, Wylie DW, Schuster SM. J. Biol. Chem. 1986;261:1914–19. [PubMed] [Google Scholar]
- 38.Oinonen C, Rouvinen J. Protein Sci. 2000;9:2329–37. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brannigan JA, Dodson G, Duggleby HJ, Moody PCE, Smith JL, et al. Nature. 1995;378:416–19. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 40.Voges D, Zwickl P, Baumeister W. Annu. Rev. Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 41.Duggleby HJ, Tolley SP, Hill CP, Dodson EJ, Dodson G, et al. Nature. 1995;373:264–68. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 42.Oinonen C, Tikkanen R, Rouvinen J, Peltonen L. Nat. Struct. Biol. 1995;2:1102–8. doi: 10.1038/nsb1295-1102. [DOI] [PubMed] [Google Scholar]
- 43.Zalkin H, Smith JL. Adv. Enzymol. Relat. Areas Mol. Biol. 1998;72:87–144. doi: 10.1002/9780470123188.ch4. [DOI] [PubMed] [Google Scholar]
- 44.Krahn JM, Kim JH, Burns MR, Parry RJ, Zalkin H, et al. Biochemistry. 1997;36:11061–68. doi: 10.1021/bi9714114. [DOI] [PubMed] [Google Scholar]
- 45.Massière F, Badet-Denisot M-A. Cell. Mol. Life Sci. 1998;54:205–22. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teplyakov A, Obmolova G, Badet B, Badet-Denisot M-A. J. Mol. Biol. 2001;313:1093–102. doi: 10.1006/jmbi.2001.5094. [DOI] [PubMed] [Google Scholar]
- 47.Bork P, Koonin EV. Proteins: Struct. Funct. Genet. 1994;20:347–55. doi: 10.1002/prot.340200407. [DOI] [PubMed] [Google Scholar]
- 48.Tesmer JJG, Klem TJ, Deras ML, Davisson VJ, Smith JL. Nat. Struct. Biol. 1996;3:74–86. doi: 10.1038/nsb0196-74. [DOI] [PubMed] [Google Scholar]
- 49.Goto M, Omi R, Miyahara I, Sugahara M, Hirotsu K. J. Biol. Chem. 2003;278:22964–71. doi: 10.1074/jbc.M213198200. [DOI] [PubMed] [Google Scholar]
- 50.MacRae IJ, Segel IH, Fisher AJ. Biochemistry. 2001;40:6795–804. doi: 10.1021/bi010367w. [DOI] [PubMed] [Google Scholar]
- 51.Miller MT, Gerratana B, Stapon A, Townsend CA, Rosenzweig AC. J. Biol. Chem. 2003;278:40996–1002. doi: 10.1074/jbc.M307901200. [DOI] [PubMed] [Google Scholar]
- 52.Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. Nat. Struct. Biol. 2001;8:684–89. doi: 10.1038/90394. [DOI] [PubMed] [Google Scholar]
- 53.Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. Proc. Natl. Acad. Sci. USA. 2002;99:14752–57. doi: 10.1073/pnas.232361199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mueller EG, Palenchar PM. Protein Sci. 1999;8:2424–27. doi: 10.1110/ps.8.11.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boehlein SK, Stewart JD, Walworth ES, Thirumoorthy R, Richards NGJ, Schuster SM. Biochemistry. 1998;37:13230–38. doi: 10.1021/bi981058h. [DOI] [PubMed] [Google Scholar]
- 56.Anand R, Hoskins AA, Stubbe J, Ealick SE. Biochemistry. 2004;43:10328–42. doi: 10.1021/bi0491301. [DOI] [PubMed] [Google Scholar]
- 57.Endrizzi JA, Kim HS, Anderson PM, Baldwin EP. Biochemistry. 2004;43:6447–63. doi: 10.1021/bi0496945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van den Heuvel RHH, Curti B, Vanoni M, Mattevi A. Cell. Mol. Life Sci. 2004;61:669–81. doi: 10.1007/s00018-003-3316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers RS, Jensen JR, Deras IL, Smith JL, Davisson VJ. Biochemistry. 2003;42:7013–22. doi: 10.1021/bi034314l. [DOI] [PubMed] [Google Scholar]
- 60.Knöochel T, Ivens A, Hester G, Gonzalez A, Bauerle R, et al. Proc. Natl. Acad. Sci. USA. 1999;96:9479–84. doi: 10.1073/pnas.96.17.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raushel F, Thoden JB, Holden HM. Acc. Chem. Res. 2003;36:539–48. doi: 10.1021/ar020047k. [DOI] [PubMed] [Google Scholar]
- 62.Huang XY, Holden HM, Raushel FM. Annu. Rev. Biochem. 2001;70:149–80. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 63.Tesson AR, Soper TS, Ciustea M, Richards NGJ. Arch. Biochem. Biophys. 2003;413:23–31. doi: 10.1016/s0003-9861(03)00118-8. [DOI] [PubMed] [Google Scholar]
- 64.Fresquet V, Thoden JB, Holden HM, Raushel FM. Bioorg. Chem. 2004;32:63–75. doi: 10.1016/j.bioorg.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Hongo S, Sato T. Arch. Biochem. Biophys. 1985;238:410–17. doi: 10.1016/0003-9861(85)90181-x. [DOI] [PubMed] [Google Scholar]
- 66.Markin RS, Luehr CA, Schuster SM. Biochemistry. 1981;20:7226–32. doi: 10.1021/bi00528a027. [DOI] [PubMed] [Google Scholar]
- 67.Rognes SE. Phytochemistry. 1975;14:1975–82. [Google Scholar]
- 68.Rose IA. Methods Enzymol. 1980;64:47–59. doi: 10.1016/s0076-6879(80)64004-x. [DOI] [PubMed] [Google Scholar]
- 69.Ibba M, Söll D. Annu. Rev. Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 70.Arnez JG, Moras D. Trends Biochem. Sci. 1997;22:211–16. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 71.Barshop BA, Wrenn RF, Frieden C. Anal. Biochem. 1983;130:134–45. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- 72.Buchanan JM. Adv. Enzymol. Relat. Areas Mol. Biol. 1973;39:91–183. doi: 10.1002/9780470122846.ch2. [DOI] [PubMed] [Google Scholar]
- 73.Bera AK, Smith JL, Zalkin H. J. Biol. Chem. 2000;275:7975–79. doi: 10.1074/jbc.275.11.7975. [DOI] [PubMed] [Google Scholar]
- 74.van den Heuvel RHH, Ferrari D, Bossi RT, Ravasio S, Curti B, et al. J. Biol. Chem. 2002;277:24579–83. doi: 10.1074/jbc.M202541200. [DOI] [PubMed] [Google Scholar]
- 75.Bera AK, Chen SH, Smith JL, Zalkin H. J. Biol. Chem. 1999;274:36498–504. doi: 10.1074/jbc.274.51.36498. [DOI] [PubMed] [Google Scholar]
- 76.Schramm VL. Annu. Rev. Biochem. 1998;67:693–720. doi: 10.1146/annurev.biochem.67.1.693. [DOI] [PubMed] [Google Scholar]
- 77.Radzicka A, Wolfenden R. Methods Enzymol. 1995;249:284–312. doi: 10.1016/0076-6879(95)49039-6. [DOI] [PubMed] [Google Scholar]
- 78.Greco A, Gong SS, Ittmann M, Basilico C. Mol. Cell. Biol. 1989;9:2350–59. doi: 10.1128/mcb.9.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gong SS, Basilico C. Nucleic Acids Res. 1990;18:3509–13. doi: 10.1093/nar/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greco A, Ittmann M, Basilico C. Proc. Natl. Acad. Sci. USA. 1987;84:1565–69. doi: 10.1073/pnas.84.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hongo S, Takeda M, Sato T. Biochem. Intern. 1989;18:661–66. [PubMed] [Google Scholar]
- 82.Colletta G, Cirafici AM. Biochem. Biophys. Res. Commun. 1992;183:265–72. doi: 10.1016/0006-291x(92)91638-7. [DOI] [PubMed] [Google Scholar]
- 83.Kilberg MS, Pan Y-X, Chen H, Leung-Pineda V. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hinnebusch AG. J. Biol. Chem. 1997;272:21661–64. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 85.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang PC, McGrath BC, Reinert J, Olsen DS, Lei L, et al. Mol. Cell. Biol. 2002;22:6681–88. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, et al. Mol. Cell. Biol. 2001;21:4347–68. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harding HP, Novoa I, Zhang YH, Zeng HQ, Wek R, et al. Mol. Cell. 2000;6:1099–108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 89.Vattem KM, Wek RC. Proc. Natl. Acad. Sci. USA. 2004;101:11269–74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu PD, Harding HP, Ron D. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. J. Biol. Chem. 2002;277:24120–27. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 92.Chen H, Pan Y-X, Dudenhausen EE, Kilberg MS. J. Biol. Chem. 2004;279:50829–39. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 93.Gong SS, Guerrini L, Basilico C. Mol. Cell. Biol. 1991;11:6059–66. doi: 10.1128/mcb.11.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hutson RG, Kilberg MS. Biochem. J. 1994;303:745–50. doi: 10.1042/bj3040745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guerrini L, Gong SS, Mangasarian K, Basilico C. Mol. Cell. Biol. 1993;13:3202–12. doi: 10.1128/mcb.13.6.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barbosa-Tessmann IP, Chen C, Zhong C, Siu F, Schuster SM, et al. J. Biol. Chem. 2000;275:26976–85. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- 97.Leung-Pineda V, Kilberg MS. J. Biol. Chem. 2002;277:16585–91. doi: 10.1074/jbc.M110972200. [DOI] [PubMed] [Google Scholar]
- 98.Zhong C, Chen C, Kilberg MS. Biochem. J. 2003;372:603–9. doi: 10.1042/BJ20030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barbosa-Tessmann IP, Pineda VL, Nick HS, Schuster SM, Kilberg MS. Biochem. J. 1999;339:151–58. [PMC free article] [PubMed] [Google Scholar]
- 100.Waye MM, Stanners CP. Cancer Res. 1981;41:3104–6. [PubMed] [Google Scholar]
- 101.Siu FY, Chen C, Zhong C, Kilberg MS. J. Biol. Chem. 2001;276:48100–7. doi: 10.1074/jbc.M109533200. [DOI] [PubMed] [Google Scholar]
- 102.Pan Y-X, Chen H, Siu F, Kilberg MS. J. Biol. Chem. 2003;278:38402–12. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- 103.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Biochem. J. 1999;339:135–41. [PMC free article] [PubMed] [Google Scholar]
- 104.Capizzi RL, Holcenberg JS. In: Cancer Medicine. Holland JF, Frei E, Bast RC Jr, Kuffe DW, Morton DL, Weichselbaum RR, editors. Lea & Febiger; Philadelphia: 1993. pp. 796–805. [Google Scholar]
- 105.Chabner BA, Loo TL. In: Cancer Chemotherapy and Biotherapy. Chabner BA, Longo DL, editors. Lippincott-Raven; Philadelphia: 1996. pp. 485–92. [Google Scholar]
- 106. Deleted in proof.
- 107.den Boer ML, Pieters R, Kazemier KM, Rottier MMA, Zwaan CM, et al. Blood. 1998;91:2092–98. [PubMed] [Google Scholar]
- 108.Hutson RG, Kitoh T, Amador DAM, Cosic S, Schuster SM, Kilberg MS. Am. J. Physiol. Cell Physiol. 1997;272:C1691–99. doi: 10.1152/ajpcell.1997.272.5.C1691. [DOI] [PubMed] [Google Scholar]
- 109.Aslanian AM, Kilberg MS. Biochem. J. 2001;358:59–67. doi: 10.1042/0264-6021:3580059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prager MD, Bachynsky N. Biochem. Biophys. Res. Commun. 1968;31:43–47. doi: 10.1016/0006-291x(68)90028-4. [DOI] [PubMed] [Google Scholar]
- 111.Andrulis IL, Argonza R, Cairney AEL. Somat. Cell Mol. Genet. 1990;16:59–65. doi: 10.1007/BF01650480. [DOI] [PubMed] [Google Scholar]
- 112.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, et al. N. Engl. J. Med. 2004;351:533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 113.Fine BM, Kaspers GJ, Ho M, Loonen AH, Boxer LM. Cancer Res. 2005;65:291–99. [PubMed] [Google Scholar]
- 114.Fenrick R, Amann JM, Lutterbach B, Wang L, Westendorf JJ, et al. Mol. Cell. Biol. 1999;19:6566–74. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramakers-van Woerden NL, Pieters R, Loonen AH, Hubeek I, van Drunen E, et al. Blood. 2000;96:1094–99. [PubMed] [Google Scholar]
- 116.Kaspers GJ, Smets LA, Pieters R, Van Zantwijk CH, Van Wering ER, Veerman AJ. Blood. 1995;85:751–56. [PubMed] [Google Scholar]
- 117.Stams WA, den Boer ML, Beverloo HB, Meijerink JP, Stigter RL, et al. Blood. 2003;101:2743–47. doi: 10.1182/blood-2002-08-2446. [DOI] [PubMed] [Google Scholar]
- 118.Krejci O, Starkova J, Otova B, Madzo J, Kalinova M, et al. Leukemia. 2004;18:434–41. doi: 10.1038/sj.leu.2403259. [DOI] [PubMed] [Google Scholar]
- 119.Stams WA, den Boer ML, Holleman A, Appel IM, Beverloo HB, et al. Blood. 2005;11:4223–25. doi: 10.1182/blood-2004-10-3892. [DOI] [PubMed] [Google Scholar]
- 120.Pui CH, Relling MV, Downing JR. N. Engl. J. Med. 2004;350:1535–48. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 121.Horowitz B, Madras BK, Old LJ, Boyce EJ, Meister A. Science. 1968;160:533–35. doi: 10.1126/science.160.3827.533. [DOI] [PubMed] [Google Scholar]
- 122.Pfeiffer NE, Mehlhaff PM, Wylie DE, Schuster SM. J. Biol. Chem. 1987;262:11565–70. [PubMed] [Google Scholar]
- 123.Bachmann BO, Li RF, Townsend CA. Proc. Natl. Acad. Sci. USA. 1998;95:9082–86. doi: 10.1073/pnas.95.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McNaughton HJ, Thirkettle JE, Zhang ZH, Schofield CJ, Jensen SE, et al. Chem. Commun. 1998;1998:2325–26. [Google Scholar]
- 125.Baggaley KH, Brown AG, Schofield CJ. Nat. Prod. Rep. 1997;14:309–33. doi: 10.1039/np9971400309. [DOI] [PubMed] [Google Scholar]
- 126.Gerlt JA, Babbitt PC. Annu. Rev. Biochem. 2001;70:209–46. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 127.O'Brien PJ, Herschlag D. Chem. Biol. 1999;6:R91–105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 128.Knowles JR. Annu. Rev. Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 129.Parr IB, Dribben AB, Norris SR, Hinds MG, Richards NGJ. J. Chem. Soc. Perkin Trans. 1. 1999;8:1029–38. [Google Scholar]
- 130.Baldwin JE, Moloney MG, North M. Tetrahedron. 1989;45:6319–25. [Google Scholar]
- 131.Parr IB, Boehlein SK, Dribben AB, Schuster SM, Richards NGJ. J. Med. Chem. 1996;39:2367–78. doi: 10.1021/jm9601009. [DOI] [PubMed] [Google Scholar]
- 132.Brooks BR, Cheatham TE. Theor. Chem. Acc. 1998;99:279–88. [Google Scholar]
- 133.Brunger AT, Adams PD, Rice LM. Curr. Opin. Struct. Biol. 1998;8:600–11. doi: 10.1016/s0959-440x(98)80152-8. [DOI] [PubMed] [Google Scholar]
- 134.Boehlein SK, Richards NGJ, Schuster SM. J. Biol. Chem. 1994;269:26789–95. [PubMed] [Google Scholar]
- 135.Boehlein SK, Walworth ES, Richards NGJ, Schuster SM. J. Biol. Chem. 1997;272:12384–92. doi: 10.1074/jbc.272.19.12384. [DOI] [PubMed] [Google Scholar]
- 136.Boehlein SK, Walworth ES, Schuster SM. Biochemistry. 1997;36:10168–77. doi: 10.1021/bi970494l. [DOI] [PubMed] [Google Scholar]
- 137.Mokotoff M, Brynes S, Bagaglio JF. J. Med. Chem. 1975;18:888–91. doi: 10.1021/jm00243a005. [DOI] [PubMed] [Google Scholar]
- 138.Ahmad A, Misra LN. Phytochemistry. 1994;37:183–86. [Google Scholar]
- 139.Romagni JG, Duke SO, Dayan FE. Plant Physiol. 2000;123:725–32. doi: 10.1104/pp.123.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Romagni JG, Duke SO, Dayan FE. Plant Physiol. 2005;137:1487. doi: 10.1104/pp.104.900143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Winum JY, Scozzafava A, Montero JL, Supuran CT. Med. Res. Rev. 2005;25:186–228. doi: 10.1002/med.20021. [DOI] [PubMed] [Google Scholar]
- 142.Kim S, Lee SW, Choi E-C, Choi SY. Appl. Microbiol. Biotechnol. 2003;61:278–88. doi: 10.1007/s00253-003-1243-5. [DOI] [PubMed] [Google Scholar]
- 143.Tao JS, Schimmel P. Exp. Opin. Investig. Drugs. 2000;9:1767–75. doi: 10.1517/13543784.9.8.1767. [DOI] [PubMed] [Google Scholar]
- 144.Boehlein SK, Nakatsu T, Hiratake J, Thirumoorthy R, Stewart JD, et al. Biochemistry. 2001;40:11168–75. doi: 10.1021/bi0155551. [DOI] [PubMed] [Google Scholar]
- 145.Nakama T, Nureki O, Yokoyama S. J. Biol. Chem. 2001;276:48387–93. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 146.Yanagisawa T, Lee JT, Wu HC, Kawakami M. J. Biol. Chem. 1994;269:24304–9. [PubMed] [Google Scholar]
- 147.Phillips DR, Uramoto M, Isono K, McCloskey JA. J. Org. Chem. 1993;58:854–59. [Google Scholar]
- 148.Moriguchi T, Asai N, Okada K, Seio K, Sasaki T, et al. J. Org. Chem. 2002;67:3290–300. doi: 10.1021/jo016176g. [DOI] [PubMed] [Google Scholar]
- 149.Moriguchi T, Yanagi T, Kunimori M, Wada T, Sekine M. J. Org. Chem. 2000;65:8229–38. doi: 10.1021/jo0008338. [DOI] [PubMed] [Google Scholar]
- 150.Filippov D, Timmers CM, Roerdink AR, van der Marcel GA, van Boom JH. Tetrahedron Lett. 1998;39:4891–94. [Google Scholar]
- 151.von der Haar F, Gabius H-J, Cramer F. Angew. Chem. Int. Ed. Engl. 1981;20:217–23. [Google Scholar]
- 152.Pope AJ, Lapointe J, Mensah L, Brown MJB, Benson N, Moore KJ. 1998;273:31691–701. doi: 10.1074/jbc.273.48.31691. [DOI] [PubMed] [Google Scholar]
- 153.Ding Y, Wang J, Schuster SM, Richards NGJ. J. Org. Chem. 2002;67:4372–75. doi: 10.1021/jo025510l. [DOI] [PubMed] [Google Scholar]
- 154.Sanchez-Martin RM, Mittoo S, Bradley M. Curr. Top. Med. Chem. 2004;4:653–69. doi: 10.2174/1568026043451113. [DOI] [PubMed] [Google Scholar]
- 155.Ramström O, Lehn J-M. Nat. Rev. Drug Discov. 2002;1:26–36. doi: 10.1038/nrd704. [DOI] [PubMed] [Google Scholar]
- 156.Bajorath J. Nat. Rev. Drug Discov. 2002;1:882–94. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- 157.Thompson LA, Ellman JA. Chem. Rev. 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 158.Brown MJB, Mensah LM, Doyle ML, Broom NJP, Osbourne N, et al. Biochemistry. 2000;39:6003–11. doi: 10.1021/bi000148v. [DOI] [PubMed] [Google Scholar]
- 159.Brown P, Richardson CM, Mensah LM, O'Hanlon PJ, Osborne NF, et al. Bioorg. Med. Chem. 1999;7:2473–85. doi: 10.1016/s0968-0896(99)00192-3. [DOI] [PubMed] [Google Scholar]
- 160.Forrest AK, Jarvest RL, Mensah LM, O'Hanlon PJ, Pope AJ, et al. Bioorg. Med. Chem. Lett. 2000;10:1871–74. doi: 10.1016/s0960-894x(00)00360-7. [DOI] [PubMed] [Google Scholar]
- 161.Teague SJ, Davis AM, Leeson PD, Oprea T. Angew. Chem. Int. Ed. Engl. 1999;38:3743–48. doi: 10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 162.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 163.Morrison JF, Walsh CT. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 164.Logusch EW, Walker DM, McDonald JF, Franz JE, Villafranca JJ, et al. Biochemistry. 1990;29:366–72. doi: 10.1021/bi00454a009. [DOI] [PubMed] [Google Scholar]
- 165.Abell LM, Villafranca JJ. Biochemistry. 1991;30:6135–41. doi: 10.1021/bi00239a008. [DOI] [PubMed] [Google Scholar]
- 166.Liaw SH, Eisenberg D. Biochemistry. 1994;33:675–81. doi: 10.1021/bi00169a007. [DOI] [PubMed] [Google Scholar]
- 167.Manning JM, Moore S, Rowe WB, Meister A. Biochemistry. 1969;8:2681–85. doi: 10.1021/bi00834a066. [DOI] [PubMed] [Google Scholar]
- 168.Weisbrod RE, Meister A. J. Biol. Chem. 1973;248:3997–4002. [PubMed] [Google Scholar]
- 169.Koizumi M, Hiratake J, Nakatsu T, Kato H, Oda J. J. Am. Chem. Soc. 1999;121:5799–800. [Google Scholar]
- 170.Kim S, Germond J-E, Pridmore D, Söll D. J. Bacteriol. 1996;178:2459–61. doi: 10.1128/jb.178.8.2459-2461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cedar H, Schwartz JH. J. Biol. Chem. 1969;244:4112–21. [PubMed] [Google Scholar]
- 172.Hinchman SK, Henikoff SA, Schuster SM. J. Biol. Chem. 1992;267:144–49. [PubMed] [Google Scholar]
- 173.Nakatsu T, Kato H, Oda J. Nat. Struct. Biol. 1998;5:15–19. doi: 10.1038/nsb0198-15. [DOI] [PubMed] [Google Scholar]
- 174.Hinchman SK, Schuster SM. Protein Eng. 1992;5:279–83. doi: 10.1093/protein/5.3.279. [DOI] [PubMed] [Google Scholar]
- 175.Sugiyama A, Kato H, Nishioka T, Oda J. Biosci. Biotechnol. Biochem. 1992;56:376–79. doi: 10.1271/bbb.56.376. [DOI] [PubMed] [Google Scholar]
- 176.Stewart JJP. J. Comput. Chem. 1989;10:209–20. [Google Scholar]
- 177.Pang SS, Guddat LW, Duggleby RG. J. Biol. Chem. 2003;278:7639–44. doi: 10.1074/jbc.M211648200. [DOI] [PubMed] [Google Scholar]
- 178.Singh V, Shi W, Evans GB, Tyler PC, Furneaux RH, et al. Biochemistry. 2004;43:9–18. doi: 10.1021/bi0358420. [DOI] [PubMed] [Google Scholar]
- 179.Kitchen DB, Decornez H, Furr J, Bajorath J. Nat. Rev. Drug Discov. 2004;3:935–49. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 180.Brooijmans N, Kuntz ID. Annu. Rev. Biophys. Biomol. Struct. 2003;32:335–73. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 181.Schneider G, Fechner U. Nat. Rev. Drug Discov. 2005;4:649–63. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 182.Gane PG, Dean PM. Curr. Opin. Struct. Biol. 2000;10:401–4. doi: 10.1016/s0959-440x(00)00105-6. [DOI] [PubMed] [Google Scholar]
- 183.Bohacek RS, McMartin C, Guida WC. Med. Res. Dev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 184.Bogdanov B, Smith RD. Mass Spectrom. Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 185.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Proc. Natl. Acad. Sci. USA. 2004;101:10000–5. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lipinski C, Hopkins A. Nature. 2004;432:855–61. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 187.Stoughton RB, Friend SH. Nat. Rev. Drug Discov. 2005;4:345–50. doi: 10.1038/nrd1696. [DOI] [PubMed] [Google Scholar]
RELATED RESOURCES
- Schröder M, Kaufman RJ. Annu. Rev. Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]