Abstract
Mammals respond to dietary nutrient fluctuations; for example, deficiency of dietary protein or an imbalance of essential amino acids activates an amino acid response (AAR) signal transduction pathway, consisting of detection of uncharged tRNA by GCN2 kinase, eIF2α phosphorylation, and ATF4 expression. In concert with heterodimerization partners, ATF4 activates specific genes via a C/EBP-ATF response element (CARE). This review outlines the ATF4-dependent transcriptional mechanisms associated with the AAR, focusing on progress during the last five years. Recent evidence suggests that maternal nutrient deprivation not only has immediate metabolic effects on the fetus, but also triggers gene expression changes in adulthood, possibly through epigenetic mechanisms. Therefore, understanding the transcriptional programs initiated by amino acid limitation is critical and timely.
The Amino Acid Response Pathway
Mammals have evolved a wide range of adaptive mechanisms to detect and respond to fluctuations in dietary nutrients. At the level of individual cells, dietary protein limitation is manifested as amino acid deprivation, which activates an amino acid response (AAR) signal transduction pathway (Figure 1). As reviewed recently [1], amino acid limitation regulates numerous steps in gene expression including chromatin structure, transcription start site, transcription rates, mRNA splicing, RNA export, RNA turnover, and translation. The following summary primarily highlights developments during the last five years that further our understanding of ATF4-mediated transcription following AAR pathway activation. We apologize to authors in advance for citations that were not included due to space and topic limitations.
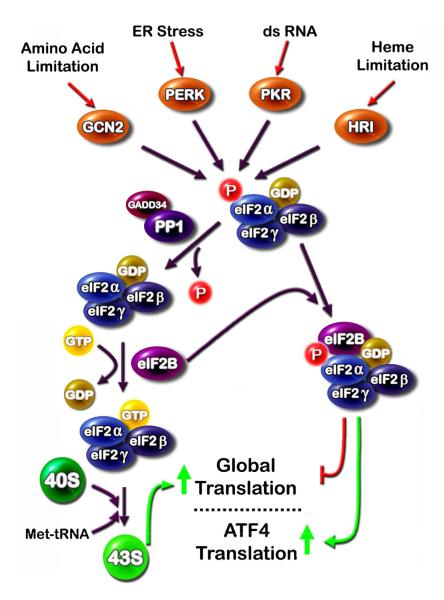
Figure 1.
Several cellular stress signals are transduced through four eukaryotic initiation factor 2α (eIF2α) kinases. (i) The eIF2α kinases general non-derepressible 2 (GCN2), double-stranded RNA-activated protein kinase (PKR), double-stranded RNA activated protein kinase-like ER kinase (PERK), and heme-regulated inhibitor kinase (HRI) are activated by a wide spectrum of cell stress signals and all phosphorylate eIF2α on serine 51. (ii) Phospho-eIF2α binds eIF2B in a non-functional complex and leads to (iii) suppression of global translation, but a paradoxical increase in translation of specific mRNA species, such as that for activating transcription factor 4 (ATF4). De-phosphorylation of eIF2α is mediated by protein phosphatase 1 (PP1) which is targeted to eIF2α by growth arrest and DNA damage-inducible 34 (GADD34).
Mechanistically, the general control non-derepressible 2 (GCN2) kinase acts as an amino acid sensor by binding uncharged tRNA, which leads to phosphorylation of the translation initiation factor 2 (eIF2α) on serine-51. The kinase activity of GCN2 is activated when the protein binds any one of the uncharged tRNA molecules. Thus, depletion at the cellular level of any individual amino acid can trigger the AAR. Phospho-eIF2α suppresses general protein synthesis, but promotes a paradoxical increase in translation of selected mRNA species. Among these are activating transcription factor 5 (ATF5) [2], growth arrest and DNA damage-inducible 34 (GADD34) [3], and ATF4 [4,5]. ATF5 has been implicated in cell growth and development, especially in the nervous system [6]. GADD34 is a component of the feedback loop that permits reactivation of translation by targeting protein phosphatase 1 (PP1) to phospho-eIF2α which is then dephosphorylated, thereby permitting translation of the up-regulated stress-responsive mRNA species [7-9]. ATF4 triggers increased transcription by binding to C/EBP-ATF response elements (CARE), so named because they are composed of a half-site for the C/EBP family and a half-site for the ATF family of transcription factors [10,11]. The products of these CARE-containing genes modulate a wide spectrum of cellular events designed to adapt to dietary stress. If adaptation is unsuccessful, ATF4-induced apoptosis can also occur.
GCN2 is only one of four known eIF2α kinases that collectively respond to a wide array of cellular stresses (Figure 1). Endoplasmic reticulum (ER) stress triggers a multi-arm pathway termed the unfolded protein response (UPR), which includes as one of the arms activation of an ER-resident kinase called double-stranded RNA activated protein kinase-like ER kinase (PERK). As an eIF2α kinase, PERK also increases ATF4 synthesis and a downstream transcriptional program. Given that the subset of genes induced by ATF4 in response to the AAR and the UPR pathways is not identical, one of the interesting questions yet to be answered is how transcriptional specificity is achieved. Some insight has been gained from one ATF4 target gene, the system A sodium-dependent neutral amino acid transporter 2 (SNAT2). SNAT2 contains a CARE site that binds ATF4 and triggers increased transcription during the AAR. However, in HepG2 human hepatoma cells, despite increased ATF4 binding to the SNAT2 gene during UPR activation, transcription activity is not enhanced [12]. Simultaneous activation of the AAR and the UPR pathways revealed that the UPR actually suppresses the increased SNAT2 transcription induced by the AAR pathway, demonstrating that the UPR pathway generates a repressive signal downstream of ATF4 binding. Further investigation will hopefully identify the mechanism of that repressive signal.
Dietary Protein Restriction
In multi-cellular organisms, amino acid homeostasis must be actively controlled, in part because of the body’s inability to synthesize approximately half of the biologically critical amino acids. Consequently, dietary protein intake must supply these molecules. Many studies have tested the biological effects of a diet reduced in or completely devoid of protein. For example, Endo et al. [13] used microarray analysis to show that total protein deprivation of rats affected classes of genes ranging across all cellular functions. Those results are consistent with data from yeast that show that amino acid limitation altered expression of 10-20% of the entire genome [reviewed in 14]. In humans, data is emerging to show that nutrient deprivation not only has immediate effects on gene expression, but also has long-term effects through novel mechanisms. The Dutch famine of 1944 provides a striking example of the effect of malnutrition during development on adult onset chronic degenerative diseases [reviewed in 15]. Briefly, adults whose mothers were exposed to famine during early gestation had a higher risk of coronary heart disease, a higher body mass index, and a more atherogenic lipid profile. One of the current theories to explain these observations is that maternal malnutrition leads to epigenetic changes in the placenta and/or fetus which, in turn, lead to stable, heritable changes in gene expression that lasts into adult life [16]. Animal studies have demonstrated that even a limited period of maternal dietary deficiency can induce fetal epigenetic changes that persist into adulthood [17]. For example, protein restriction in utero causes genome wide changes in fetal hepatic DNA methylation, potentially altering the regulation of important genes in offspring [18]. Thus, accumulating evidence indicates that maternal malnutrition, including protein limitation, causes fetal epigenetic changes that alter gene expression after birth. How these epigenetic changes are triggered represents an important question from the perspective of both basic knowledge and global health care.
Dietary Amino Acid Imbalance
When animals are presented with a diet containing a deficiency in a single essential amino acid, that imbalance is rapidly recognized and food intake is suppressed [reviewed in19]. In fact, a decline in the blood level for the deficient amino acid is correlated with a decrease in feeding [20], and studies have identified the anterior pyriform cortex region of the brain as the location where protein quality is detected. GCN2 kinase-deficient mice illustrate that recognition of an amino acid-imbalanced diet by the brain requires uncharged tRNA sensing by GCN2 [21,22]. Microarray analysis of ATF4-deficient fibroblasts [23] and hepatoma cells [24] have documented that ATF4 regulates a wide array of genes involved in amino acid transport, metabolism, oxidation status, and energy management. However, it is also clear that individual tissues, particularly developing bone, use ATF4 signaling to regulate tissue-specific genes as well [25]. As an illustration of inter-tissue homeostasis, feeding GCN2 knockout mice a leucine-deficient diet resulted in muscle loss, but a concurrent sparing of liver mass, whereas in wild-type mice, the leucine deficiency caused muscle and hepatic weight loss [26].
Interestingly, a dietary amino acid imbalance alters metabolism beyond that for amino acids. For example, induction of genes involved in cholesterol biosynthesis occurs after feeding rats a protein diet based on wheat gluten, which is deficient in lysine and threonine [13]. However, feeding rats a diet completely devoid of protein also resulted in a decrease in expression of these same genes. Paradoxically, but consistent with previous reports, serum cholesterol levels were reduced by both diets. As further evidence for a link between dietary amino acid and lipid metabolism, GCN2 is required for inhibiting fatty acid synthesis and mobilizing lipid stores that occurs in response to leucine deprivation in mice [27]. Collectively, these results provide evidence that dietary amino acid availability alters metabolic pathways beyond protein homeostasis. To understand these relationships at the cellular and organismal levels, additional studies in the laboratory and in the clinic are needed. Central to these studies will be the transcriptional program initiated by increased levels of ATF4.
Translational Control of ATF4 Synthesis
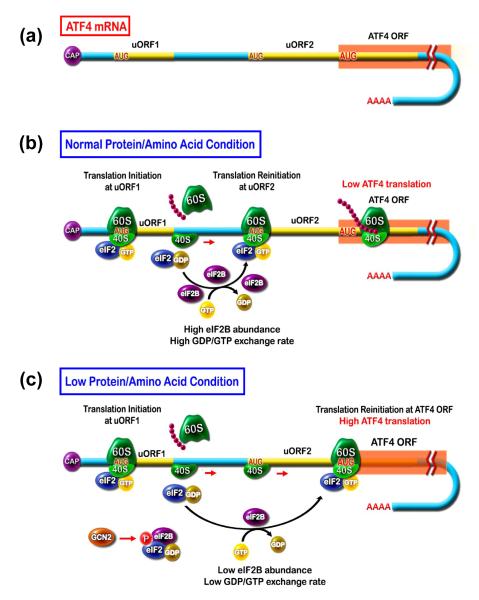
Although best characterized in yeast [28], it is accepted that amino acid limitation in mammalian cells also increases uncharged tRNA accumulation, causing GCN2 kinase to phosphorylate the α-subunit of the translation initiation factor eIF2α (Figure 1). Phospho-eIF2α functions as an inhibitor of eIF2B [29], which catalyzes the exchange of GDP for GTP during the activation of the eIF2 complex that is necessary for assembly of the 43S ribosome. The ATF4 mRNA contains two upstream open reading frames, uORF1 and uORF2, that are located 5′ to the ATF4 coding sequence, and both are translated in the non-stressed condition to the exclusion of ATF4 itself (Figure 2). The uORF2 overlaps with that for ATF4, but is out of frame. During amino acid limitation, as a consequence of phospho-eIF2α inhibition of eIF2B and the corresponding decrease in functional eIF2 complex, ribosome scanning bypasses the uORF2 and translation re-initiation occurs at the ATF4 coding region. Thus, synthesis of ATF4 protein is selectively elevated in response to amino acid deficiency.
Figure 2.
Synthesis of activating transcription factor 4 (ATF4) is increased in response to cell stress. (a) The mRNA for ATF4 contains two short upstream open reading frames (uORF). (b) In normal protein/amino acid conditions, ribosomal scanning leads to translation of uORF1 and re-initiation at uORF2, which is out-of-frame and overlaps with the ATF4 coding sequence. Thus, little or no ATF4 is synthesized. (c) During low protein/amino acid conditions, general control non-derepressible 2 (GCN2) phosphorylates eIF2α, which in turn depletes the available GDP-GTP exchange factor, eukaryotic initiation factor 2B (eIF2B), by non-functional binding. Consequently, there is a reduction in active eIF2-GTP and a slower rate of re-initiation after uORF1 translation. As a result, ribosome assembly occurs after the AUG in uORF2 and coincident with translation of the ORF encoding ATF4. In this way, increased ATF4 protein production occurs only during periods of cellular stress.
The Self-Limiting Model for ATF4 Action on CARE Containing Genes
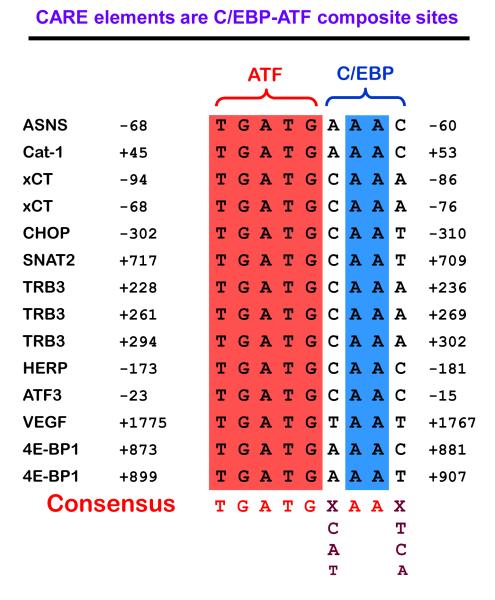
A comprehensive review of ATF4 has been published by Ameri and Harris [30]. As one of its actions, ATF4 activates transcription by binding to CARE sequences, likely as heterodimers with members of the C/EBP family, although the identity and properties of these proposed heterodimers have not been studied extensively. Figure 3 illustrates some of the genes that have been identified to contain functional CARE sequences. Consistent with ATF4’s role as the primary activating factor in the AAR pathway, the ATF half-site is conserved, whereas the C/EBP half-site is divergent. It is possible that different ATF4 dimerization partners bind to the C/EBP half-site to provide transcriptional specificity to the ATF4 signal that is generated by four independent eIF2α kinases, each responding to a variety of different stimuli.
Figure 3.
The genomic C/EBP-ATF (CCAAT-enhancer binding protein-activating transcription factor) composite sites that function as ATF4-responsive C/EBP-ATF response elements (CARE) are composed of half sites for C/EBP and ATF family members. Shown are some examples of genes for which CARE activity has been demonstrated and the gene location for the C/EBP-ATF sites. The genes are as follows: ASNS, asparagine synthetase; Cat-1, cationic amino acid transporter 1; xCT, sodium-independent aspartate/glutamate/cystine transporter; CHOP, C/EBP homology protein; SNAT2, System A neutral amino acid transporter 2; TRB3, Tribbles 3; HERP, homocysteine-induced ER protein; ATF3, activating transcription factor 3; VEGF, vascular endothelial growth factor; 4E-BP1, eukaryotic initiation factor 4E binding protein 1.
Among the ATF4-activated genes are the transcription factors C/EBPβ [31], ATF3 [32,33], and CHOP [34], which then act as ATF4 counter-regulatory signals. C/EBPβ can heterodimerize with ATF4 at CARE sites [35,36]. Increased C/EBP mRNA content and C/EBP functional activity, as measured by in vitro DNA binding activity, is increased in animals fed a low protein diet [31,37]. Both in vitro and in vivo [31,38] studies have documented increased C/EBPβ binding to the asparagine synthetase (ASNS) CARE site after amino acid limitation, but the functional consequences remain unclear. ATF3 is rapidly induced in response to diverse stress signals [39]. Pan et al. [40] and Jiang et al. [32] demonstrated that ATF3 expression is increased by the AAR and UPR pathways, and these inductions required the eIF-2α kinases GCN2 and PERK, respectively [32] (Figure 1). The UPR-associated ATF3 mRNA increase is reduced in both GCN2−/− [32] and ATF4−/− fibroblasts [23]. Mechanistically, both transcription [33] and mRNA stabilization [41] can contribute to induction of ATF3 expression. Interestingly, the ATF3 gene exhibits extensive alternative splicing, and amino acid availability regulates exon selection such that different ATF3 isoforms with opposite activities are synthesized [40]. Full-length ATF3 antagonizes ATF4 action, whereas ATF3ΔZIP3, an isoform with a truncated leucine-zipper domain, further enhances ATF4 activation at CARE sites. While the physiologic consequences of these contrary activities have yet to be explored fully, differential expression of these isoforms may provide further modulation and/or gene specificity to the ATF4 signal.
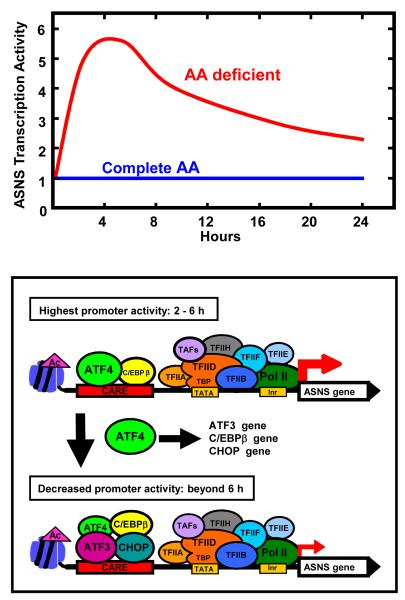
Using the ASNS gene, a working model of ATF4 activation via a CARE site was proposed by Chen et al. [38] in which ATF4 activation of C/EBPβ and ATF3 expression leads to a self-limiting cycle of ATF4-dependent transcription, and more recent studies [42] have added CHOP as a negative regulator of ATF4 action as well (Figure 4). Studying the CARE-containing CHOP gene, Fawcett et al. [11] showed that at 2 h of arsenite treatment ATF4 activates transcription, whereas after 6 h the ATF3 protein abundance was increased and counteracted ATF4 function. Those authors used electrophoresis mobility shift analysis to document that ATF3 binding activity to CARE sites increased at the same time as the binding activity and transcriptional stimulatory action of ATF4 declined. Chen et al. [38] extended those studies using chromatin immunoprecipitation (ChIP) analysis of the ASNS gene to illustrate that ATF4 binding to the CARE site occurs as early as 30-45 minutes after amino acid deprivation and elevated ATF4 binding continues for about 4 hours. In conjunction with ATF4 binding, histone acetylation and the recruitment of the general transcription machinery, including RNA polymerase II, is increased. After 4-6 h of amino acid limitation, and subsequent to the ATF4-dependent induction of C/EBPβ, CHOP, and ATF3 expression, there is a gradual decline in ATF4 binding to the CARE site, an increase in C/EBPβ, ATF3, and CHOP recruitment, and a parallel decrease in ASNS transcription activity. While this self-limiting cycle of ATF4 action has been demonstrated for a number of CARE-containing genes [33] as described below, there are clearly documented, but poorly understood, mechanistic differences between some of these genes. Also, how C/EBPβ, CHOP, and ATF3 antagonize ATF4 action is not well understood. Given that ChIP data represent an average of the cells in a given assay, whether or not ATF4 is completely displaced at an individual CARE site or whether there are protein compositional changes in the existing ATF4 complexes has not been conclusively established. Likewise, whether or not ATF4 is subject to post-translational modification during this period is also unknown. These and other details of the protein complexes that assemble at CARE sites will need to be determined before further mechanistic issues can be addressed.
Figure 4.
(a) Model for activating transcription factor 4 (ATF4) self-limiting regulation of the asparagine synthetase (ASNS) gene. (b) During the initial 2-6 h of amino acid limitation, enhanced translational control of ATF4 protein production results in increased binding of ATF4 to the AARE. A low level of CCAAT-enhancer binding protein β (C/EBPβ) is constitutively bound. In parallel with ATF4 binding, histone acetylation occurs and the general transcription machinery is recruited to the promoter. Beyond 6 h, ATF4 action has caused increased expression of C/EBPβ and ATF3 proteins, and their binding to the C/EBP-ATF response element (CARE) increases coincident with a decline in the transcription activity of the ASNS gene. The C/EBP homology protein (CHOP) gene represents a variation on this general theme given that it also requires phospho-ATF2 and p300/CBP associated factor (PCAF). Ac, acetylated (histone); CHOP, C/EBP homology protein Inr, initiator sequence; Pol II, RNA polymerase II; TAF, TBP associated factors; TBP, TATA binding protein; TATA, TBP binding sequence; TFIIA/B/D/E/F/H, RNA Pol II associated general transcription factors.
Examples of Target Genes Activated by the AAR
Using the transcriptional mechanisms outlined above, cells adapt to a low protein or amino acid imbalanced diet by regulating expression of specific genes involved in transport, intermediary metabolism, oxidative stress, and energy metabolism. Although hundreds of genes have been identified by microarray analysis [14,24,43], the following examples are among those most extensively investigated with regard to transcriptional mechanisms.
ASNS
ASNS transcription is induced during either the AAR or UPR pathways through a bipartite regulatory unit in the promoter that contains nutrient sensing response elements I and II (NSRE-I and II) [44]. NSRE-I is a CARE sequence (Figure 3), and NSRE-II (5′-GTTACA-3′) is located 11-bp downstream of NSRE-1 [45]. Together these two elements function as an enhancer and mediate activation of the ASNS gene in response to ATF4, regardless of whether the initial stimulus was amino acid limitation or ER stress [38]. Interestingly, when cells are incubated in amino acid complete medium, the “fed” state, transcription from the ASNS gene starts at multiple sites between the CARE sequence at −68/−60 of the proximal promoter and the nucleotide often designated as +1, with no clear preference for a single start site. In contrast, after induction of the ASNS gene by amino acid limitation, transcription starts preferentially at the +1 site [46]. This shift in RNA polymerase II alignment to a preferred nucleotide coincides with the increased transcription rate and may reflect the recruitment and assembly of the general transcription machinery in response to ATF4 binding.
CAT1
Expression of the cationic amino acid transporter CAT-1 is increased by amino acid deprivation by several mechanisms involving both transcription and post-transcriptional events [47,reviewed in48]. First, the CAT-1 gene contains a CARE sequence in the first exon (Figure 3) and the ATF4 self-limiting model was confirmed for CAT-1 transcription [36]. Second, CAT-1 mRNA contains an internal ribosome entry sequence (IRES) that promotes translation during amino acid limitation [49]. It is proposed that translation of a uORF in CAT-1 mRNA causes the unfolding of an inhibitory structure in the mRNA, resulting in a conformation that yields an active IRES. Third, during amino acid deprivation, the cytoplasmic content of the RNA binding protein HuR is increased and CAT-1 mRNA is stabilized by HuR binding to the 3′UTR [50]. In both HepG2 and Huh7 human hepatoma cells, CAT-1 mRNA content is similar, but the abundance of CAT-1 protein is much less in Huh7 cells due to translational repression by the microRNA miR-122 [51]. Following amino acid deprivation of Huh7 cells, the translational repression is relieved by CAT-1 mRNA release from cytoplasmic processing bodies in a HuR-dependent manner. These microRNA studies are significant as they document an additional level of translational control of gene expression by amino acid availability. The extent to which this mechanism applies to other amino acid-regulated mRNA species has yet to be tested; also, it is not clear if miR-122, which is not expressed in HepG2 and other cell types, is the only microRNA that can mediate this control.
CHOP
Amino acid control of CHOP expression occurs by transcriptional and post-transcriptional mechanisms [52,53], playing a critical role in a variety of cell stresses. The CHOP promoter contains a CARE (Figure 3), and its association with ATF4, C/EBPβ, and ATF3 appears similar to many other ATF4 responsive genes [33]. However, the CHOP gene is distinct from ASNS in that CHOP activation following amino acid limitation requires phosphorylated ATF2 and p300/CBP-associated factor (PCAF), whereas these two factors do not appear to play a critical role in ASNS regulation [34,54]. Interestingly, ATF4 and CHOP cooperate to activate the TRB3 gene [55] and to inhibit transcription from the ASNS gene [42] (Figure 4). Simultaneous binding of CHOP and ATF4 also occurs at the CARE sites for TRB3, SNAT2, and VEGF, but the transcriptional consequences vary [42]. These differences may depend on the differences in the C/EBPβ half-site of the CARE or additional sequences within each gene that modulates the ATF4-CHOP signal. The recent data illustrating the multifaceted regulation of the CHOP gene and its role as either an activator or inhibitor of transcription is consistent with earlier work revealing the complexity of its actions [56,57].
SNAT2
System A neutral amino acid transporter 2 (SNAT2), which contributes to whole body nitrogen homeostasis [58,59], is induced by amino acid deprivation [60-62]. The SNAT2 transporter represents an important and highly regulated process by which cells actively take up neutral amino acids to adapt to limiting extracellular amino acids. Therefore, understanding the transcriptional mechanisms by which SNAT2 activity is controlled will provide insight into how cells and tissues adapt to dietary protein insufficiency. The first intron of the SNAT2 gene contains a CARE that is about 700 bp downstream of the transcription start site [63,64]. Amino acid limitation leads to ATF4 binding specifically at the CARE and subsequently, recruitment of the general transcription machinery to the promoter. The question arises how this distal CARE site interacts with the general transcription machinery.
During amino acid limitation in yeast, the Mediator transcription complex functions as a bridge between GCN4, the yeast counterpart to ATF4, and the general transcription machinery [65]. The Mediator complex has been proposed to function as a necessary component of the RNA polymerase II (Pol II) general transcription machinery [66]. However, in yeast, Fan et al. [67] showed that there is not always a correlation between Pol II and Mediator recruitment to many highly active genes. They concluded that the yeast Mediator complex is recruited to genes by specific enhancer binding proteins and there may be a preference for Mediator action at genes induced by environmental stress or sub-optimal growth conditions. These results led to the investigation of Mediator’s role in amino acid control of mammalian gene expression [68]. For the SNAT2 gene, there was little or no recruitment of Mediator to the gene during amino acid limitation, and siRNA knockdown of numerous Mediator subunits caused no significant decrease in activated SNAT2 transcription. These results argue against the hypothesis that mammalian Mediator is necessary for all Pol II-dependent transcription or that Mediator is required for transcription of genes activated by sub-optimal growth conditions.
TRB3
TRB3 is the human homolog of Drosophila tribbles, which functions as a regulator of cell growth [55,69]. The TRB3 gene contains three tandemly-arranged CARE sites [55], and in vitro assays illustrate the potential for ATF4 binding to these sites [55,70]. Interestingly, ATF4 and TRB3 physically interact [71,72], and TRB3 antagonizes the cell growth inhibitory and cytotoxic effects of ATF4 [69,71]. Although CHOP was initially considered to be an inhibitor of transcription due to its non-productive binding of other C/EBP family members [56], induction of TRB3 after amino acid limitation is CHOP-dependent [55]. CHOP binds to the TRB3 gene, and over-expression of CHOP further enhances TRB3 expression induced by the AAR [42]. These results again underscore the complex feedback relationships between ATF4 and its downstream effectors. For example, ATF4 increases transcription from both the CHOP and TRB3 genes, and CHOP is required for TRB3 activation. However, TRB3 [69,71] and CHOP [42] subsequently antagonize further ATF4 action. Additional investigation is necessary to fully understand these interrelationships and how they impact cellular responses to amino acid stress.
Interaction between Amino Acid Responsive (AAR) Pathways
Relatively, little is known about the interplay between the AAR and other stress signaling pathways, but there appear to be significant connections to the mitogen activated protein kinase (MAPK) pathways. For example, inhibition of c-Jun N-terminal kinase (JNK) signaling induces ATF4 synthesis in the absence of other cellular stress [73,74], possibly because JNK inhibits PP1 activity [75], which is responsible for eIF2α dephosphorylation [9]. Amino acid limitation increased MAPK/ERK kinase (MEK)-dependent extracellular signal-regulated kinase (ERK) phosphorylation [76,77] in a GCN2 and phospho-eIF2α dependent manner [73], and leads to an increase in SNAT2-mediated neutral amino acid transport that is blocked by inhibition of ERK phosphorylation [76]. Conversely, Thiaville et al. [73] demonstrated that increased phosphorylation of eIF2α and ATF4 synthesis following amino acid limitation is blocked by MEK-ERK pathway-specific inhibitors. Thus, bi-directional cross-talk exists between the MEK-ERK signaling pathway and the AAR pathway, but the molecular mechanisms by which these two pathways exchange information has not been established.
In many ways, the mTOR signaling pathway functions in opposition to the AAR pathway, in that the mTOR complex monitors amino acid sufficiency and promotes protein translation and cell growth [78]. It would be assumed that counter-regulatory mechanisms exist to coordinate the action of the AAR pathway and the mTOR pathway, but these processes are poorly understood. However, recent data demonstrates that cellular stresses that trigger eIF2α phosphorylation cause induction of REDD1 (regulated in development and DNA damage responses 1) protein expression in an ATF4-dependent manner [74,79]. REDD1 acts as a negative regulator of mTOR activity [78]. Thus, during amino acid limitation, increased ATF4 content will coordinately activate the AAR program of genes and concurrently suppress mTOR activity by inducing REDD1synthesis.
Future Directions
Despite the significant progress during the last five years in characterizing the cellular and molecular transcriptional response to dietary protein or amino acid deficiency, many questions remain. Among these are the mechanisms by which ATF4 signaling, triggered by four different eIF2α kinases (Figure 1), becomes selective for the appropriate subset of genes. This question may be related to the fact that besides ATF4, many other transcription factors are induced by protein/amino acid limitation. What are their roles and what are the target genes that fulfill these roles? Also, microarray analysis of amino acid deprived cells indicates GCN2-independent mechanisms for increased transcription [43], suggesting amino acid sensors other than GCN2. What is the identity of these sensors and how do they function? The concept of amino acid transporters serving as sensors has been put forth as one possibility [80]. How does tissue specificity modulate the response to protein or amino acid deficiency? These are just a few examples of the remaining questions that make this area of investigation a rich opportunity for future research that will require the disciplines of nutrition, cell biology, biochemistry, and genomics.
Acknowledgements
The authors wish to thank other members of the laboratory for helpful discussion. Funding is acknowledged from the National Institutes of Health (DK-52064, DK-70647).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kilberg MS, et al. Nutritional control of gene expression: How mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou D, et al. Phosphorylation of eIF2 Directs ATF5 Translational Control in Response to Diverse Stress Conditions. J. Biol. Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 3.Lee YY, et al. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J. Biol. Chem. 2009;284:6661–6673. doi: 10.1074/jbc.M806735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu PD, et al. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green LA, et al. The transcription factor ATF5: role in neurodevelopment and neural tumors. J. Neurochem. 2009;108:11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novoa I, et al. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 2001;153:1011–1021. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brush MH, et al. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novoa I, et al. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfgang CD, et al. gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fawcett TW, et al. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 1999;339:135–141. [PMC free article] [PubMed] [Google Scholar]
- 12.Gjymishka A, et al. Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity is repressed in HepG2 cells. J. Biol. Chem. 2008;283:27736–27747. doi: 10.1074/jbc.M803781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo Y, et al. Dietary protein quantity and quality affect rat hepatic gene expression. J. Nutr. 2002;132:3632–3637. doi: 10.1093/jn/132.12.3632. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roseboom TJ, et al. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Mol. Cell Endocrinol. 2001;185:93–98. doi: 10.1016/s0303-7207(01)00721-3. [DOI] [PubMed] [Google Scholar]
- 16.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu. Rev. Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 17.Waterland RA, Garza C. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am. J. Clin. Nutr. 1999;69:179–197. doi: 10.1093/ajcn/69.2.179. [DOI] [PubMed] [Google Scholar]
- 18.Lillycrop KA, et al. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 19.Gietzen DW, Rogers QR. Nutritional homeostasis and indispensable amino acid sensing: a new solution to an old puzzle. Trends Neurosci. 2006;29:91–99. doi: 10.1016/j.tins.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Koehnle TJ, et al. Diets deficient in indispensable amino acids rapidly decrease the concentration of the limiting amino acid in the anterior piriform cortex of rats. J. Nutr. 2004;134:2365–2371. doi: 10.1093/jn/134.9.2365. [DOI] [PubMed] [Google Scholar]
- 21.Hao S, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 22.Maurin AC, et al. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 2005;1:273–277. doi: 10.1016/j.cmet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 24.Lee JI, et al. HepG2/C3A cells respond to cysteine-deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics. 2008;33:218–229. doi: 10.1152/physiolgenomics.00263.2007. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J. Biol. Chem. 2004;279:47109–47114. doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- 26.Anthony TG, et al. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J. Biol. Chem. 2004;279:36553–36561. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- 27.Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 29.Kimball SR, Jefferson LS. Role of amino acids in the translational control of protein synthesis in mammals. Semin. Cell Dev. Biol. 2005;16:21–27. doi: 10.1016/j.semcdb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Ameri K, Harris AL. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Thiaville MM, et al. Deprivation of protein or amino acid induces C/EBPbeta synthesis and binding to amino acid response elements, but its action is not an absolute requirement for enhanced transcription. Biochem. J. 2008;410:473–484. doi: 10.1042/BJ20071252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang HY, et al. Activating Transcription Factor 3 Is Integral to the Eukaryotic Initiation Factor 2 Kinase Stress Response. Mol. Cell. Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan YX, et al. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherasse Y, et al. The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP. Nucleic Acids Res. 2007;35:5954–5965. doi: 10.1093/nar/gkm642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siu F, et al. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 36.Lopez AB, et al. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem. J. 2007;402:163–173. doi: 10.1042/BJ20060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marten NW, et al. Effect of dietary protein restriction on liver transcription factors. Biochem. J. 1996;317:361–370. doi: 10.1042/bj3170361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, et al. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J. Biol. Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 39.Hai T, et al. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Y-X, et al. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple ATF3 mRNA species which, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J. Biol. Chem. 2003;278:38402–38412. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- 41.Pan YX, et al. Interaction of RNA-binding Proteins HuR and AUF1 with the Human ATF3 mRNA 3′-Untranslated Region Regulates Its Amino Acid Limitation-induced Stabilization. J. Biol. Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su N, Kilberg MS. C/EBP Homology Protein (CHOP) Interacts with Activating Transcription Factor 4 (ATF4) and Negatively Regulates the Stress-dependent Induction of the Asparagine Synthetase Gene. J. Biol. Chem. 2008;283:35106–35117. doi: 10.1074/jbc.M806874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deval C, et al. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 2009;276:707–718. doi: 10.1111/j.1742-4658.2008.06818.x. [DOI] [PubMed] [Google Scholar]
- 44.Barbosa-Tessmann IP, et al. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 2000;275:26976–26985. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- 45.Zhong C, et al. Characterization of the nutrient sensing response unit in the human asparagine synthetase promoter. Biochem. J. 2003;372:603–609. doi: 10.1042/BJ20030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Kilberg MS. Alignment of the transcription start site coincides with increased transcriptional activity from the human asparagine synthetase gene following amino acid deprivation of HepG2 cells. J. Nutr. 2006;136:2463–2467. doi: 10.1093/jn/136.10.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatzoglou M, et al. Regulation of cationic amino acid transport: The story of the CAT-1 transporter. Annu. Rev. Nutr. 2004;24:377–399. doi: 10.1146/annurev.nutr.23.011702.073120. [DOI] [PubMed] [Google Scholar]
- 48.Verrey F, et al. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 2004;447:532–542. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- 49.Yaman I, et al. The Zipper Model of Translational Control. A Small Upstream ORF Is the Switch that Controls Structural Remodeling of an mRNA Leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 50.Yaman I, et al. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharyya SN, et al. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 52.Bruhat A, et al. Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J. Biol. Chem. 1997;272:17588–17593. doi: 10.1074/jbc.272.28.17588. [DOI] [PubMed] [Google Scholar]
- 53.Abcouwer SF, et al. Glutamine deprivation induces the expression of GADD45 and GADD153 primarily by mRNA stabilization. J. Biol. Chem. 1999;274:28645–28651. doi: 10.1074/jbc.274.40.28645. [DOI] [PubMed] [Google Scholar]
- 54.Averous J, et al. Induction of CHOP Expression by Amino Acid Limitation Requires Both ATF4 Expression and ATF2 Phosphorylation. J. Biol. Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 55.Ohoka N, et al. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Gene. Devel. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 57.Wang X-Z, et al. Identification of novel stress-induced genes downstream of chop. EMBO J. 1998;17:3619–3630. doi: 10.1093/emboj/17.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hyde R, et al. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem. J. 2003;373:1–18. doi: 10.1042/bj20030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch. 2004;447:784–795. doi: 10.1007/s00424-003-1117-9. [DOI] [PubMed] [Google Scholar]
- 60.Ling R, et al. Involvement of transporter recruitment as well as gene expression in the substrate-induced adaptive regulation of amino acid transport system A. Biochim. Biophys. Acta. 2001;1512:15–21. doi: 10.1016/s0005-2736(01)00310-8. [DOI] [PubMed] [Google Scholar]
- 61.Gazzola RF, et al. The adaptive regulation of amino acid transport system A is associated to changes in ATA2 expression. FEBS Lett. 2001;490:11–14. doi: 10.1016/s0014-5793(01)02126-3. [DOI] [PubMed] [Google Scholar]
- 62.Bain PJ, et al. The mechanism for transcriptional activation of the human ATA2 transporter gene by amino acid deprivation is different than that for asparagine synthetase. J. Nutr. 2002;132:3023–3029. doi: 10.1093/jn/131.10.3023. [DOI] [PubMed] [Google Scholar]
- 63.Palii SS, et al. Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 2004;279:3463–3471. doi: 10.1074/jbc.M310483200. [DOI] [PubMed] [Google Scholar]
- 64.Palii SS, et al. Characterization of the amino acid response element within the human SNAT2 system A transporter gene. Biochem. J. 2006;395:517–527. doi: 10.1042/BJ20051867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Natarajan K, et al. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 66.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J. Biol. Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 67.Fan X, et al. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 68.Thiaville MM, et al. Activated transcription via mammalian amino acid response elements does not require enhanced recruitment of the Mediator complex. Nucleic Acids Res. 2008;36:5571–5580. doi: 10.1093/nar/gkn538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ord D, et al. TRB3 protects cells against the growth inhibitory and cytotoxic effect of ATF4. Exp. Cell Res. 2007;313:3556–3567. doi: 10.1016/j.yexcr.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 70.Ord D, Ord T. Characterization of human NIPK (TRB3, SKIP3) gene activation in stressful conditions. Biochem. Biophys. Res. Commun. 2005;330:210–218. doi: 10.1016/j.bbrc.2005.02.149. [DOI] [PubMed] [Google Scholar]
- 71.Ord D, Ord T. Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp. Cell Res. 2003;286:308–320. doi: 10.1016/s0014-4827(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 72.Bowers AJ, et al. SKIP3, a novel Drosophila tribbles ortholog, is overexpressed in human tumors and is regulated by hypoxia. Oncogene. 2003;22:2823–2835. doi: 10.1038/sj.onc.1206367. [DOI] [PubMed] [Google Scholar]
- 73.Thiaville MM, et al. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J. Biol. Chem. 2008;283:10848–10857. doi: 10.1074/jbc.M708320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin HO, et al. SP600125 negatively regulates the mammalian target of rapamycin via ATF4-induced Redd1 expression. FEBS Lett. 2009;583:123–127. doi: 10.1016/j.febslet.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 75.Monick MM, et al. Active ERK contributes to protein translation by preventing JNK-dependent inhibition of protein phosphatase 1. J. Immunol. 2006;177:1636–1645. doi: 10.4049/jimmunol.177.3.1636. [DOI] [PubMed] [Google Scholar]
- 76.Franchi-Gazzola R, et al. Adaptive increase of amino acid transport System A requires ERK1/2 activation. J. Biol. Chem. 1999;274:28922–28928. doi: 10.1074/jbc.274.41.28922. [DOI] [PubMed] [Google Scholar]
- 77.Leung-Pineda V, et al. Induction of p21 and p27 Expression By Amino Acid Deprivation of HepG2 Human Hepatoma Cells Involves mRNA Stabilization. Biochem. J. 2004;379:79–88. doi: 10.1042/BJ20031383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 79.Whitney ML, et al. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem. Biophys. Res. Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor PM. Amino acid transporters: eminences grises of nutrient signalling mechanisms? Biochem. Soc. Trans. 2009;37:237–241. doi: 10.1042/BST0370237. [DOI] [PubMed] [Google Scholar]