Abstract
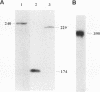
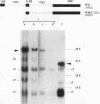
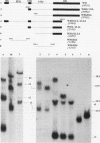
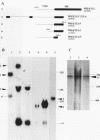
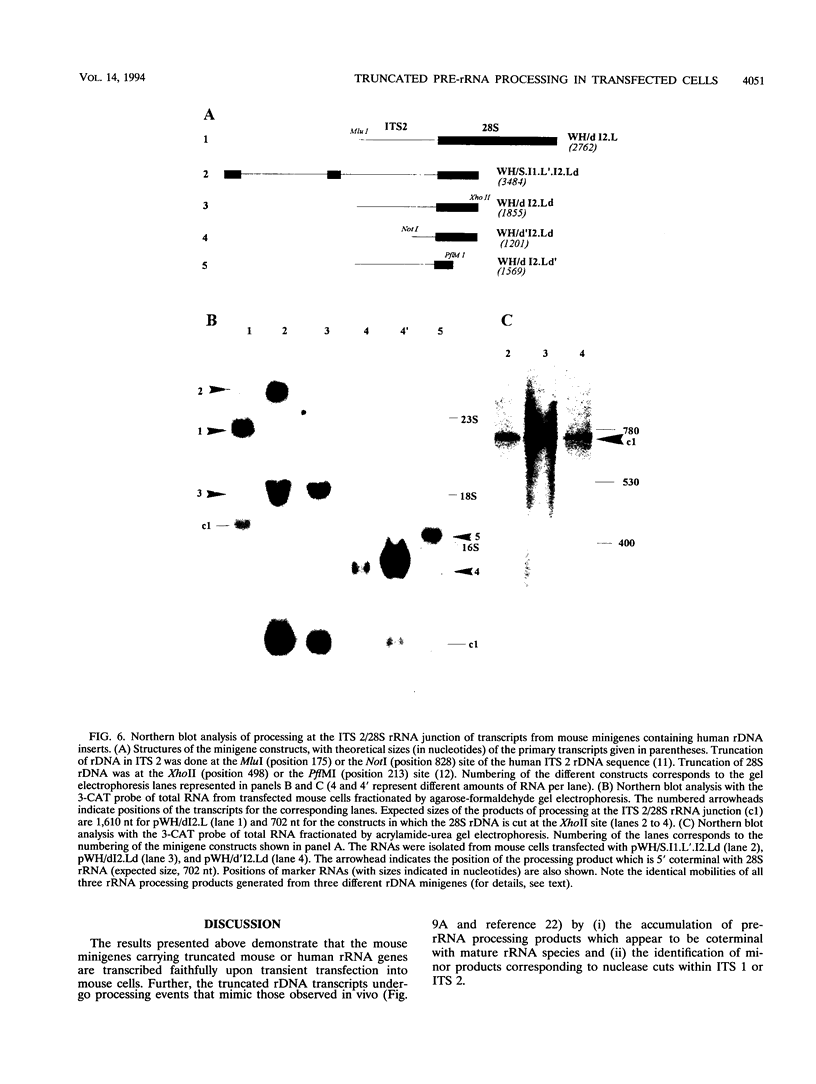
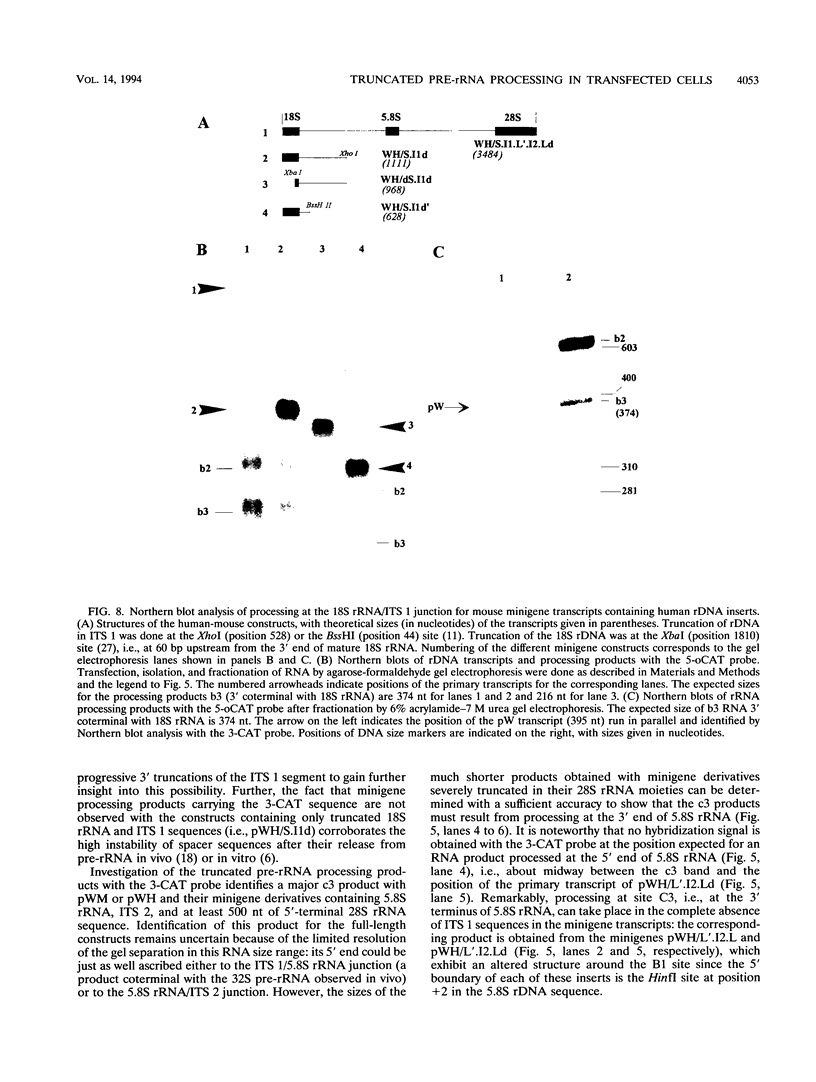
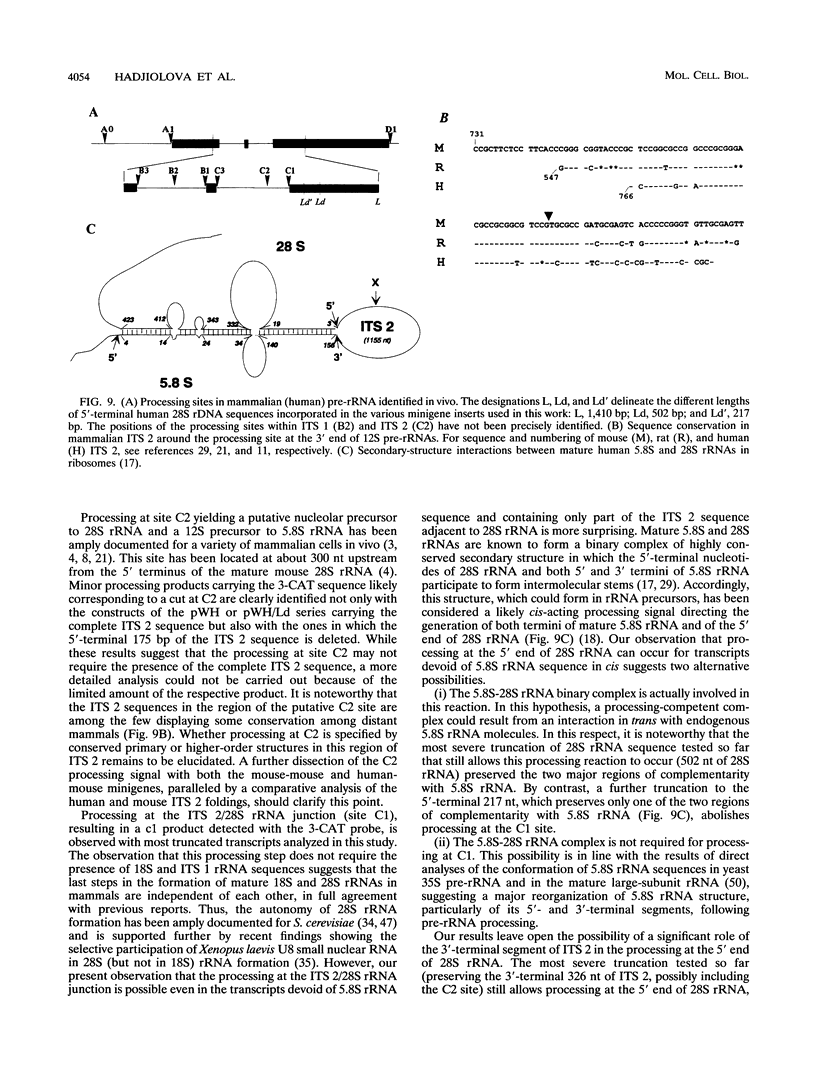
The processing of pre-rRNA in eukaryotic cells involves a complex pattern of nucleolytic reactions taking place in preribosomes with the participation of several nonribosomal proteins and small nuclear RNAs. The mechanism of these reactions remains largely unknown, mainly because of the absence of faithful in vitro assays for most processing steps. We have developed a pre-rRNA processing system using the transient expression of ribosomal minigenes transfected into cultured mouse cells. Truncated mouse or human rRNA genes are faithfully transcribed under the control of mouse promoter and terminator signals. The fate of these transcripts is analyzed by the use of reporter sequences flanking the rRNA gene inserts. Both mouse and human transcripts, containing the 3' end of 18S rRNA-encoding DNA (rDNA), internal transcribed spacer (ITS) 1, 5.8S rDNA, ITS 2, and the 5' end of 28S rDNA, are processed predominantly to molecules coterminal with the natural mature rRNAs plus minor products corresponding to cleavages within ITS 1 and ITS 2. To delineate cis-acting signals in pre-rRNA processing, we studied series of more truncated human-mouse minigenes. A faithful processing at the 18S rRNA/ITS 1 junction can be observed with transcripts containing only the 60 3'-terminal nucleotides of 18S rRNA and the 533 proximal nucleotides of ITS 1. However, further truncation of 18S rRNA (to 8 nucleotides) or of ITS 1 (to 48 nucleotides) abolishes the cleavage of the transcript. Processing at the ITS 2/28S rRNA junction is observed with truncated transcripts lacking the 5.8S rRNA plus a major part of ITS 2 and containing only 502 nucleotides of 28S rRNA. However, further truncation of the 28S rRNA segment to 217 nucleotides abolishes processing. Minigene transcripts containing most internal sequences of either ITS 1 or ITS 2, but devoid of ITS/mature rRNA junctions, are not processed, suggesting that the cleavages in vivo within either ITS segment are dependent on the presence in cis of mature rRNA sequences. These results show that the major cis signals for pre-rRNA processing at the 18S rRNA/ITS 1 or the ITS2/28S rRNA junction involve solely a limited critical length of the respective mature rRNA and adjacent spacer sequences.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachellerie J. P., Qu L. H. Direct ribosomal RNA sequencing for phylogenetic studies. Methods Enzymol. 1993;224:349–357. doi: 10.1016/0076-6879(93)24026-q. [DOI] [PubMed] [Google Scholar]
- Bourbon H., Michot B., Hassouna N., Feliu J., Bachellerie J. P. Sequence and secondary structure of the 5' external transcribed spacer of mouse pre-rRNA. DNA. 1988 Apr;7(3):181–191. doi: 10.1089/dna.1988.7.181. [DOI] [PubMed] [Google Scholar]
- Bowman L. H., Goldman W. E., Goldberg G. I., Hebert M. B., Schlessinger D. Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol. 1983 Aug;3(8):1501–1510. doi: 10.1128/mcb.3.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L. H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981 Oct 10;9(19):4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc Natl Acad Sci U S A. 1987 Feb;84(3):629–633. doi: 10.1073/pnas.84.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Sequence organization and RNA structural motifs directing the mouse primary rRNA-processing event. Mol Cell Biol. 1991 Jan;11(1):458–467. doi: 10.1128/mcb.11.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Hadjiolova K. V., Kermekchiev M. B., Stanchev B. S., Hadjiolov A. A. A 12 S precursor to 5.8 S rRNA associated with rat liver nucleolar 28 S rRNA. Biochim Biophys Acta. 1983 Jan 20;739(1):79–84. doi: 10.1016/0167-4781(83)90047-7. [DOI] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Chambers C., Gorski J. L., Stambolian D., Schmickel R. D., Sylvester J. E. Sequence and structure correlation of human ribosomal transcribed spacers. J Mol Biol. 1990 Mar 5;212(1):27–35. doi: 10.1016/0022-2836(90)90302-3. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I. Mapping of a mouse ribosomal DNA promoter by in vitro transcription. Nucleic Acids Res. 1981 Nov 25;9(22):6093–6102. doi: 10.1093/nar/9.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney T., Jr Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/- 100 nucleotides from the 5' end and the second 500 +/- 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985 Jul 11;13(13):4905–4919. doi: 10.1093/nar/13.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S-like) ribosomal RNA sequences presented in a secondary structure format. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2319–2330. doi: 10.1093/nar/18.suppl.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova K. V., Georgiev O. I., Nosikov V. V., Hadjiolov A. A. Localization and structure of endonuclease cleavage sites involved in the processing of the rat 32S precursor to ribosomal RNA. Biochem J. 1984 May 15;220(1):105–116. doi: 10.1042/bj2200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova K. V., Georgiev O. I., Nosikov V. V., Hadjiolov A. A. Mapping of the major early endonuclease cleavage site of the rat precursor to rRNA within the internal transcribed spacer sequence of rDNA. Biochim Biophys Acta. 1984 Jun 16;782(2):195–201. doi: 10.1016/0167-4781(84)90024-1. [DOI] [PubMed] [Google Scholar]
- Hadjiolova K. V., Nicoloso M., Mazan S., Hadjiolov A. A., Bachellerie J. P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem. 1993 Feb 15;212(1):211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Normann A., Bartsch I., Grummt I. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 1988 May;7(5):1497–1502. doi: 10.1002/j.1460-2075.1988.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor G. R., Mogg A. E., Burke J. F., Caskey C. T. Histochemical staining of clonal mammalian cell lines expressing E. coli beta galactosidase indicates heterogeneous expression of the bacterial gene. Somat Cell Mol Genet. 1987 May;13(3):253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- McCallum F. S., Maden B. E. Human 18 S ribosomal RNA sequence inferred from DNA sequence. Variations in 18 S sequences and secondary modification patterns between vertebrates. Biochem J. 1985 Dec 15;232(3):725–733. doi: 10.1042/bj2320725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Structure of mouse rRNA precursors. Complete sequence and potential folding of the spacer regions between 18S and 28S rRNA. Nucleic Acids Res. 1983 May 25;11(10):3375–3391. doi: 10.1093/nar/11.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P. Secondary structure of the 5' external transcribed spacer of vertebrate pre-rRNA. Presence of phylogenetically conserved features. Eur J Biochem. 1991 Feb 14;195(3):601–609. doi: 10.1111/j.1432-1033.1991.tb15743.x. [DOI] [PubMed] [Google Scholar]
- Michot B., Despres L., Bonhomme F., Bachellerie J. P. Conserved secondary structures in the ITS2 of trematode pre-rRNA. FEBS Lett. 1993 Feb 1;316(3):247–252. doi: 10.1016/0014-5793(93)81301-f. [DOI] [PubMed] [Google Scholar]
- Musters W., Boon K., van der Sande C. A., van Heerikhuizen H., Planta R. J. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J. 1990 Dec;9(12):3989–3996. doi: 10.1002/j.1460-2075.1990.tb07620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R. J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989 Feb;9(2):551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis B. A., Steitz J. A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993 Jun 18;73(6):1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Raziuddin, Little R. D., Labella T., Schlessinger D. Transcription and processing of RNA from mouse ribosomal DNA transfected into hamster cells. Mol Cell Biol. 1989 Apr;9(4):1667–1671. doi: 10.1128/mcb.9.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumard C. M., Torres C., Eichler D. C. In vitro processing at the 3'-terminal region of pre-18S rRNA by a nucleolar endoribonuclease. Mol Cell Biol. 1990 Aug;10(8):3868–3872. doi: 10.1128/mcb.10.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J. A., Ohrlein A., Grummt I. In vitro mutagenesis and transcriptional analysis of a mouse ribosomal promoter element. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2137–2141. doi: 10.1073/pnas.81.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Mougey E. B. News from the nucleolus: rRNA gene expression. Trends Biochem Sci. 1991 Feb;16(2):58–62. doi: 10.1016/0968-0004(91)90025-q. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Stoyanova B. B., Hadjiolov A. A. Alterations in the processing of rat-liver ribosomal RNA caused by cycloheximide inhibition of protein synthesis. Eur J Biochem. 1979 May 15;96(2):349–356. doi: 10.1111/j.1432-1033.1979.tb13046.x. [DOI] [PubMed] [Google Scholar]
- Vance V. B., Thompson E. A., Bowman L. H. Transfection of mouse ribosomal DNA into rat cells: faithful transcription and processing. Nucleic Acids Res. 1985 Oct 25;13(20):7499–7513. doi: 10.1093/nar/13.20.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989 Jun;53(2):256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990 Jun;2(3):521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Yeh L. C., Lee J. C. Higher-order structure of the 5.8 S rRNA sequence within the yeast 35 S precursor ribosomal RNA synthesized in vitro. J Mol Biol. 1991 Feb 20;217(4):649–659. doi: 10.1016/0022-2836(91)90523-9. [DOI] [PubMed] [Google Scholar]
- van der Sande C. A., Kwa M., van Nues R. W., van Heerikhuizen H., Raué H. A., Planta R. J. Functional analysis of internal transcribed spacer 2 of Saccharomyces cerevisiae ribosomal DNA. J Mol Biol. 1992 Feb 20;223(4):899–910. doi: 10.1016/0022-2836(92)90251-e. [DOI] [PubMed] [Google Scholar]