Dear Editor,
Karrikins and strigolactones (SL) are two classes of butenolide compounds that control many aspects of plant physiology. Karrikins, originally found in the smoke of forest fires, have the ability to stimulate seed germination1. SL are endogenous plant hormones that mediate responses to low levels of soil nutrients, such as phosphate and nitrate2. They promote root branching to increase nutrient uptake while inhibiting shoot branching to reduce nutrient demand. SL are also secreted from roots to stimulate symbiotic associations with arbuscular mycorrhizal fungi for increased nutrient uptake, signals that are exploited to induce seed germination in parasitic weeds, including Striga and Orobanche, which are major causes of crop losses2.
Genetic studies in Arabidopsis have identified KAI2 and MAX2 as two key players in karrikin signal transduction3. KAI2 has high sequence similarity to the bacterial signaling protein RbsQ4 (Supplementary information, Figure S1), which encodes an α/β hydrolase. MAX2 is the F-box component of SCF E3 ubiquitin ligase. SL signaling is also mediated by MAX2 and by a paralog of KAI2, D142. Moreover, a D14 homolog from petunia, DAD2, has hydrolytic activity toward the synthetic SL, GR24, and this activity has been proposed to be essential for SL perception5. Similarly, hydrolysis of the butenolide moiety of karrikins by KAI2 has been proposed as a part of the karrikin signaling mechanism6. However, the mechanisms of catalysis as well as the molecular bases of the signaling specificity of KAI2 and D14 toward karrikins and SL remain unclear.
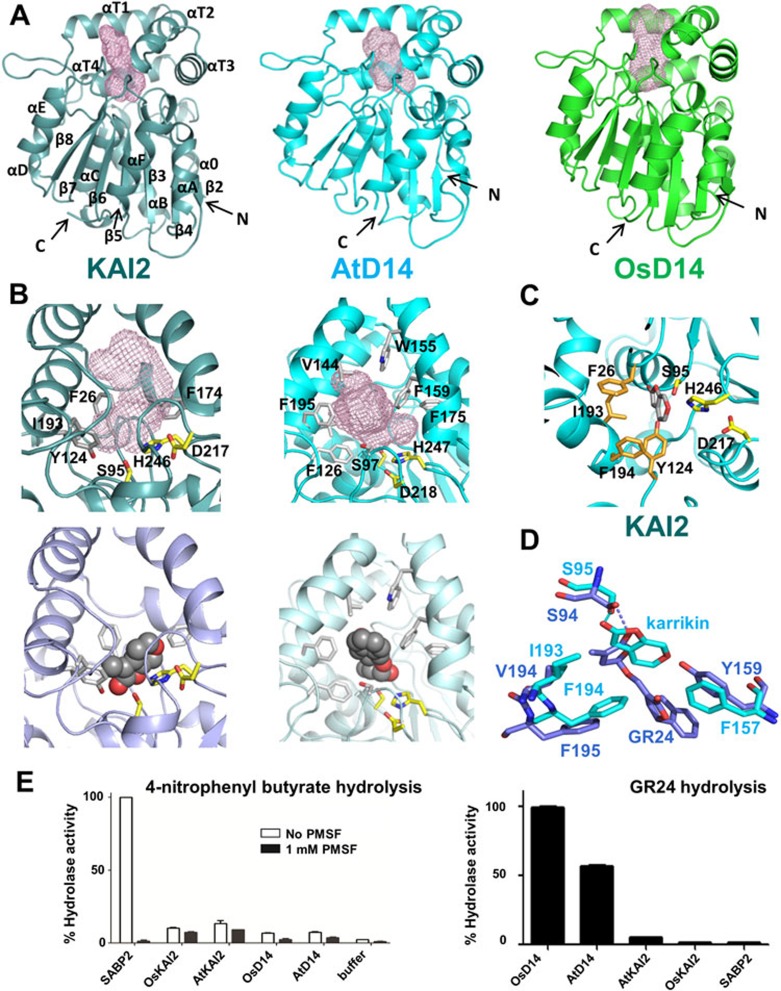
To further understand their functions, we crystallized KAI2 and D14 proteins from Arabidopsis thaliana (AtKAI2 and AtD14) and D14 from rice, Oryza sativa (OsD14), and determined their structures (see Supplementary information, Data S1 and Table S1). The overall structures confirm that they are members of the α/β hydrolase superfamily. All 3 structures share a common fold of a seven-stranded β-sheet (β2-β8) surrounded by five helices (α0, αB-αE) at one side and two (αA and αF) at the other, and a top domain displaying a double layer V-shaped helical fold containing four helices (αT1-αT4) that harbor a substrate-binding pocket (Figure 1A). The two D14 structures are almost superimposable, as well as the recently solved structure of petunia DAD25 (Supplementary information, Figure S3; rmsd < 0.85 Å), confirming that all three proteins are D14 orthologs.
Figure 1.
Structures and activities of KAI2 and D14 proteins. (A) Structure overview of apo KAI2, AtD14, and OsD14. Ligand-binding pockets are indicated as mesh. (B) KAI2 and D14 ligand-binding pockets (top) and docked ligands (bottom). (C) Close-up view of the KAI2 catalytic triad with docked ligand. (D) Structural alignment of ligands and key ligand specificity-conferring residues. (E) Hydrolase activity of KAI2, D14, and SABP2 toward a generic small substrate (left) and GR24 (right).
The catalytic triad residues of the D14 proteins are S97, H247 and D218, and those of KAI2 are S95, H246 and D217, all of which are located at the bottom of the hydrophobic substrate-binding pocket and on the loops following the β4, β7 and β6 strands, respectively (Figure 1A-C). The triad serine residue of hydrolases functions as highly reactive nucleophile that binds to and hydrolyzes substrates, while the histidine and aspartate residues form a charge relay network to increase the nucleophilicity of the serine and to function as acceptor of the serine hydroxyl proton.
Molecular docking of karrikins into the KAI2 pocket (Figure 1B, 1C and Supplementary information, Figure S2) and of GR24 into the AtD14 pocket (Figure 1B) indicated a snug fit of these compounds in pockets lined by bulky aromatic side chains. The lactone carboxyl groups of the docked karrikin and SL are in close proximity to the hydroxyl groups of the triad serine residues (2.9 Å and 2.5 Å, respectively) to allow a nucleophilic attack, supported by the histidine and aspartate charge relay network, that would result in the hydrolysis of the butenolide rings (Figure 1B, 1C and Supplementary information, Figure S4). Ultra performance liquid chromatography (UPLC) combined with ES-MS confirmed the hydrolysis of the GR24 butenolide ring and identified the GR24 ABC-ring and lactone D-ring as final hydrolysis products (Supplementary information, Figures S4 and S6).
When we tried to co-crystallize OsD14 with GR24, we obtained the crystal structure of D14 covalently bound to a GR24 degradation intermediate, 2,4,4,-trihydroxy-3-methyl-3-butenal, which is clearly revealed by the electron density map (Supplementary information, Figure S4A-S4C). The transition state captured in this structure shows the hydroxyl group of S97 attached to C1 of the intermediate, which is further stabilized by water-mediated hydrogen bonds with H247 and Y159 (Supplementary information, Figure S4B). This intermediate suggests that the initial nucleophilic attack causes an electron shift, followed by the addition of a water molecule, to lead to the release of the ABC ring product and the formation of a S97-stabilized open lactone. The latter then converts to the intermediate found in the crystal structure by 1,4-addition of a water molecule to the two conjugated double bonds. The C1 enol tautomer of the transition product can then undergo an intra-molecular Michael addition of the C1 hydroxyl to the carbonyl group at position 4 to form the final closed lactone ring (Supplementary information, Figure S4D) identified by MS (Supplementary information, Figure S6).
Despite that KAI2 has high sequence and structure similarity to the two D14 proteins (Supplementary information, Figure S1), KAI2 is not able to mediate the strigolactone branching signaling, and the D14 proteins do not promote seed germination in response to karrikin exposure7. While the overall structures of these hydrolases are very similar (rmsd ≤ 1.15 Å), their substrate binding pockets differ significantly. KAI2 has a relatively small pocket of 279 Å3 compared to the larger pockets of AtD14 (357 Å3) and OsD14 (432 Å3). In addition, the pocket of KAI2 is constricted in the middle by the inward shift of helix αT4, whose bulky residues I193 (V194 in both D14 proteins) and F194 face into the pocket (Figure 1B, 1D). Overlay of the apo structures of KAI2 and D14 indicates that these two residues of KAI2 would clash with the position of GR24 as docked into the D14 binding pocket. Conversely, the larger binding pocket of the D14 proteins cannot accommodate karrikin as docked into the KAI2 pocket because the KAI2 ligand-binding residue F157 is replaced with a tyrosine in D14, whose hydroxyl group clashes with the oxygen atom of the 6-member pyran ring of karrikin (Figure 1D). Therefore, the positions of 3 bulky hydrophobic residues, I193/IV194, F194/F195, and F157/Y159 are likely the main determinants of ligand specificity.
To biochemically assess hydrolase activity and specificity, we first determined the activities of KAI2 and the D14 proteins towards a small generic hydrolase substrate, 4-nitrophenyl butyrate, and compared them to the activity of SABP2, a methyl salicylate-cleaving α/β hydrolase involved in systemic acquired resistance signaling8,9. Our biochemical assays demonstrated that SABP2 is able to hydrolyze 4-nitrophenyl butyrate, and its enzymatic activity can be entirely blocked by PMSF, a commonly used hydrolase inhibitor (Figure 1E). Despite the structural similarity to SABP2, KAI2 and D14 do not show significant esterase activity for 4-nitrophenyl butyrate, either in the presence or absence of PMSF, and the low activity of KAI2 and D14 are not inhibited by PMSF (Figure 1E). The crystal structure of SABP2 in complex with salicylic acid8 revealed a small binding pocket of 167 Å3, whose entrance is covered by a cap domain (Supplementary information, Figure S5), which can enhance the capture of the substrate during catalysis. In contrast, both KAI2 and D14 have a rigid open entrance to their substrate binding pockets where a bound substrate can leave before being hydrolyzed, which may explain why KAI2 and D14 have much lower activity than SABP2. In contrast to the activities toward 4-nitrophenyl butyrate, only the D14 proteins were able to hydrolyze GR24, demonstrating hormone substrate specificity for this class of signaling hydrolases. However, the GR24 hydrolase activity is extremely low, with a turnover rate of only one GR24 molecule per D14 molecule per three minutes.
D14 and KAI2 are absolutely required for SL and karrikin signaling, respectively, and are functionally linked to the same F-box protein, AtMAX2/OsD3. This pathway is reminiscent of gibberellin signal perception and transduction, in which the catalytically inactive α/β-hydrolase GID1 functions as the hormone receptor to mediate hormone-dependent complex formation between GID1, DELLA transcriptional repressors that function as coreceptors, and the GID2 F-box protein10,11,12. F-box-binding mediated degradation of the DELLA repressors leads then to induction of gibberellin-responsive genes. While SL and karrikins might be precursors of the actual signaling molecules, thought to be generated by D14- and KAI2-catalyzed hydrolysis, the extremely low hydrolase activity and the apparent lack of signaling activity of GR24 hydrolytic products5 suggest that D14 and KAI2 may instead function as hormone receptors in analogy to GID1. Moreover, while endogenous SL occur at low concentrations, D14 alone exhibits only weak substrate-binding affinity, consistent with a possible coreceptor requirement for high-affinity binding. We speculate that D14 and KAI2, which unlike SABP2 lack moveable lids to control access to the ligand-binding pocket, likely associate with coreceptors in the ligand-bound state to retain the ligands in the pocket, thereby increasing ligand/substrate affinity. In analogy with GID1, and consistent with a GR24-mediated Y2H interaction between petunia D14 and MAX25, a ternary receptor-hormone-coreceptor complex might recruit SCFD3/MAX2 for coreceptor ubiquitination and degradation. Association and possible degradation of SL- and karrikin-specific corepressors would therefore provide a simple explanation for D14 and KAI2 signaling through the same F-box protein.
In summary, the structures reported in this study revealed unique pocket topologies as a basis for karrikin and SL signaling specificities, identified the pathway and mechanism of D14-catalyzed GR24 hydrolysis, and further support the likely roles of KAI2 and D14 in karrikin and SL perception.
Acknowledgments
This work was supported by the Jay and Betty Van Andel Foundation, Amway (China), the National Natural Science Foundation of China (NSFC 91217311) and US National Institute of Health (R01GM102545). The atomic coordinates have been deposited in the Protein Data Bank with accession codes listed in Supplementary information, Table S1. We thank staff members at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U and the Life Science Collaborative Access Team of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of APS was supported by the Office of Science of the US Department of Energy, under Contract No. DE-AC02-06CH11357.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Sequence alignment of KAI2 and D14 proteins from rice (Os) and from Arabidopsis (At) with the petunia (Ph) D14 ortholog DAD2, bacterial RbsQ, and SABP2 from tobacco.
Comparison of the docking of karrikin 1 (A) and karrikin 2 (B) into the ligand binding pocket of KAI2.
Structure overlay of the D14 proteins from petunia (red), rice (green), and Arabidopsis (cyan).
The D14 hydrolase mechanism.
Comparison of the conformations of the cap domain of SAPB2 (left, with the part different to D14 colored in magenta), with the top domains of KAI2 (middle, with the part different to SAPB2 colored in green), and AtD14 (right, with the same part in yellow).
UPLC/MS analysis of D14-catalyzed GR24 hydrolysis.
Wild type OsD14 (residues 51–318) was expressed as an N-terminal His6-Sumo fusion protein from pSUMO (LifeSensors).
Statistics of data collection and structure refinement.
References
- Flematti GR, Ghisalberti EL, Dixon KW, et al. Science. 2004. p. 977. [DOI] [PubMed]
- Brewer PB, Koltai H, Beveridge CA.Mol Plant 2012. Nov 15. doi: 10.1093/mp/sss130 [DOI] [PubMed]
- Nelson DC, Scaffidi A, Dun EA, et al. Proc Natl Acad Sci USA. 2011. pp. 8897–8902. [DOI] [PMC free article] [PubMed]
- Kaneko T, Tanaka N, Kumasaka T. Protein Sci. 2005. pp. 558–565. [DOI] [PMC free article] [PubMed]
- Hamiaux C, Drummond RS, Janssen BJ, et al. Curr Biol. 2012. pp. 2032–2036. [DOI] [PubMed]
- Scaffidi A, Waters MT, Bond CS, et al. Bioorg Med Chem Lett. 2012. pp. 3743–3746. [DOI] [PubMed]
- Waters MT, Nelson DC, Scaffidi A, et al. Development. 2012. pp. 1285–1295. [DOI] [PubMed]
- Forouhar F, Yang Y, Kumar D, et al. Proc Natl Acad Sci USA. 2005. pp. 1773–1778. [DOI] [PMC free article] [PubMed]
- Kumar D, Klessig DF. Proc Natl Acad Sci USA. 2003. pp. 16101–16106. [DOI] [PMC free article] [PubMed]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, et al. Nature. 2008. pp. 520–523. [DOI] [PubMed]
- Murase K, Hirano Y, Sun TP, et al. Nature. 2008. pp. 459–463. [DOI] [PubMed]
- Sun TP. Plant Physiol. 2010. pp. 567–570. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of KAI2 and D14 proteins from rice (Os) and from Arabidopsis (At) with the petunia (Ph) D14 ortholog DAD2, bacterial RbsQ, and SABP2 from tobacco.
Comparison of the docking of karrikin 1 (A) and karrikin 2 (B) into the ligand binding pocket of KAI2.
Structure overlay of the D14 proteins from petunia (red), rice (green), and Arabidopsis (cyan).
The D14 hydrolase mechanism.
Comparison of the conformations of the cap domain of SAPB2 (left, with the part different to D14 colored in magenta), with the top domains of KAI2 (middle, with the part different to SAPB2 colored in green), and AtD14 (right, with the same part in yellow).
UPLC/MS analysis of D14-catalyzed GR24 hydrolysis.
Wild type OsD14 (residues 51–318) was expressed as an N-terminal His6-Sumo fusion protein from pSUMO (LifeSensors).
Statistics of data collection and structure refinement.