Abstract
Neuropeptidomics refers to a global characterization approach for the investigation of neuropeptides, often under specific physiological conditions. Neuropeptides comprise a complex set of signaling molecules that are involved in regulatory functions and behavioral control in the nervous system. Neuropeptidomics is inherently challenging because neuropeptides are spatially, temporally and chemically heterogeneous, making them difficult to predict in silico from genomic information. Mature neuropeptides are produced from intricate enzymatic processing of precursor proteins/prohormones via a range of post-translational modifications, resulting in multiple final peptide products from each prohormone gene. Although there are several methods for targeted peptide studies, mass spectrometry (MS), with its qualitative and quantitative capabilities, is ideally suited to the task. MS provides fast, sensitive, accurate, and high-throughput peptidomic analyses of neuropeptides without requiring prior knowledge of the peptide sequences. Aided by liquid chromatography (LC) separations and bioinformatics, MS is quickly becoming a leading technique in neuropeptidomics. This chapter describes several LC-MS analytical methods to identify, characterize and quantify neuropeptides, while emphasizing the sample preparation steps so integral to experimental success.
Keywords: sample preparation, liquid chromatography (LC), mass spectrometry (MS), peptide identification, stable isotope labeling, quantitation
1. Introduction
Acting as signaling molecules, neuropeptides participate in many regulatory and behavioral functions, such as feeding, sleeping, and learning. Neuropeptidomics is the comprehensive analysis of neuropeptides in cells and tissues in the nervous system. Because of the variations inherent to neuropeptide prohormone processing, the presence of post-translational modifications (PTMs), and the intrinsic low relative abundance of neuropeptides, neuropeptidomics methodologies generally demand high sensitivity, great selectivity and a wide dynamic range of detection (1-6). Assisted by the development of enhanced sample preparation techniques and high performance liquid chromatography (HPLC), mass spectrometry (MS) has proven to be particularly useful for neuropeptidomics, primarily due to its capability to unambiguously characterize peptides in complex biological samples without requiring prior knowledge of the peptide sequences (6-7). Since its early success in characterizing thyrotropin-releasing hormone (8), MS has been used to characterize hundreds of putative signaling peptides in a range of animals (9-17). In addition, several MS-based measurement approaches have been developed that enable relative quantitation of peptide levels in biological samples, including correlating peptide level changes to specific traits, conditions and even behaviors (18-20).
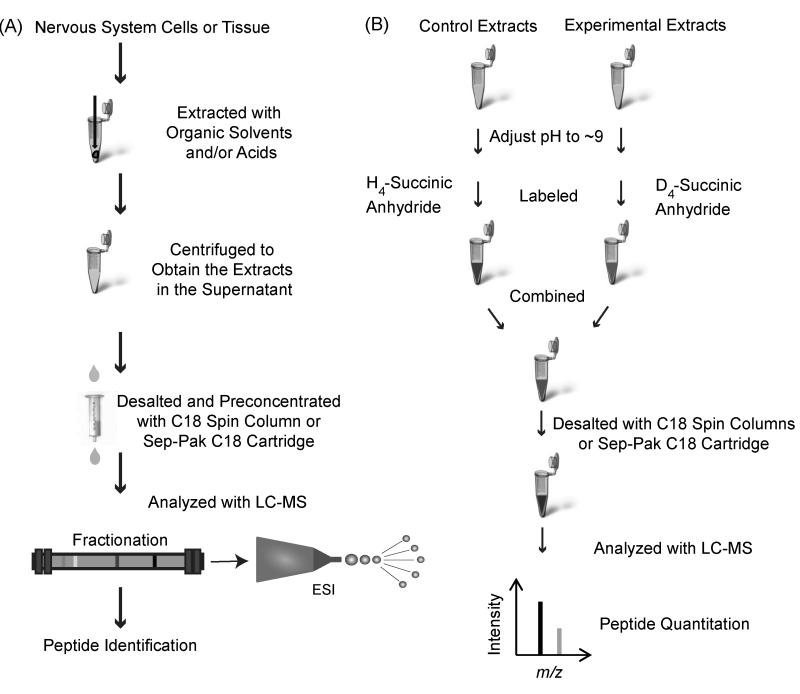
By implementing the appropriate sample preparation protocols and LC-MS platforms, MS-based neuropeptidomics analysis can provide a wealth of information about the cells/tissues under investigation: identification and characterization (21-22), distribution and location (23-24), temporal and spatial release dynamics (25), and relative level changes (18-20, 26-28). Specifically, this chapter describes generalized MS-based approaches to achieve peptide identification and relative quantitation, while allowing flexibility in sample type and size. The protocol steps describe the mechanical disruption of cells or tissue by homogenization, extraction of peptides with organic solvents and/or acids, desalting and pre-concentration, and LC fractionation followed by peptide measurement, sequencing, identification (Fig. 1A and Fig. 2), and quantitation (Fig. 1B and Fig. 3).
Fig. 1.
Workflow of sample processing (A) prior to LC-MS data analysis in a typical MS-based neuropeptidomics approach for peptide identification, and (B) sample processing and succinic anhydride labeling prior to LC-MS data analysis for MS-based neuropeptide quantitation.
Fig. 2.
Typical LC-ESI-IT MS results using the animal model Aplysia californica. Left: a peptide was identified from the existing prohormone database via MS/MS. Right: de novo sequencing of unassigned MS/MS spectra facilitate the refinement of the existing prohormone database. The sequences in italics indicate the signal peptide of the prohormone.
Fig. 3.

LC-ESI-Q-TOF MS spectra of bradykinin standards labeled with isotopic succinic anhydride tags (molecular weight, 1059.55; masses after labeling 1159.62(H)/1163.66(D); charge state +2). (A). Forward labeling is within 4% of the expected ratio, 2:1. (B). Reverse labeling is within 18% of the expected ratio, 1:2.
Scrupulous sampling, optimized LC separation, and appropriate MS detection are essential to successful neuropeptidomics analyses. We emphasize sampling because the detection of neuropeptides in complex biological matrices can be compromised by the presence of salts, lipids and numerous protein degradation products. Protocols specific to LC-MS characterization will also be described here (see Chapter 6 for additional details on HPLC peptide separation). Although there are a number of LC-MS platforms available, we describe capillary LC coupled to electrospray ionization (ESI)-ion trap (IT) MS for peptide measurement, sequencing, and identification. ESI-IT MS is commonly used for peptide identification due to its high sensitivity (29).
With the advent of effective labeling reagents (26-27, 30), quantitative analysis of peptide levels became possible. Using stable isotopic labeling, neuropeptides from experimental and control samples are chemically modified with the addition of structurally identical functional groups that incorporate different isotopes of the same element; thus, after labeling, neuropeptides from the control and sample treatments are distinguished by mass (Fig. 1B), and the relative change in peak heights between these conditions are obtained for each peptide pair. Common isotopic reagents target the N-terminus and the ε-amino group of Lys (e.g., d0 or d6 acetic anhydride, d0 or d4 succinic anhydride, and d0 or d9 trimethylammoniumbutyrate) (26-27), or the C-terminus (e.g., d0- or d3-methanol) (30). We elaborate on a labeling protocol that uses the commercially available reagent succinic anhydride for peptide quantitation because it produces stable labeling products with measurable mass differences between light and heavy forms (although other reagents are available (7, 26-28)). Nano-scale UPLC coupled to ESI quadrupole time-of-flight (Q-TOF) MS is used to measure the relative peptide levels. This approach has been selected because the high resolution and speed of ESI-Q-TOF enhances the ability to differentiate the labeled peptides (29) in complex biological samples.
2. Materials
2.1 Peptide Extraction
HPLC grade acetone (Fisher Scientific, Pittsburgh, PA).
Deionized water from a Milli-Q filtration system (Millipore, Bedford, MA).
Hydrochloric acid (HCl) (Fisher Scientific).
Acidified acetone (acetone/water/hydrochloric acid = 40/6/1, v/v/v).
Homogenizer: pellet pestle cordless motor and pellet pestles with microtubes (Kimble Chase, Vineland, NJ).
Centrifuge 5804R with rotor, 30 × 3.75G (Eppendorf, Hauppauge, NY).
Savant SpeedVac Vacuum Concentrator (Thermo Scientific, Waltham, MA).
LC-MS grade acetonitrile (ACN) with 0.1% formic acid (FA) and 0.01% trifluoroacetic acid (TFA) (Sigma-Aldrich, St. Louis, MO).
LC-MS grade water (H2O) with 0.1% FA and 0.01% TFA (Sigma-Aldrich).
Reconstitution solution: ACN/H2O (2/98, v/v) with 0.1% FA and 0.01% TFA.
2.2 Peptide Desalting and Pre-Concentration
Microcon centrifugal filter unit YM-10 membrane, NMWCO 10 kDa (Millipore).
Deionized water (e.g., from a Milli-Q filtration system (Millipore)).
C18 Spin columns (Pierce, Rockford, IL) or Sep-Pak C18 cartridges (Waters, Milford, MA).
Activating solution: ACN/H2O (50/50, v/v) with 0.1% FA and 0.01% TFA.
Equilibrating and desalting solution: ACN/H2O (5/95, v/v) with 0.1% FA and 0.01% TFA.
Eluting solution: ACN/H2O (70/30, v/v) with 0.1% FA and 0.01% TFA.
2.3 Peptide Fractionation and Identification
HPLC system: Micromass™ CapLC system (Micromass, Manchester, UK) with manual injector (Valco Instruments Co. Inc., Houston, TX) and OPTI-PAK® Trap Column 0.5 μL C18 (Optimize Technologies, Inc., Oregon City, OR).
HPLC column: LC Packings™ 300 μm i.d. × 15 cm, C18 PepMap100, 3 μm particle size, 100 Å pores (LC Packings, San Francisco, CA).
Solvent A: ACN/H2O (5/95, v/v) with 0.1% FA and 0.01% TFA as mobile phase.
Solvent B: ACN/H2O (95/5, v/v) with 0.1% FA and 0.01% TFA as mobile phase.
Solvent C: ACN/H2O (2/98, v/v) with 0.1% FA and 0.01% TFA as loading solvent.
ESI-IT MS system: HCTultra PTM Discovery System™ (Bruker Daltonics, Billerica, MA).
Software: Bruker Daltonics Hystar to integrate the HPLC and the ESI-IT MS, and to establish the HPLC method.
Software: Bruker Daltonics EsquireControl to set up the ESI-IT MS acquisition method and control the ESI-IT MS.
Software: Bruker Daltonics DataAnalysis to generate the ESI-IT MS and tandem MS (MS/MS) spectra for peptide identification.
Online search engine: Mascot (www.matrixscience.com, Matrix Science, London, UK) to search the MS/MS spectra against peptide sequences in the selected database.
Software: Peaks Studio (Bioinformatics Solutions Inc., Waterloo, ON, Canada) as an alternative algorithm for database searching and automatic de novo sequencing.
2.4 Peptide Quantitation with Succinic Anhydride (SA) Labeling
Peptide extraction materials (see Subheading 2.1).
Solution of 1 M NaOH in deionized water.
Solution of 1 M phosphate buffer in deionized water (pH 9.5).
Labeling reagents: 2 M H4-succinic anhydride (SA-H) in dimethyl sulfoxide (DMSO), and 2 M D4-succinic anhydride (SA-D) in DMSO (Sigma Aldrich).
pH paper.
Solution of 2.5 M glycine in deionized water.
Solution of 2 M hydroxylamine in DMSO.
Sample clean-up and pre-concentration materials (see Subheading 2.2).
UPLC system: nanoAcquity UPLC with autosampler (Waters).
UPLC column: nanoAcquity UPLC Atlantis dC18 Column, 75 μm i.d. × 15 cm, 3 μm particle size, 100 Å pores (Waters).
Solvent A: H2O with 0.1% FA and 0.01% TFA as mobile phase.
Solvent B: ACN with 0.1% FA and 0.01% TFA as mobile phase.
ESI-Q-TOF MS system: API Premier US™ (Waters).
Software: MassLynx (Waters) to set up both UPLC separation and MS acquisition methods, control the UPLC-ESI-Q-TOF MS system, and generate MS spectra.
3. Methods
Mass spectrometry-based qualitative and quantitative analyses for neuropeptidomics includes several steps. The first involves peptide extraction from a pool of many cells or a tissue region, with the goal of having a sample large enough to ensure the peptides are detectable. However, larger samples do not always ensure success. In some cases, the peptides are highly concentrated and localized into a small region within the tissue. Another important factor in sample preparation is the reduction of peptides attributable to degradation of high-abundance proteins. To prevent postmortem protein degradation, extraction procedures have been developed to deactivate the protein proteases. Protease activity can be reduced by using a strong, acidified organic solution for extraction (21), by microwaving the samples immediately after the dissection (28), or by sacrificing the animals with focused microwave irradiation (3). Here, we describe a peptide extraction protocol using fresh tissues placed in an acidified acetone solution. Larger proteins in the extracts are filtered out based on molecular size. The filtrates are further desalted and pre-concentrated with a reversed solid phase column before being analyzed by LC-ESI-IT MS and MS/MS. Software and search engines containing either public precursor protein databases or in-house prohormone databases are used specifically for neuropeptide/hormone identification. The advantage of a using a tailored prohormone database is that more targeted results are obtained, especially if likely prohormone PTMs are included in the library and thus can be predicted using expert systems such as NeuroPred (31). For quantitative analysis, we detail the peptide labeling reaction with succinic anhydride, and introduce UPLC-ESI-Q-TOF MS analysis for characterizing relative peptide level changes.
3.1. Peptide Extraction
Homogenize the biological samples (e.g., isolated cells or tissue from the nervous system) in acidified acetone (see Notes 1 and 2).
Centrifuge the homogenate at 14,000 rpm at ambient temperature in a 1.5 mL microcentrifuge tube for at least 5 min or until all the solid debris is precipitated.
Transfer the supernatant into a different microcentrifuge tube.
Vacuum dry the supernatant in a SpeedVac concentrator.
Reconstitute the samples in 20--40 μL of ACN/H2O (2/98, v/v) with 0.1% FA and 0.01% TFA, and vortex well.
Centrifuge the reconstituted samples at 14,000 rpm at ambient temperature for at least 5 min or until the solid debris is precipitated, and save the supernatant.
3.2. Peptide Desalting and Pre-Concentration
Filter the supernatant through a Microcon YM-10 cut-off filter to remove proteins larger than 10 kDa from the extract (see Note 3).
Desalt the samples with C18 Spin columns or Sep-Pak C18 cartridges following the manufacturer’s instructions for the corresponding products (see Note 4).
Vacuum dry the elution solution from the desalting step in a SpeedVac concentrator.
Reconstitute the samples in 20--25 μL of ACN/H2O (2/98, v/v) containing 0.1% FA and 0.01% TFA, vortex well, and centrifuge.
Place the samples in a 4°C refrigerator for short-term storage or in a −20°C freezer for long-term storage.
3.3. Peptide Fractionation and Identification
Thaw the frozen samples at ambient temperature if necessary.
Vortex the samples for 5 min.
Centrifuge the samples at 14,000 rpm at ambient temperature for 5 min.
Prepare the LC-MS system by first washing the injection loop a few times with an organic solution (e.g., ACN or MeOH), followed by deionized water.
Set up the HPLC method: set the isocratic flow of Solvent C at a flow rate of 8 μL/min for 5 min to load the sample onto the C18 trap column, and the separation gradient of the mobile phases at a flow rate of 2.5 μL/min, starting from 2% to 80% of solvent B in 40--80 min (determined by the sample complexity), staying at 80% of solvent B for 5--10 min to elute any hydrophobic compounds, then ramping back to 2% of B, and maintaining at 2% of B for 10--15 min to equilibrate the column (see Note 5). If UV detection is available prior to MS, the wavelength is generally set up at 214 nm for peptide detection.
Equilibrate the HPLC system with a blank run (no sample injection) to make sure the column and lines are clean.
Inject the samples at a volume not greater than the HPLC injection loop size into the HPLC system with a syringe.
Separate the samples using a reversed phase column with the established HPLC method.
The Hystar software triggers the ESI-IT MS acquisition at the same time as the HPLC separation. Data-dependent MS/MS acquisition is activated once the peptide ion peaks in the MS scan reach a certain threshold, as indicated in the MS acquisition method in the EsquireControl. The MS acquisition method also includes the selection of 2--3 precursor ions with highest intensities for MS/MS acquisitions within 1 min, and dynamic exclusion of already fragmented precursor ions after 2--3 iterations within 1 min. The MS scan is generally in the range of 300--2000 m/z, and MS/MS is typically performed in the range of 50--2000 m/z. The preferred mass-to-charge ratio (m/z) and charge state (z) can also be defined.
Use the Bruker Daltonics DataAnalysis software to display the MS and MS/MS spectra, and to find compounds for identification by selection and deconvolution of the MS/MS spectra. A typical LC-MS result is shown in Fig. 2. The processed MS/MS data can be further converted to a universal format (e.g., mgf directly through DataAnalysis, or mzMXL found at http://ionsource.com/functional_reviews/Compass Xport /CompassXport.htm).
Import the converted data files to the selected search engine (e.g., Mascot or PEAKS Studio).
Set the search parameters, such as mass tolerance (see Note 6) and PTMs (see Note 7), in the search engine for both database searching and automatic de novo sequencing, if available.
Search against public or in-house databases and initiate automatic de novo sequencing at the same time, if available (Fig. 2).
Manually check all of the positive search results on peptide identities for correct peak assignments, reasonable cleavage sites, and PTMs (see Notes 6--8).
For MS/MS data that do not result in positive sequence assignments via database search or automatic de novo sequencing, conduct manual de novo sequencing to generate sequence tags by matching the space between two fragment peaks to an amino acid with or without neural losses (e.g., −18 Da for H2O loss and −17 Da for NH3 loss). The sequence tags can be used to search against genome databases with the assistance of bioinformatics (9, 32), as shown in Fig. 2.
3.4. Peptide Quantitation with Succinic Anhydride (SA) Labeling
Peptide extraction is conducted for all the samples (see Subheading 3.1 and Note 9).
Adjust the pH of the control and the experimental sample solutions to ~9 using 1 M phosphate buffer and 1 M NaOH.
Add equal amounts of the labeling reagents (2 M SA-H and SA-D) to the both the control and the experimental samples (see Note 10).
Vortex the mixed reactants and centrifuge.
Incubate the reactants at ambient temperature for 10 min.
Adjust the pH of the sample solutions to ~9 with the 1 M NaOH solution.
Repeat steps 3 through 6 at least five times for each sample.
Quench extra SA in the samples with the 2.5 M glycine solution for 40 min at room temperature before combining the control and the experimental samples (see Note 11).
Use a Microcon YM-10 cut-off filter to remove proteins larger than 10 kDa from the combined sample solutions (see Note 3).
Adjust the pH of the combined sample solutions to ~9 using the 1 M NaOH solution.
Add 2 M hydroxylamine solution into the combined sample solutions (see Note 11).
Incubate the solutions at ambient temperature for 30 min.
Desalt and pre-concentrate the labeled samples with C18 Spin columns or Sep-Pak C18 cartridges (see Subheading 3.2, Steps 2--5).
Analyze the samples with UPLC-ESI-Q-TOF. The UPLC method is similar to the HPLC method described above in Subheading 3.3, Step 5 with autosampler; here, inject the samples using Solvent A at a flow rate of 400 nL/min to load the samples directly onto the nano-column, and a mobile phase set to a flow rate of 250 nL/min to separate the samples. MS scans are acquired in the range of 300--2000 m/z.
Use MassLynx to display the MS spectra; the original peptide mass is determined by converting the experimentally measured mass of the labeled peptide into that expected from an unlabeled peptide (see Eq. 1). The peak assignment is performed by matching the calculated original peptide mass with the known peptide mass previously identified under Subheading 3.3 and using peptide elution orders to confirm the assignment.
Generate extracted ion chromatograms for the MS spectra of both the light and the heavy forms of the labeled peptides using MassLynx, and sum the intensities of the mass spectra over the entire elution time for the paired peptides, respectively, for quantitation. The relative intensities or areas of paired peaks in the MS scan reflect the relative amounts of peptides in the control and the experimental samples. With SA-H/SA-D as labeling reagents (Fig. 3), light and heavy isotope-labeled peak pairs are separated by 4 Da for singly charged ions and 2 Da for doubly charged ions.
Perform statistical analysis (e.g., Student’s t-test) to identify significant differences between the control and experimental samples and evaluate data consistency between technical/biological replicates.
Notes
Alternative extraction solutions that work well for peptides include acidified methanol (methanol/H2O/HAc = 90/9/1, v/v/v) (9), (methanol/H2O/HAc = 90/1/9, v/v/v) (33) or 10 mM HCl (26-28). To improve the efficiency of peptide extraction, thorough homogenization of biological samples and the combination of supernatants from a multi-stage approach using the same or different extraction solutions can be employed (15-16).
It is recommended that a minimum volume of extraction solution be used to enable complete homogenization without over-diluting the samples. For example, Hatcher et al. (21) used 40 μL of acidified acetone to homogenize SCN punches in rats, and Che et al. (28) suggested 100 μL of 10 mM HCl per pituitary be used for peptide extraction in mice.
The cut-off filter should be rinsed with deionized water or other buffers before use, as stated in the manufacturer’s instructions, to prevent discharge of polymers into the samples. Also, the filter needs to be washed multiple times during sample filtration, followed by combining the individual filtrate fractions together to improve sample recovery.
Desalting and pre-concentration with C18 Spin columns or Sep-Pak C18 cartridges requires activation and equilibration of the sorbent, loading the samples multiple times to improve sample recovery, washing away the salts, and eluting out the peptides. Attention should be paid to prevent drying of the packing materials in the spin columns and/or cartridges.
Two-dimensional LC may be needed for complicated samples, in which different separation principles (e.g., ion exchange or reversed phase), different mobile phases (e.g., ACN/water or methanol/water), and different ion pairing reagents (FA or acetic acid) can be used (9, 33-34).
Mass tolerance is determined by both the MS platform and calibration selected. It is generally suggested to set the mass tolerance at 0.5 Da for both ESI-IT MS and MS/MS.
Common modifications include C-terminal amidation, N-terminal pyroglutamate formation from glutamine or glutamic acid, and disulfide bonding. Other modifications such as methionine oxidation, N-terminal acetylation, tyrosine sulfation, and phosphorylation of tyrosine, threonine, and serine can be considered as they are sometimes encountered with neuropeptides.
Neuropeptide precursor processing involves the basic site cleavages of the prohormones, although many basic sites are not cleaved (31). Therefore, putatively identified neuropeptides that form through the cleavages of the prohormone at KR, RR, RK, KK, or RXnR (n = 0, 2, 4, or 6), or other predicted cleavages, are more likely to be correct assignments (1, 4, 6, 33). In addition, at least three consecutive amino acid matches are required for a positive identification.
At least three biological replicates are required to obtain statistics for quantitative experiments. Generally, simultaneous forward (Sample A with SA-H and Sample B with SA-D) and reverse labeling (Sample B with SA-H and Sample A with SA-D) are recommended. It is essential to simultaneously extract all the samples using the same extraction approach, and minimize the sample loss for more consistent results.
Usually, 1--2 μL of labeling reagent is added at a time. The desired amount of labeling reagent for complete reactions is ~1000 times more than the amount of peptides in the samples.
The total moles of glycine or hydroxylamine used here should equal that of added SA in the samples.
| Eq. 1 |
M is the calculated original peptide mass; (m/z)H and (m/z)D are the monoisotopic mass-to-charge ratios of the light and heavy form peptides, respectively; z is the ion charge; L is the number of labeled tags.
Acknowledgments
This material is based upon work supported by the National Science Foundation (NSF) under Award No. CHE-0526692, by Award No. NS031609 from the National Institute of Neurological Disorders and Stroke (NINDS) and Award No. DA018310 from the National Institute On Drug Abuse (NIDA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF, NIDA, NINDS or National Institutes of Health.
References
- 1.Hook V, Funkelstein L, Lu D, et al. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen H, Engelbrecht J, Brunak S, et al. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Svensson M, Sköld K, Svenningsson P, et al. Peptidomics-based discovery of novel neuropeptides. J Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 4.Fricker LD. Neuropeptide-processing enzymes: applications for drug discovery. AAPS J. 2005;7:E449–455. doi: 10.1208/aapsj070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakubowski JA, Kelley WP, Sweedler JV. Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: an important component of conotoxin discovery. Toxicon. 2006;47:688–699. doi: 10.1016/j.toxicon.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 7.Fricker LD, Lim J, Pan H, et al. Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 8.Burgus R, Dunn TF, Desiderio D, et al. [Molecular structure of the hypothalamic hypophysiotropic TRF factor of ovine origin: mass spectrometry demonstration of the PCA-His-Pro-NH2 sequence] C R Acad Sci Hebd Seances Acad Sci D. 1969;269:1870–1873. [PubMed] [Google Scholar]
- 9.Hummon AB, Richmond TA, Verleyen P, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314:647–649. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Kelley WP, Billimoria CP, et al. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 11.Romanova EV, Roth MJ, Rubakhin SS, et al. Identification and characterization of homologues of vertebrate beta-thymosin in the marine mollusk Aplysia californica. J Mass Spectrom. 2006;41:1030–1040. doi: 10.1002/jms.1060. [DOI] [PubMed] [Google Scholar]
- 12.Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 13.Predel R, Wegener C, Russell WK, et al. Peptidomics of CNS-associated neurohemal systems of adult Drosophila melanogaster: a mass spectrometric survey of peptides from individual flies. J Comp Neurol. 2004;474:379–392. doi: 10.1002/cne.20145. [DOI] [PubMed] [Google Scholar]
- 14.Fogli A, Bulet P. Peptidomics analysis of lymphoblastoid cell lines. Methods Mol Biol. 2010;615:247–257. doi: 10.1007/978-1-60761-535-4_19. [DOI] [PubMed] [Google Scholar]
- 15.Sturm RM, Dowell JA, Li L. Rat brain neuropeptidomics: tissue collection, protease inhibition, neuropeptide extraction, and mass spectrometric analysis. Methods Mol Biol. 2010;615:217–226. doi: 10.1007/978-1-60761-535-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bora A, Annangudi SP, Millet LJ, et al. Neuropeptidomics of the supraoptic rat nucleus. J Proteome Res. 2008;7:4992–5003. doi: 10.1021/pr800394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamanaka Y, Park D, Yin P, et al. Transcriptional orchestration of the regulated secretory pathway in neurons by the bHLH protein DIMM. Curr Biol. 2010;20:9–18. doi: 10.1016/j.cub.2009.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Che FY, Vathy I, Fricker LD. Quantitative peptidomics in mice: effect of cocaine treatment. J Mol Neurosci. 2006;28:265–275. doi: 10.1385/JMN:28:3:265. [DOI] [PubMed] [Google Scholar]
- 19.Brockmann A, Annangudi SP, Richmond TA, et al. Quantitative peptidomics reveal brain peptide signatures of behavior. Proc Natl Acad Sci U S A. 2009;106:2383–2388. doi: 10.1073/pnas.0813021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che FY, Eipper BA, Mains RE, et al. Quantitative peptidomics of pituitary glands from mice deficient in copper transport. Cell Mol Biol. 2003;49:713–722. [PubMed] [Google Scholar]
- 21.Hatcher NG, Atkins N, Jr., Annangudi SP, et al. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci U S A. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernay B, Gaillard M-C, Guryča V, et al. Discovering new bioactive neuropeptides in the striatum secretome using in vivo microdialysis and versatile proteomics. Mol Cell Proteomics. 2009;8:946–958. doi: 10.1074/mcp.M800501-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman TA, Rubakhin SS, Romanova EV, et al. MALDI mass spectrometric imaging using the stretched sample method to reveal neuropeptide distributions in Aplysia nervous tissue. Anal Chem. 2009;81:9402–9409. doi: 10.1021/ac901820v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatcher NG, Richmond TA, Rubakhin SS, et al. Monitoring activity-dependent peptide release from the CNS using single-bead solid-phase extraction and MALDI TOF MS detection. Anal Chem. 2005;77:1580–1587. doi: 10.1021/ac0487909. [DOI] [PubMed] [Google Scholar]
- 26.Che FY, Fricker LD. Quantitation of neuropeptides in Cpe(fat)/Cpe(fat) mice using differential isotopic tags and mass spectrometry. Anal Chem. 2002;74:3190–3198. doi: 10.1021/ac015681a. [DOI] [PubMed] [Google Scholar]
- 27.Che FY, Fricker LD. Quantitative peptidomics of mouse pituitary: comparison of different stable isotopic tags. J Mass Spectrom. 2005;40:238–249. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- 28.Che FY, Lim J, Pan H, et al. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Kinter M, Sherman NE. Protein sequencing and identification using tandem mass spectrometry. John Wiley & Sons, Inc.; New York: 2000. [Google Scholar]
- 30.Goodlett DR, Keller A, Watts JD, et al. Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun Mass Spectrom. 2001;15:1214–1221. doi: 10.1002/rcm.362. [DOI] [PubMed] [Google Scholar]
- 31.Southey BR, Amare A, Zimmerman TA, et al. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006;34:W267–272. doi: 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amare A, Hummon AB, Southey BR, et al. Bridging neuropeptidomics and genomics with bioinformatics: Prediction of mammalian neuropeptide prohormone processing. J Proteome Res. 2006;5:1162–1167. doi: 10.1021/pr0504541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowell JA, Heyden WV, Li L. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res. 2006;5:3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 34.Holm A, Storbråten E, Mihailova A, et al. Combined solid-phase extraction and 2D LC–MS for characterization of the neuropeptides in rat-brain tissue. Anal Bioanal Chem. 2005;382:751–759. doi: 10.1007/s00216-005-3146-z. [DOI] [PubMed] [Google Scholar]