Abstract
This paper reports on the development of a lens-less and image-sensor-less micro-electro-fluidic (MEF) approach for real-time monitoring of the locomotion of microscopic nematodes. The technology showed promise for overcoming the constraint of the limited field of view of conventional optical microscopy, with a relatively low cost, good spatial resolution, and high portability. The core of the device was microelectrode grids formed by orthogonally arranging two identical arrays of microelectrode lines. The two microelectrode arrays were spaced by a microfluidic chamber containing a liquid medium of interest. As a nematode (e.g., Caenorhabditis elegans) moved inside the chamber, the invasion of its body parts into some intersection regions between the microelectrodes caused changes in electrical resistance of these intersection regions. The worm's presence at or absence from a detection unit was determined by a comparison between the measured resistance variation of this unit and a pre-defined threshold resistance variation. An electronic readout circuit was designed to address all detection units and read out their individual electrical resistance. By this means, it was possible to obtain the electrical resistance profile of the whole MEF grids, and thus, the physical pattern of the swimming nematode. We studied the influence of a worm's body on the resistance of an addressed unit. We also investigated how the full-frame scanning and readout rate of the electronic circuit and the dimensions of a detection unit posed an impact on the spatial resolution of the reconstructed images of the nematode. Other important issues, such as the manufacturing induced initial non-uniformity of the grids and the electrotaxic behaviour of nematodes, were also studied. A drug resistance screening experiment was conducted by using the grids with a good resolution of 30 × 30 μm2. The phenotypic differences in the locomotion behaviours (e.g., moving speed and oscillation frequency extracted from the reconstructed images with the help of software) between the wild-type (N2) and mutant (lev-8) C. elegans worms in response to different doses of the anthelmintic drug, levamisole. The locomotive parameters obtained by the MEF grids agreed well with those obtained by optical microscopy. Therefore, this technology will benefit the whole-animal assays by providing a structurally simple, potentially cost-effective device capable of tracking the movement and phenotypes of important nematodes in various microenvironments.
Introduction
The microscopic nematode C. elegans is an important genetic model to address fundamental questions in developmental biology, neurobiology, and behavioural biology.1,2 The locomotion behaviours of C. elegans are under complex neuronal regulation and affected by a plethora of factors such as chemicals, temperature, light, electric field, and age.3 Tracking and analysis of the locomotive parameters (e.g., travelled distance, speed, amplitude, and oscillation frequency) of C. elegans and other important microscopic nematodes is crucial to provide the mechanistic correlation between the genotype and phenotype of the nematodes under various environmental conditions.4 Optical microscopic imaging technique is essential for observing and extracting movement information from live nematodes.5–11 Conventional bench-top microscope, along with a digital camera and tracking software program, is a popular means of detecting the locomotive parameters of microscopic nematodes. The optical imaging system and the worm test vials or plates are independent of each other. This microscope-camera setup can provide very detailed information about the behaviours of nematodes. But, the relatively high cost and large footprint of the setup are not well suited for integration into a portable system-on-chip device. Besides, the limited field of view of the objective lens in the microscope makes it often difficult to simultaneously monitor multiple experiments in multi-well culture plates without an automated, high-resolution motorized moving stage.12 This may become problematic when quantitative measurements are needed on a large number of nematodes at a single nematode resolution. Therefore, there is a great need of developing a cost-effective and structurally simple detection mechanism that potentially will have no any limitation to the field of view.
Microfluidics is an attractive technology with the potential to streamline workflows and processes in the biomedical and health sciences.13–15 Due to the small size of microscopic nematodes such as C. elegans, there is growing interest in studying the nematodes in the area of microfluidics and lab on a chip.16–41 Many promising microfluidic technologies have been reported for culturing, manipulating, and analysing nematodes, including an automated microfluidic compact disc cultivation system,19 an artificial soil substrate for rapid delivery of fluid-borne stimuli to worms,20 a microfluidic device for directing worm movement and sorting worms based on the electrotaxic effect,21,22 a microdroplet technology for encapsulating individual worms for toxicology bioassays,23,24 a miniature worm clamping device for facilitating imaging and laser-mediated microsurgery,25–32 a microfluidic device for engineering various microenvironments for study of the sensory neuron and behavioral activity of nematodes,33–36 a force sensing pillar array for biomechanical measurements of nematodes,37 a micro-maze for examining the behaviours of nematodes,38 a microchamber array for behaviour-based chemical screening with precise temporal control of stimuli,39 and a microfluidic device capable of recording electrophysiological signals of multiple worm immobilized inside microchannels.40 Recently, we also developed an optofluidic device for detecting muscular force generation of nematodes in response to various chemical environments.41 On the other hand, in order to monitor dynamic behaviours of live microorganisms with low cost and high throughput, several on-chip optical imaging technologies have recently been developed.42–45 A remarkable technology is the optofluidic microscope (OFM).42–44 The microorganism sample of interest is transferred into a channel and then imaged by a complementary metal-oxide-semiconductor (CMOS) or charge-coupled-device (CCD) image sensor chip. An array of small circular apertures is located on top of the image sensor chip, spanning across the whole channel. When the channel is illuminated by an external light source, the sample casts a shadow on the image sensor and the time resolved transmission signal is converted into the spatial information of the sample. Some variations of the OFM device have been developed such as the sub-pixel resolving OFM,45 and the fluorescent OFM with a high submicron resolution.46–48 Another excellent optical imaging technique for microfluidic applications is the digital in-line holography (DILH).49–55 In this technology, a channel device contains the microorganism of interest and is placed between the light source and the recording plane of a CMOS/CCD image sensor. The light source is spatially filtered by a pinhole to increase the coherence length. The scattering wave from the sample interferes with a reference wave from the light source. Thus, a hologram or interference pattern is formed by the superposition of the two wave fronts for digital recording. Another representative technique for optical imaging of microorganisms is the lens-less, wide-field monitoring array based on the shadow imaging principle, while also relies on using a CMOS/CCD image sensor.56,57 The device has been shown to detect and count thousands of individual cells in real time. Despite the considerable progress in the development of compact on-chip imaging-based tracking systems for microfluidic applications, almost all of the existing technologies essentially utilize the optical effects, such as shadow and interference patterns due to the presence of microorganisms, and thus, inevitably require a sophisticated imaging sensor chip, along with a light source, for observation.
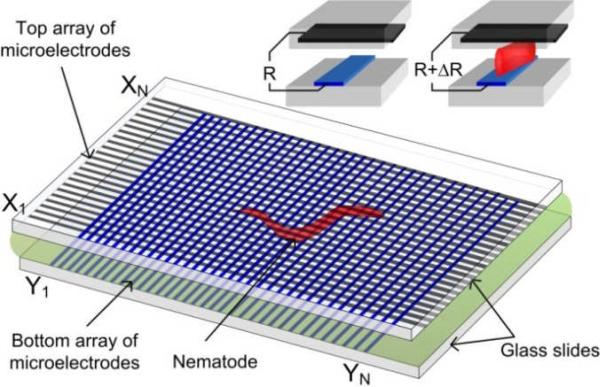
In this paper, we report on the development of a non-optical, integrated device for on-chip detection of the locomotion behaviours of nematodes, with a cost-effective and simple architecture, and fair spatial resolution. The present technology requires no lens or image sensor chip. The core of the device consists of two identical linear arrays of thin-film microelectrodes arranged orthogonally on two glass slides. Each microelectrode array has N periodically spaced electrode strips. Thus, N × N intersection regions are formed between the upper and lower microelectrode arrays. A microfluidic chamber is created between the two glass slides, where a worm under test will swim freely (see a video clip in Electronic Supporting Information). As the nematode moves inside the microfluidic chamber, its body may appear in several intersection regions of the microelectrodes. Because the electrical resistivity of the worm's body is often different from that of the surrounding medium, a resistance change is detected at an intersection region that indicates the presence of part of the worm's body near the intersection region. By electrically addressing all intersection regions and probing their individual resistances in a short time period (before the worm changes its posture) using an electronic scanning and readout circuit, it is possible to get the electrical resistance profile, and thus, the physical pattern, of the moving nematode. Generally, microscopic nematodes such as C. elegans move relatively slow and exhibit a sinusoidal pattern induced by alternating dorsal and ventral muscle contraction. C. elegans swims with an oscillation frequency (fworm) ranging from a fraction of one Hz to several Hz. As discussed later, by increasing the full-frame scanning frequency (fsc) of the electronic circuit, the swimming nematode under test could be treated as a static object during a measurement cycle of tsc = 1 / fsc. This enabled us to obtain the movement patterns or behavioural information of the nematode in a quasi-real time manner. To prevent the electrotaxic behaviour of the worms (moving toward the cathode) and the electrokinetic effects (electrophoresis of the worms and electro-osmosis of the surrounding fluid) within the MEF grids, the electric field applied between two orthogonal microelectrodes for the resistance measurement was set to be lower than the threshold field strength of the electrostatic and electrokinetic effects, which will be discussed later.
Methods and Experimental Details
Theoretical estimation
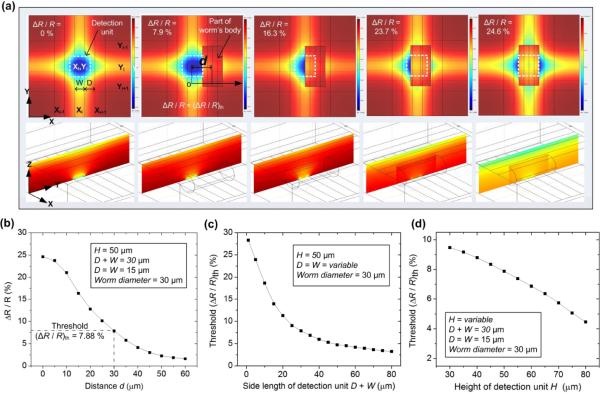
We first theoretically estimated how the presence of part of the worm's body affected the electrical resistance of an addressed intersection region of the microelectrodes. The three-dimensional finite element analysis (FEA) method based on the commercial software COMSOL was employed for this simulation. The variables W and D denote the width of microelectrode and the spacing between two neighbouring microelectrodes, respectively (Fig. 2a). A detection unit or an image pixel was constructed by extending D / 2 from each side of an intersection region (see the white dashed line square in Fig. 2a), covering a square area with the side length of D + W. Thus, the centreline between any two neighbouring microelectrodes on the glass substrate served as the boundary of detection units. A microfluidic chamber was formed between the upper and bottom glass slides. The depth or height of the chamber was set to be H = 50 μm because generally, L1–L4 stage C. elegans larvae is no more than 50 μm in diameter. The side length of detection unit was set to be D + W = 30 μm. For simplification of the FDTD analysis, D = W = 15 μm.
Fig. 2.
(a) Simulated electric field distributions at the X-Y plane (upper row) and Z-Y plane (lower row) of the MEF grids as the worm's body part appears at different locations relative to an addressed detection unit. d is denoted as the planar centre-to-centre distance d between the worm's body part and the intersection region. From left to right: d = ∞, 30 μm, 15 μm, 5 μm, and 0. For simplification, part of a nematode's body was modelled by a 60 μm long, 30 μm diameter cylinder laid on side within the grids. The height or depth of the grids was set to be H = 50 μm. The side length of detection unit was set to be D + W = 30 μm with D = W = 15 μm. The field distributions in the upper row were observed at the planar central plane H / 2 above the lower substrate of the device. The field distributions in the lower row were observed at the Z-Y plane across the spatial centre of the unit (Xi, Yi). (b) Simulated relative resistance variation ΔR / R as a function of d. (c) Simulated threshold relative resistance variation (ΔR / R)th as a function of a side length D + W of a detection unit with D = W and H = 50 μm. (d) Simulated (ΔR / R)th as a function of a depth of the grids or H with D + W = 30 μm and D = W.
Modelling a whole nematode flexibly moving inside the MEF grids was fairly difficult due to the changing body posture of the nematode over time. Here, we used a 60 μm long and 30 μm diameter cylinder (laid on side within the grids) to represent just part of the worm's body interacting with a selected detection unit. The planar coverage area of the cylinder (30 μm × 60 μm) was thus twice that of the detection unit (30 μm × 30 μm). We found that the simulation results with a cylindrical object longer than 60 μm was almost the same as those with the 60 μm long cylinder used here. The electrical resistivity of the worm's body and of the M9 buffer (or the medium in the microfluidic chamber) was ~350 Ω·cm and ~104 Ω·cm, respectively. Fig. 2a shows that as the worm's body part moved toward the spatial centre of the selected detection unit, the electric field distributions inside and outside of this unit significantly changed and the electrical resistance R measured at this unit increased (ΔR > 0). When the body part completely overlapped the intersection region, the simulated maximal relative resistance variation ΔR / R reached 24.6 %. However, to distinguish between the worm's presence at and absence from a detection unit, we defined the threshold relative resistance variation (ΔR / R)th as the critical ΔR / R detected when the worm reached the boundary of the selected unit (see the 2nd panel of Fig. 2a). The simulated value of (ΔR / R)th was ~7.9 %, below which the body part could be interpreted to be outside of the unit, and above which the body part was considered to be inside the unit. As we can see later, there inevitably existed a certain level of the manufacturing induced initial resistance variation between all detection units. Therefore, only if the absolute value of (ΔR / R)th or |(ΔR / R)th| was greater than the original resistance variation, the MEF grids device would be able to detect the presence of a worm's body part at a detection unit. Figs. 2c and 2d show the influence of the side length D + W and the height H of the detection unit on (ΔR / R)th. The results indicate that having a small D + W would lead to a high (ΔR / R)th. This could make it easier and more accurate to detect the presence/absence of a worm's body part. Also, as the depth of the detection unit increased from 30 μm to 80 μm, the simulated (ΔR / R)th was found to significantly decrease from 9.4 % to 4.3 %. Although using a low H was preferable, the minimum H was determined by the body diameter of the worm under test. It is also important to point out that the proposed lens-less, image-sensor-less detection approach can work not only for larval stage (L1–L4) C.elegans, but for young adult and older ones (that can get as thick as ~100 μm) by increasing H.
Fabrication for microelectrode grids
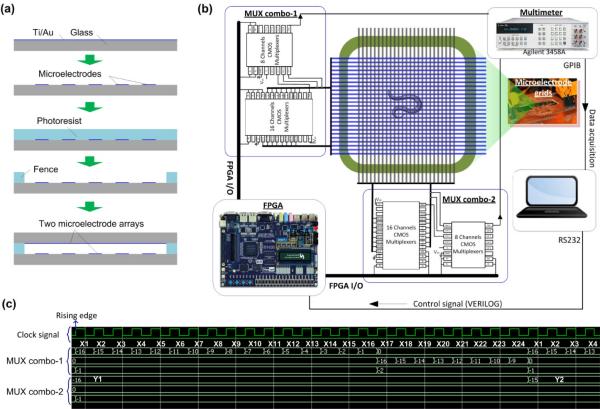
To fabricate a microelectrode array, a 10 nm thick titanium (Ti) layer and a 250 nm thick gold (Au) layer were deposited on a glass slide using e-beam evaporation. The Ti layer was used to improve adhesion between the Au layer and the glass slide. The Ti-Au composite layers were then patterned by conventional photo-lithography, followed by chemical wet etching with Au etchant (GE-8148, Transene). Two identical microelectrode arrays were formed on two respective glass slides by using this method. To control the depth of a microfluidic chamber between the two arrays, SU-8 photoresist (SU-8-50, MicroChem) was used to form a fence along the edges of one glass slide. Lastly, the MEF grids were formed by placing the two microelectrode arrays face-to-face and orthogonal to each other (Fig. 3a).
Fig. 3.
(a) Fabrication processes for the microelectrode grids: deposition of Ti and Au → patterning of microelectrodes → patterning of photoresist fence → assembly of the device by orthogonally stacking two identical microelectrode arrays under microscope. (b) Architecture of the electronic circuit for the grids. (c) Representative digital signals for addressing detection units on the chip.
Full-frame scanning and readout electric circuit
Fig. 3b shows the architecture of the electronic circuit designed for the fabricated MEF grids (24 rows and 24 columns). A field programmable gate array or FPGA (Altera Cyclone II) was used and programmed to respectively control a vertical and a horizontal multiplexer (MUX) combo for addressing individual detection units. The electrical resistance of an addressed detection unit was readout by a multimeter (3458A, Agilent) with 100000 readings s−1. The multimeter communicated with data acquisition and analysis software through the parallel general purpose interface bus (GPIB) transceiver. To adjust timing with the multimeter, the FPGA was interfaced with the software using the series RS232 communication protocol.
To verify the timing properties of the hardware design, the timing simulation was performed on the ModelSim platform. Fig. 3c shows part of the timing result as an example. The horizontal microelectrode Y1 was selected by the MUX combo-2. The vertical microelectrodes from X1 t o X24 were sequentially selected by the MUX combo-1 at each raising edge of the clock. Thus, the 24 detection units ((X1, Y1), (X2, Y1), …, (X24, Y1)) in row Y1 were addressed one by one. This allowed the multimeter to read out their electrical resistance. To obtain the full-frame resistances, this process described above needed to repeat 24 times, taking a period time of tsc = 24 × 24 × TCLK where TCLK was the clock period. We note that TCLK can be adjusted by the hardware (FPGA) and software (Verilog hardware description language). The typical value of TCLK was 10 μs, obtained by dividing the on-board oscillator frequency fosc (Cyclone II FPGA has fosc = 50 MHz) by an integer m (m = 500 was chosen here) via coding in Verilog.
Image reconstruction and parameter extraction
The electrical resistance matrix Ri,j of order N × N was obtained by reading out the resistances of all detection units, where Ri,j(t) (i, j = 1, 2, …, N) represented the resistance of the detection unit at the intersection of line i and row j at time t. Then, the resistance variation matrix ΔRi,j(t) was calculated by ΔRi,j(t) = Ri,j(t) − Ri,j(t0), where Ri,j(t0) was the original resistance matrix at t0 = 0 without worm introduced. Lastly, the relative resistance variation matrix ΔRi,j(t) / Ri,j(t) was obtained for the image reconstruction. To convert the matrix ΔRi,j(t) / Ri,j(t) to a grayscale image, we used the function “mat2gray” in the MATLAB image processing toolbox to apply a linear scaling and offset to the matrix. The minimum value was mapped to 0 (black) and the maximum value was mapped to 1 (white). Other values in the matrix were linearly scaled into corresponding grayscale values. Thus, the grayscale image of the matrix ΔRi,j(t) / Ri,j(t) was formed. To convert the matrix ΔRi,j(t) / Ri,j(t) to a binary image, each element value in the matrix was compared with the threshold (ΔR/R)th. When ΔRi,j(t) / Ri,j(t) < (ΔR / R)th, a binary 0 (black) appeared at the pixel of the reconstructed image. In contrast, when Ri,j(t) / Ri,j(t) > (ΔR/R)th, a binary 1 (white) appeared at the pixel. To display a pseudocolour map of the matrix for an input data set, the function “corrmap.m” of the MATLAB was used.
The motility parameters were analysed by using the worm tracking program we previously developed for a microscope-camera tracking system.41,58 Briefly, the binary images were sequenced and compressed into the Audio Video Interleave (.avi) video format. The .avi video was post-processed by a worm tracking program that was able to extract track signatures and locomotion (e.g., number and duration of stops, and cut-off region) of individual and/or worms. The program analysed a large number of images to recognize a moving object (here worm), and then, extract motility parameters such as amplitude, wavelength, oscillation frequency, body postures, path traversed by the worm, and average moving velocity.
Optical tracking system for comparison purposes
To compare the reconstructed images based on the MEF grids approach and their corresponding optical images, we used a stereo microscope (MZ205, Leica) equipped with a video camera (QICamera) to capture a series of digital images (1392 × 1040 pixels) at a specific time interval of 66.7 ms. The motility parameters of worms were extracted by using the worm tracking programmed aforementioned.
Nematodes and liquid medium
C. elegans worms (wild-type N2 and levamisole resistant ZZ15 lev-8) were obtained from the Caenorhabditis Genetics Centre at University of Minnesota (St. Paul, MN). They were cultivated at 25 °C on Nematode Growth Medium plates seeded with Escherichia coli OP50 bacteria. For the experiments, the worms were picked using a sterilized platinum wire and transferred to the microfluidic chamber through an opening in the top glass slide of the MEF grids.
To test how nematodes changed their locomotion behaviours in the anthelmintic drug levamisole. The levamisole solutions with different concentrations were prepared by dissolving levamisole stock solution with appropriate amounts of the M9 buffer solution. The recipe for the M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 mL 1 M MgSO4, and H2O to 1 L) was a standard recipe taken from the Wormbook.59
Resistivity of nematodes
In the FEA simulation mentioned above, the electrical resistivity of nematodes was set to be 350 Ω·cm. This resistivity value was estimated by inversely calculating from the measured total resistance of a detection unit when the worm's body was positioned at the planar spatial centre of the detection unit (as we will see later in Fig. 5). Specifically, different values of electrical resistivity for the worm's body were fed into the FEA software COMSOL to approach the measured total resistance of the detection unit. When the discrepancy between the simulated and experimental resistance of the detection unit was less than ± 1 %, the corresponding resistivity of the worm used in the simulation was regarded as the actual resistivity of the worm. It should be noted that no obvious change was found in the body resistivity of the worms at different stages (L1–L4).
Fig. 5.

(a) Experimental result of ΔR / R as a function of d. d is the planar centre-to-centre distance between the worm's body part and the intersection region. Two insets show the close-ups of a nematode swimming inside the MEF grids. The blue dashed lines outline part of the worm's body parts. X and Y error bars represent standard deviations of the mean value of d and ΔR / R, respectively, from 60 resistance data obtained at 5 detection units. (b) Histogram of the distribution of (ΔR / R)th in the MEF grids. The distribution was plotted based on the value of (ΔR / R)th obtained at 226 grids.
Results and Discussion
Fig. 4 shows the original distribution of the electrical resistance of the 24 × 24 MEF grids with the M9 buffer as a medium but with no worm introduced. The microelectrodes used here were 15 μm wide and 15 μm spaced. The readout time for this whole resistance matrix was tsc = 5.76 ms at fsc = 174 Hz. The result shows that the initial relative variation between the maximum and minimum resistance was ~3.3 %, reflecting the manufacturing-induced non-uniformity of the microelectrode geometry and dimensions.
Fig. 4.

Initial electrical resistance distribution of the fabricated 24 × 24 grids with W = D = 15 μm. No worm was introduced to the grids.
To obtain an actual value of (ΔR / R)th for a nematode C. elegans swimming in the M9 buffer solution, a L3-stage worm (~30 μm diameter, and ~591 μm long) was introduced to the MEF grids operating at fsc = 174 Hz. Particularly, we examined the electrical resistances detected at different detection units near the nematode at a time point. The distance between an addressed unit and certain body part of the worm was measured by using the optical image recorded by the stereo microscope at the same time point. Fig. 5a shows that when the body edge of the worm was on the boundary line of the unit, the experimental value of (ΔR / R)th was found to be 8.1 ± 1.7 % (mean ± standard deviation, obtained from 60 resistance data obtained at 5 detection units). It is also obvious that as the nematode invaded deep into an addressed detection unit, the measured ΔR / R of this unit increased. The maximum value of ΔR / R = 22.4 ± 1.8 % occurred when the detection unit was completely covered by the worm. Compared to the simulated plot of the ΔR / R vs. distance given in Fig. 2b, the experimental result showed a relatively good agreement with the simulated result. The discrepancy was probably caused by the simplified model used in the FEA study and the measurement error in the locations of the worm's body part and its distance to an addressed unit. The histogram of the distribution of (ΔR / R)th over 226 grids is given in Fig. 5b, showing a Gaussian-like distribution between 6.5 % and 10.5 %. We note that although there were a total of 24 × 24 = 576 grids in the MEF device, it was almost impossible for a worm to swim over with all grids.
It should be pointed out that the experimental (ΔR / R)th of 8.1 % was much higher than the manufacturing induced maximum original resistance variation of 3.3 %. Thus, we used (ΔR / R)th = 8.1 % to distinguish between the presence and absence of a nematode relative to a detection unit during reconstructing a binary image of the nematode. In fact, one could also subtract the manufacturing-induced “background” profile (Fig. 4) from the actual resistance profile of the grids to blank out the heterogeneity of the original resistance. This, in principle, would allow for a better representation of the reconstructed nematode image. Here, because the value of (ΔR / R)th was much higher than the maximum original resistance variation of the grids, we did not do subtraction operation in this work. It should be also pointed out that the reconstruction of the grayscale and pseudo colour images of a nematode was independent of the actual value of (ΔR / R)th.
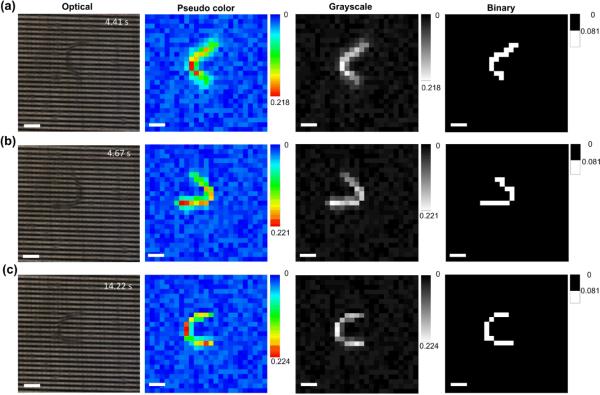
Fig. 6 shows the time-lapse optical images and corresponding reconstructed pseudo colour, grayscale, and binary images of a moving C. elegans (L3) in the MEF grids. The device operated at fsc = 174 Hz and tracked the changes in shape and position of the worm over time. The reconstructed images were then analysed by the worm tracking program. It was found that the oscillation frequency and moving speed of the worm was fworm = 1.8 ± 0.3 Hz and vworm = 270 ± 21 μm s−1, respectively (mean ± standard deviation, n = 12 worms). For comparison purposes, the optical microscope-camera setup was used to track the nematodes of the same developmental stage swimming in a 1-inch Petri dish containing the same M9 buffer. The optical measurement revealed that fworm = 1.9 ± 0.3 Hz and vworm = 266 ± 17 μm s−1 (mean ± standard deviation, n = 12 worms), which were almost the same as those obtained by the MEF grids. Therefore, the present device was able to obtain the locomotion information for reconstructing the real-time images of nematodes for post-analysis of locomotive parameters.
Fig. 6.
Optical images and corresponding reconstructed pseudo colour, grayscale, and binary images of a C. elegans swimming in the MEF grids. The full-frame scanning frequency was fsc = 174 Hz. The pseudo colour, grayscale and binary images were obtained from the electrical measurements. Scale bars represent 100 μm.
The accuracy of detecting the position and shape of a moving nematode highly depended on the full-frame scanning rate fsc of the electronic circuit designed. Suppose that the device has N × N grids and the multimeter used in the system has a reading speed of n readings s−1. Thus, the allowable maximum fsc is n / (N × N) Hz or the electronic circuit takes (N × N) / n seconds to read out all N × N resistances. Furthermore, suppose that the nematode under test has the moving speed of vworm and the body wavelength of λ that can maximally crosses x number of microelectrodes in a direction. Thus, λ = x (D + W). As a result, the electronic circuit spends tact = (x / N) × (N × N / n) = x × N / n seconds on reading out the resistances of the detection units across over the nematode. Within the time period of tact, the worm may shift a maximum distance of Δl = vworm × tact = (vworm × λ × N) / (n × (D + W)). It is obvious that the lower the value of Δl, the higher the accuracy of detecting the position and shape of the moving object. By using a smaller number of microelectrodes N in the grids, the electronic circuit can complete reading the N × N resistance matrix faster, thus decreasing the value of Δl. However, it is noted that the minimum number of microelectrodes on each glass slide is determined by λ of the nematodes. Generally, larval stage C. elegans has the total body length of L ≤ 1 mm and moves at a speed of vworm ≤ 500 μm s−1. The present device was designed by taking half the total body length as the body wavelength or λ ≤ ~500 μm. Therefore, by choosing N = 24 and D = W = 15 μm, the grids could cover an area of 720 × 720 μm2 that allowed larval stage C. elegans to move within the grids. Because the multimeter used here had n = 100000 readings s−1, the maximum fsc was set to be 174 Hz, resulting in a theoretical maximum value of Δl = ~2 μm. We note that it was quite hard to obtain experimental Δl in a short time of tact = x × N / n = (500 / (15 + 15)) × 24 / 100000 = 4 ms. According to Fig. 5, a lateral shift of ~2 μm from the boundary of a detection unit may cause an erroneous variation in ΔR / R of ~0.4 %, which was much lower than the experimental (ΔR / R)th = 8.1 %. Therefore, by appropriately setting the operation frequency of the device, the body movement induced erroneous variation during a full scanning cycle had little influence on the accuracy of tracking the nematodes, which was already demonstrated in Fig. 6. In fact, for a given electronic readout circuit, operating the device at the maximum fsc = n / (N × N) Hz could be easily achieved, simply by programming the hardware language (Verilog) on the computer. Fig. 7 demonstrates that the quality of reconstructed images varied with changing fsc. We found that when fsc < 10 × fworm, the nematode was displayed as the scattered white spots in the reconstructed binary image (Fig. 7a). This is because the nematode moved considerably during a full scanning cycle. Thus, no valuable information about worm activities was obtained. Increasing fsc to 70~80 × fworm allowed these distributed spots to get closer, but the reconstructed image of the nematode was still fragmented (Fig. 7b). When fsc increased to be more than 80 × fworm, a continuous body shape of the nematode was obtained (Fig. 7c).
Fig. 7.
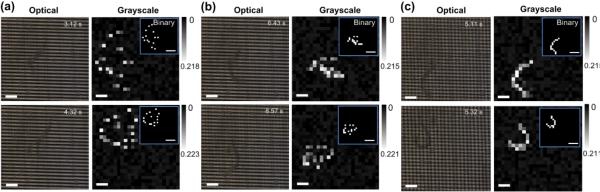
Optical images and reconstructed grayscale, and binary images of a C. elegans swimming in the MEF grids at two different time points with different full-frame scanning frequency fsc = 8 Hz (a), 70 Hz (b), and 100 Hz (c). The grayscale and binary images were obtained from the electrical measurements. Scale bars in the optical and grayscale images represent 100 μm. Scale bars in the binary images (see insets) represent 200 μm.
The pixel resolution of the reconstructed images decreased with increasing the value of D + W. Here, we designed another MEF grids device with D = W = 50 μm, 24 microelectrodes on each glass slide, and 50 μm gap between the two glass slides. The device still operated at fsc = 174 Hz. A L3-stage C. elegans was introduced in the microfluidic chamber containing the M9 buffer. Similarly, by using the same method mentioned above, (ΔR / R)th was found to be 3.8 ± 0.9 % in this case, which was close to the simulated ~3.5 % and greater than the manufacturing induced intrinsic resistance variation of ~3 % of this device (D = W = 50 μm). Thus, this device could determine the presence/absence of the nematode in the detection units as shown in Fig. 8b. However, although the movement of the nematode was detected, the use of the large-size detection unit resulted in the coarse images with only 5–7 pixels to outline the nematode in each image. Detailed information about the shape of the nematode was lost. We note that with the relatively low spatial resolution, the worm tracking program could identify the position of the worm and analyse its average moving speed, but was difficult to derive important locomotive parameters such as oscillation frequency, amplitude, and wavelength.
Fig. 8.
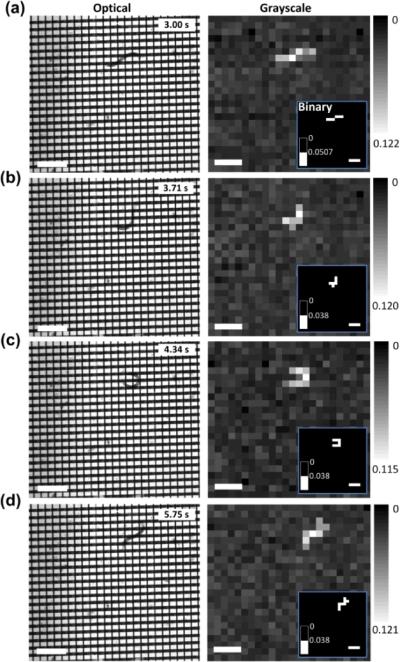
Optical images and corresponding reconstructed grayscale and binary (inset) images of a C. elegans (L3) moving in the grids. The detection frequency was set to be at fsc = 174 Hz. The grayscale and binary images were obtained from the electrical measuremen. Scale bars represent 400 μm.
Other research has revealed that C. elegans at different developmental stages respond differently to electric field [21,22]. For example, early stage (L1 and L2) wild type C. elegans was little responsive to electric field with strength less than 13 V cm−1 before electrokinetic flows occurred. Whereas, L3, L4, and adult stage worms respond to electric field robustly starting from the threshold field strength of 4, 4, and 2 V cm−1, respectively, by swimming towards the cathode. Thus, in order to use the present device for different larval stage C. elegans without physiological and behavioral side effects, we adjusted the test voltage of the multimeter (the actual voltage applied to the microelectrodes was 8 mV), by using external series connected resistors and a potentiometer. This could ensure the electric field applied between the top and bottom microelectrodes to be as low as ~1.5 V cm−1. Furthermore, we verified the responses of L1–L4 wild-type C. elegans to this electric field strength. Specifically, one of the bottom microelectrodes was set to be grounded, and also, an upper microelectrode was selected. Ten nematodes of each stage were respectively flowed to the microfluidic chamber. No directed swimming of the nematodes was observed at all five different stages. Fig. 9 shows fworm and vworm of the L1–L4 nematodes detected by both the present grids and the optical microscope-camera setup. The locomotive parameters were also derived and analysed by the worm tracking program. Since the two methods provided almost the same result, it was confirmed that the worms moving inside the grids were not affected by the applied low electric field and that the present electrical resistance measurement approach was effective and reliable to detect the locomotion behaviors of the nematodes.
Fig. 9.
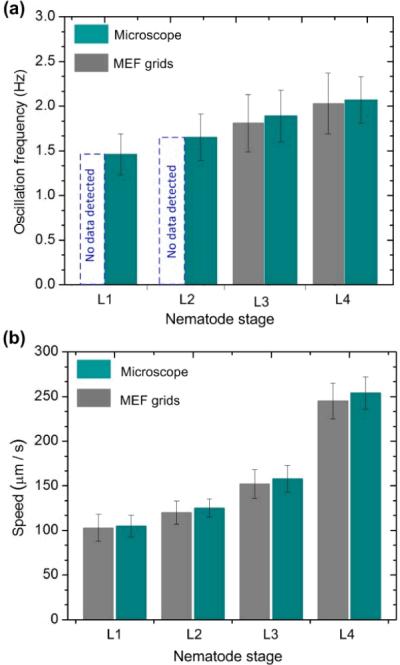
Oscillation frequency (a) and moving speed (b) of wild-type C. elegans worms at different developmental stages (L1–L4) detected by using the present MEF grids and optical microscope. Due to the large D and W (D = W = 15 μm), the device was not able to detect the oscillation frequency of the L1–L2 stage worms. Error bars represent standard deviations of the mean oscillation frequency (a) and moving speed (b), respectively, from 12 worms.
To demonstrate further the workability of the present device, a drug resistance screening experiment was conducted by using the device with D = W = 15 μm and operating at fsc = 174 Hz. In this experiment, the microfluidic chamber of the device was pre-filled by the anthelmintic levamisole with specific concentrations. The L3-stage wild-type N2 and mutant lev-8 C.elegans was respectively introduced into the chamber. Similarly, to verify the dose responses obtained by the MEF grids, the optical imaging-based locomotion tracking experiment was conducted in a Petri dish containing the same levamisole concentration solution. It is noted that even as the concentration of levamisole solution increased to a considerably high value of 500 μM, the electrical resistivity of the worm and levamisole solution was little changed. Thus, we still used (ΔR / R)th = 8.1 % to distinguish the presence/absence of the worm's body part in a detection unit.
Fig. 10 displays the responses of the two different nematodes as a function of levamisole dose in terms of fworm and vworm. The result demonstrates that the response curves obtained by the present device were almost the same as those obtained by the microscope-camera method. Specifically, at the levamisole concentrations lower than 1.0 μM for N2 and 5.0 μM for lev-8 nematodes, little reduction in fworm or vworm was observed. Increasing levamisole concentration caused to restrict their movement. The cut-off concentration was 105.4 μM for N2 and 155.3 μM for lev-8, at which almost all worms were paralyzed. The EC50 value (defined as the concentration that provokes a response half way between the basal and maximal response, of the agonist and compare for differences between isolates or strains) for levamisole was found to be ~20.2 μM for N2 worms, and ~38.7 μM for lev-8 worms by the fitting curves in Figs. 11c–d. Therefore, the present grids device can play a useful role in many phenotypic bioassays that require real-time observation of the behavioural patterns of some microorganisms.
Fig. 10.
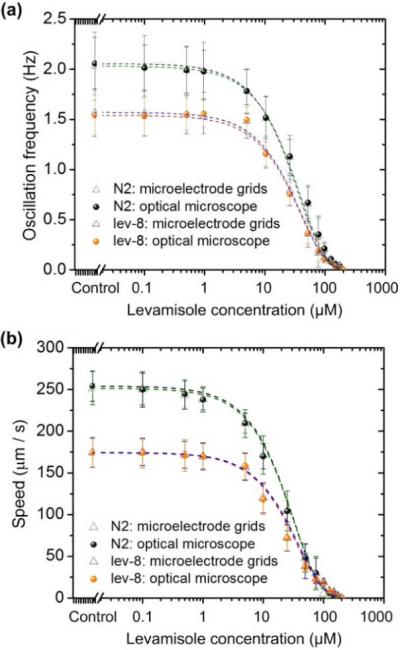
Oscillation frequency (a) and moving speed (b) of L3-stage wild-type N2 and mutant lev-8 C. elegans. Error bars represent standard deviations of the mean oscillation frequency (a) and moving speed (b), respectively, from 15 worms.
There is much room to improve the performances of the MEF grids device. At the moment of this work, the obtained spatial detection resolution of the device was at the level of 30 × 30 μm2, limited by our fabrication capability. The worm tracking program was able to extract the behavioural parameters such as moving speed and oscillation frequency based on the reconstructed images obtained over time. But, due to the limited spatial resolution, it was relatively difficult to extract other important parameters such as nematode's body amplitude. As a result, no accurate waveform of nematodes was obtained by this present device. Scaling down the grids will enable us to obtain higher resolution constructed worm images. It is interesting to address that using smaller detection units can result in a higher value of (ΔR / R)th, making it easier and more accurate to detect the presence and absence of a worm's body. Specifically, by manufacturing the microelectrodes with the submicron feature size, our locomotion behaviour detection technology could overcome the optical diffraction limit in conventional optical microscopy. Furthermore, due to the limited area coverage by a small number of microelectrodes, the current version of the MEF grids device allowed a single worm to move freely inside the grids. Obviously, layout of more microelectrodes will allow for simultaneous abd high-throughput monitoring of multiple worms with no body interference between worms. However, in order to incorporate a large number of finer (e.g., submicron feature) microelectrodes into the next version of the device (with an aim of achieving a higher spatial resolution and larger capacity for detecting multiple worms), the electronic detection system needs a faster processing speed to read out more electrical resistance elements in parallel. The device presented here used a multimeter, in conjunction with the addressing circuit, to read the electrical resistance matrix of the MEF grids. A large-scale resistance readout circuit specific for the grids device is under development. Generally, it is possible to measure the electrical resistance of a resistor on a ~ns time scale by using integrated circuit technology. As revealed in Fig. 7, the whole-frame scanning frequency of fsc ≥ 80 Hz was required to track a moving nematode. It is thus possible to extend the number of microelectrodes on each glass slide to be N = ~ (109 / fsc)1/2 = ~ 3500. For example, a scaled-up MEF grids device with spatial resolution of 5 × 5 μm2 will cover a large detection area of 1.75 × 1.75 cm2. Moreover, thanks to the simplicity of the electrical measurement approach, we can further increase the detection area by using multiple of such MEFs in parallel. Finally, we believe it is also possible to detect nematode locomotion information by finding out electrical capacitance profile of the MEF grids. The details of the results will be reported in another article.
Conclusions
In summary, we have demonstrated the MEF method of tracking the movement of nematode C. elegans based on the electrical resistance measurement for all grids. The 24 × 24 orthogonal grids were realized by conventional microfabrication techniques. Each microelectrode was 15 μm wide and each detection unit was 30 × 30 μm2 across. We found that as a worm moved closer to an addressed detection unit, the electrical resistance of this unit increased. The presence of part of the nematode's body at the selected detection unit caused a minimal electrical resistance change of ~8.1 % (vs. the simulated result of ~7.9 %), which was much higher than the manufacturing induced initial resistance variation of 3.3 % between all detection units. We designed an electronic circuit to address individual detection units and reading out their electrical resistance at the full-frame scanning frequency of 174 Hz. The circuit allowed tracking the time-varying shape and position of the worms and analysed the locomotive parameters of the nematode (e.g., oscillation frequency and moving speed) with the help of the worm tracking program. We also found that the shape and position of the nematodes could be identified when the full-frame scanning frequency of the electronic circuit increased to be more than 80 times the nematode's oscillation frequency. Furthermore, as the size of the detection unit increased, the spatial resolution of the reconstructed images decreased. In addition, we verified that no electrotaxic effect occurred at the worms since the electric field strength applied was set to be as low as ~1.5 V cm−1. Lastly, the workability of the device was demonstrated by performing a drug resistance screening experiment on the MEF device. The device was able to differentiate the locomotion behaviours of the wild-type N2 and mutant lev-8 C. elegans in response to a wide dose range of levamisole. We believe that the present MEF device can give potential to provide a cost-effective, potentially high throughput solution to tracking the behavioural phenotype changes of important nematodes for different bioassays on a chip level and large scale analysis.
Supplementary Material
Fig. 1.
Schematic of the proposed MEF grids for detecting the locomotion behaviours of microscopic nematodes.
Acknowledgements
This work is supported by the National Science Foundation under award no. ECCS-1102354, the McGee-Wagner Research Fund, the NIH grant: R56 AI047194-11 National Institute of Allergy and Infectious Diseases, and the China Scholarship Council. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases. The authors thank all members in the Laboratory for the NEMS and Lab-Chips for helpful discussion.
Footnotes
Electronic Supplementary Information (ESI) available: a video clip showing the movement of a wild-type C. elegans (L3) within the micro-electro-fluidic grids.
Notes and references
- 1.Guarente L, Kenyon C. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 2.Kaletta T, Hengartner MO. Nat. Rev. Drug Discovery. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 3.Tsechpenakis G, Bianchi L, Metaxas DN, Driscoll M. IEEE Trans. Biomed. Eng. 2008;55:1539–1549. doi: 10.1109/TBME.2008.918582. [DOI] [PubMed] [Google Scholar]
- 4.Sznitman R, Gupta M, Hager GD, Arratia PE, Sznitman J. PLoS ONE. 2010;5:e11631. doi: 10.1371/journal.pone.0011631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Sun Y, Dixon SJ, Alexander M, Roy PJ. J. Lab. Autom. 2009;14:269–276. [Google Scholar]
- 6.Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronin CJ, Feng Z, Schafer WR. Methods Mol. Biol. 2006;351:241–251. doi: 10.1385/1-59745-151-7:241. [DOI] [PubMed] [Google Scholar]
- 8.Potsaid B, Finger FP, Wen JT. IEEE Trans. Autom. Sci. Eng. 2009;6:525–535. [Google Scholar]
- 9.Cronin CJ, Mendel JE, Mukhtar S, Kim YM, Stirbl RC, Bruck J, Sternberg PW. BMC Genet. 2005;6:5. doi: 10.1186/1471-2156-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonetta SH, Golombek DA. J. Neurosci. Methods. 2007;161:273–280. doi: 10.1016/j.jneumeth.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Geng W, Cosman P, Berry CC, Feng Z, Schafer WR. IEEE Trans. Biomed. Eng. 2004;51:1811–1820. doi: 10.1109/TBME.2004.831532. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Zheng G, Lee LM. Lab Chip. 2012;12:3566–3575. doi: 10.1039/c2lc40517b. [DOI] [PubMed] [Google Scholar]
- 13.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 14.deMello AJ. Nature. 2006;442:394–402. doi: 10.1038/nature05062. [DOI] [PubMed] [Google Scholar]
- 15.Psaltis D, Quake SR, Yang CH. Nature. 2006;442:381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 16.Crane MM, Chung K, Stirman J, Lu H. Lab Chip. 2010;10:1509–1517. doi: 10.1039/b927258e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung KH, Crane MM, Lu H. Nat. Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 18.Chronis N. Lab Chip. 2010;10:432–437. doi: 10.1039/b919983g. [DOI] [PubMed] [Google Scholar]
- 19.Kim N, Dempsey CM, Zoval JV, Sze J-Y, Madou MJ. Sens. Actuators, B. 2007;122:511. [Google Scholar]
- 20.Lockery SR, Lawton KJ, Doll JC, Faumont S, Coulthard SM, Thiele TR, Chronis N, McCormick KE, Goodman MB, Pruitt BL. J. Neurophysiol. 2008;99:3136–3143. doi: 10.1152/jn.91327.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezai P, Siddiqui A, Selvaganapathy PR, Gupta BP. Lab Chip. 2010;10:220–226. doi: 10.1039/b917486a. [DOI] [PubMed] [Google Scholar]
- 22.Rezai P, Salam S, Selvaganapathy PR, Gupta BP. Lab Chip. 2012;12:1831–1840. doi: 10.1039/c2lc20967e. [DOI] [PubMed] [Google Scholar]
- 23.Shi WW, Qin J, Ye N, Lin B. Lab Chip. 2008;8:1432–1435. doi: 10.1039/b808753a. [DOI] [PubMed] [Google Scholar]
- 24.Clausell-Tormos J, Lieber D, Baret J-C, El-Harrak A, Miller OJ, Frenz L, Blouwolff J, Humphry KJ, Koster S, Duan H, Holtze C, Weitz DA, Griffiths AD, Merten CA. Chem. Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Hulme SE, Shevkoplyas SS, McGuigan AP, Apfeld J, Fontana W, Whitesides GM. Lab Chip. 2010;10:589–597. doi: 10.1039/b919265d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, Whitesides GM. Lab Chip. 2007;7:1515–1523. doi: 10.1039/b707861g. [DOI] [PubMed] [Google Scholar]
- 28.Chokshi TV, Ben-Yakar A, Chronis N. Lab Chip. 2009;9:151–157. doi: 10.1039/b807345g. [DOI] [PubMed] [Google Scholar]
- 29.Zeng F, Rohde CB, Yanik MF. Lab Chip. 2008;8:653–656. doi: 10.1039/b804808h. [DOI] [PubMed] [Google Scholar]
- 30.Chung K, Lu H. Lab Chip. 2009;9:2764–2766. doi: 10.1039/b910703g. [DOI] [PubMed] [Google Scholar]
- 31.Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- 32.Guo SX, Bourgeois F, Chokshi T, Durr NJ, Hilliard MA, Chronis N, Ben-Yakar A. Nat. Methods. 2008;5:531–533. doi: 10.1038/nmeth.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chronis N, Zimmer M, Bargmann CI. Nat. Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 34.Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Lu H, Bargmann CI. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 36.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 37.Doll JC, Harjee N, Klejwa N, Kwon R, Coulthard SM, Petzold B, Goodman MB, Pruitt BL. Lab Chip. 2009;9:1449–1454. doi: 10.1039/b818622g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin JH, Wheeler AR. Lab Chip. 2007;7:186–192. doi: 10.1039/b613414a. [DOI] [PubMed] [Google Scholar]
- 39.Chung K, Zhan M, Srinivasan J, Sternberg PW, Gong E, Schroeder FC, Lu H. Lab Chip. 2011;11:3689–3697. doi: 10.1039/c1lc20400a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lockery SR, Hulme SE, Roberts WM, Robinson KJ, Laromaine A, Lindsay TH, Whitesides GM, Weeks JC. Lab Chip. 2012;12:2211–2220. doi: 10.1039/c2lc00001f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P, Mao D, Martin RJ, Dong L. Lab Chip. 2012;12:3458–2466. doi: 10.1039/c2lc40459a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heng X, Erickson D, Baugh LR, Yaqoob Z, Sternberg PW, Psaltis D, Yang C. Lab Chip. 2006;6:1274–1276. doi: 10.1039/b604676b. [DOI] [PubMed] [Google Scholar]
- 43.Cui X, Lee LM, Heng X, Zhong W, Sternberg PW, Psaltis D, Yang C. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10670–10675. doi: 10.1073/pnas.0804612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee LM, Cui X, Yang C. Biomed. Microdevices. 2009;11:951–958. doi: 10.1007/s10544-009-9312-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng G, Lee SA, Yang S, Yang C. Lab Chip. 2010;10:3125–3129. doi: 10.1039/c0lc00213e. [DOI] [PubMed] [Google Scholar]
- 46.Schonbrun E, Abate AR, Steinvurzel PE, Weitz DA, Crozier KB. Lab Chip. 2010;10:852–856. doi: 10.1039/b923554j. [DOI] [PubMed] [Google Scholar]
- 47.Heng X, Hsiung F, Sadri A, Patt P. Anal. Chem. 2011;83:1587–1593. doi: 10.1021/ac102408g. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Cui X, Lee LM, Yang C. Opt. Express. 2008;16:15595–15602. doi: 10.1364/oe.16.015595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coskun AF, Su T-W, Ozcan A. Lab Chip. 2010;10:824–827. doi: 10.1039/b926561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mudanyali O, Tseng D, Oh C, Isikman SO, Sencan I, Bishara W, Oztoprak C, Seo S, Khademhosseini B, Ozcan A. Lab Chip. 2010;10:1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isikman SO, Bishara W, Mavandadi S, Yu SW, Feng S, Lau R, Ozcan A. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7296–7301. doi: 10.1073/pnas.1015638108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isikman SO, Bishara W, Sikora U, Yaglidere O, Yeah J, Ozcan A. Lab Chip. 2011;11:2222–2230. doi: 10.1039/c1lc20127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su T-W, Isikman SO, Bishara W, Tseng D, Erlinger A, Ozcan A. Optics Express. 2010;18:9690–9711. doi: 10.1364/OE.18.009690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh C, Isikman SO, Khadem B, Ozcan A. Optics Express. 2010;18:4717–4726. doi: 10.1364/OE.18.004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isikman SO, Sencan I, Mudanyali O, Bishara W, Oztoprak C, Ozcan A. Lab Chip. 2010;10:1109–1112. doi: 10.1039/c001200a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coskun AF, Su T-W, Ozcan A. Lab Chip. 2010;10:824–827. doi: 10.1039/b926561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozcan A, Demirci U. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 58.Carr JA, Parashar A, Gibson R, Robertson AP, Martin RJ, Pandey S. Lab Chip. 2011;11:2385–2396. doi: 10.1039/c1lc20170k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaham S, WormBook, editor. The C. elegans Research Community. WormBook; WormBook: Methods in Cell Biology (Jan. 02, 2006) doi/10.1895/wormbook.1.49.1, http://www.wormbook.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.