Abstract
Low-grade prostate cancers (Gleason pattern 3 [G3]) detected on needle biopsies are generally viewed as indolent and suitable for conservative management with only interval repeat biopsies to monitor by watchful wating. Higher grade tumors eventually emerge in 20–30% of these cases, but this process may only reflect incomplete sampling on the initial biopsy, such that it remains unknown how generally G3 tumors progress to higher grades. In this study, we examined a series of adjacent G3 and Gleason pattern 4 (G4) tumors in radical prostatectomy specimens and found that all were concordant for the TMPRSS2:ERG gene fusion. Using hybrid-capture and deep sequencing in four fusion-positive cases, we found that adjacent laser-capture microdissected G3 and G4 tumors had identical TMPRSS2:ERG fusion breakpoints, confirming their clonal origin. Two of these G3 tumors had deletion of a single PTEN gene that was also deleted in the adjacent G4, while the G4 tumors in two cases had additional PTEN losses. These findings establish that a subset of G3 tumors progress to G4, or emerge from a common precursor. Further, they show that G3 tumors which progress to G4 may have molecular features distinguishing them from G3 tumors that do not progress. Thus, determining the spectrum of these genetic or epigenetic features may allow for the identification of G3 tumors on needle biopsies that are truly indolent, versus those that have the potential to progress, or that may already be associated with a G4 tumor that was not sampled at the initial biopsy, therefore requiring more aggressive surveillance or intervention.
Keywords: prostate cancer, Gleason pattern, progression, active surveillance, gene fusion
Introduction
Prostate cancer (PCa) screening using serum prostate-specific antigen has led to marked increases in prostate needle biopsies and the detection of cancers that are low grade (Gleason pattern 3, Gleason score 3+3=6), and it has become clear that many or most of these low grade tumors will have indolent behavior (1, 2). Indeed, a large retrospective study found that only a small fraction of patients with Gleason score 6 tumors who underwent radical prostatectomy (RP) subsequently relapsed, and the relapses that did occur were almost invariably associated with the identification of higher Gleason grade tumors (G4 or G5) in the surgical specimen (3). The realization that many or most Gleason score 6 tumors are indolent has led to an increased willingness to defer surgery or radiation therapy, and to instead follow patients for evidence of progression (watchful waiting or active surveillance, AS). These patients with low grade tumors on biopsy who do not undergo immediate surgery or radiation therapy and instead chose AS also have low cancer-specific mortality rates, but there are obvious concerns about missing the opportunity to cure some small subset of patients who may progress to lethal disease (4, 5). Optimal approaches to follow these patients remain unclear, but AS patients generally undergo interval repeat biopsies to rule out potentially more aggressive disease. Higher grade tumors are detected upon subsequent biopsies in about 20–30% of these patients, who may then go on to surgery or radiation therapy. This tumor upgrading on subsequent biopsies appears to reflect primarily incomplete sampling on the original biopsies, and it remains unclear and controversial whether some G3 tumors have the capacity to progress to higher grades (4–8). In either case, concerns that more aggressive tumors may have been missed on initial biopsies, or that G3 tumors may progress to metastatic G4 or G5 tumors during surveillance, lead many patients to choose surgery or radiation over AS.
G4 glands are frequently found intermixed with or adjacent to G3 cancers, but PCa is commonly multifocal and it has not been determined whether these are clonally related (9–12). Moreover, while many studies have focused on genes that may predict aggressive behavior of Gleason score 7 tumors based on tumor recurrence after RP, molecular features that may distinguish truly-indolent G3 tumors detected on biopsies from those that have the potential for grade progression or are associated with higher grades have not been explored. This study was undertaken to determine whether a subset of G3 tumors progress to G4, or evolve from a common precursor, as characterization of these tumors may allow for the more precise identification of patients with potentially aggressive G3 tumors on biopsy who would benefit from more intense surveillance or intervention. Further characterization of clonal G3 and G4 tumors could also reveal mechanisms of progression and identify subsets of G4 tumors with distinct biological behaviors.
Materials and Methods
Prostate Tissues and Immunohistochemistry
Tissue from RPs was collected and deidentified in accordance with BIDMC IRB protocol #2010-P-000254/01. Tissues were fixed in formalin or PaxGene (Qiagen, Valencia, CA) and embedded in paraffin. Sections were stained with H&E, anti-ERG (EPR3864, Epitomics, Burlingame, CA), and PIN-4 antibodies (cocktail of anti-p63, anti-cytokeratin 5, anti-cytokeratin 14, and anti-AMACR, Biocare Medical, Concord, CA) according to manufacturer’s protocols. Gleason pattern was determined independently by at least two pathologists using the 2005 modified Gleason grading system (13).
Statistical Analysis
A Chi-Square test to evaluate the likelihood of concordant positive and negative ERG staining in adjacent G3 and G4 glands (assuming equal variance) was performed using Stata 12 software with three degrees of freedom.
Laser Capture Microdissection
Slides containing adjacent G3 and G4 glands were stained with anti-ERG to confirm fusion-status and PIN-4 antibodies to distinguish invasive cancer from PIN and intraductal carcinoma. Six-micron serial sections were cut onto polyethylene naphthalate metal frame slides (Molecular Machines & Industries, Zurich, Switzerland) and lightly stained with the Paradise Plus Staining Kit (Life Technologies, Foster City, CA). Target regions were identified using stained consecutive sections as references, and approximately 50,000 cells per sample were captured onto caps using 20-micron infrared pulses and excised from the adjacent tissue using the ultraviolet laser on an ArcturusXT Nikon Eclipse Ti-E microdissection system. G3 and G4 glands from each case were collected on separate caps. Each captured region was checked against the reference slides for accuracy prior to DNA and RNA extraction.
Library Preparation, Sequencing, and Analysis
DNA was amplified (Ovation WGA FFPE System, NuGEN, San Carlos, CA), converted to double-stranded DNA (Encore ds-DNA Module, NuGEN), and fragmented to 200–250 bp. Multiplex library construction, hybrid capture with biotinylated baits tiled across the entire PTEN, AR, TMPRSS2, and ERG loci, and library amplification were performed using the SureSelectXT Target Enrichment System (Agilent). The indexed libraries were sequenced on a HiSeq 2000 (Illumina, San Diego, CA) with 50 cycles paired end (50×50) and 7 additional indexing cycles. Alignment, mutation-calling, breakpoint analysis and SNP genotyping was performed using NextGENe Software version 2.16 (Softgenetics, State College, PA). Further details are in the Supplementary Materials and Methods. Sequencing data have been deposited with the NCBI Sequence Read Archive, accession number SRA049738.1.
Results
TMPRSS2:ERG gene fusions resulting in high-level expression of ERG occur in approximately half of PCa’s, with breakpoints clustering in a ~ 25-kb region between exons 1 and 4 of TMPRSS2 and ~75-kb region in intron 3 of ERG (14, 15). We initially used ERG immunostaining to address whether adjacent G3 and G4 tumors in RP specimens were concordant for the TMPRSS2:ERG fusion, as ERG protein expression is extremely low in fusion negative tumors (16, 17). To minimize inter-observer variations in Gleason grading and to get clean microdissections, we selected cases containing substantial clearly evident cribriform G4 glands and avoided cases with ill-defined G4 glands. In a series of 52 RP specimens, adjacent or intermixed G3 and G4 tumors were concordant for ERG expression in all cases (Supplementary Table S1). As only ~50% concordance would be expected by chance, and previous studies have shown that multifocal tumors are commonly discordant for TMPRSS2:ERG fusion (18, 19), this result suggested that the adjacent tumors were clonally related.
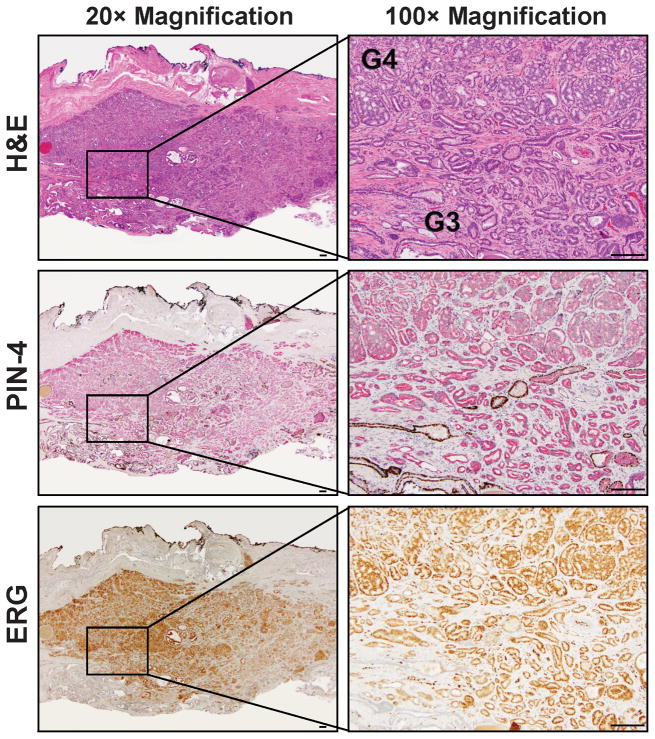
To assess clonality directly, we next mapped TMPRSS2 and ERG genomic breakpoints in adjacent fusion-positive G3 and G4 tumors from four patients. Figure 1 shows the histology and immunostaining (PIN-4 and ERG) of the tumor from one of these patients (Patient 2). PIN-4 is cocktail of antibodies recognizing basal epithelial cells (anti-p63, cytokeratin 5 and 14) with a brown chromagen and anti-AMACR recognizing neoplastic epithelium in prostatic intraepithelial neoplasia (PIN) lesions and PCa cells (red chromagen). Loss of basal cells is a defining feature of PCa, so glands expressing AMACR with no associated basal cells are cancer, while those with a discontinuous basal cell layer are PIN. Supplementary Figure 1 shows the other three patients, and the pathological features for all four are summarized in Table 1.
Figure 1.
H&E, PIN4, and ERG stains on consecutive sections of cancer tissue from Patient 2. Left: low magnification (20×). Right: Boxed target region in left panel viewed at 200× magnification showing adjacent G3 and G4 glands used for microdissection. H&E, PIN-4 (red chromogen: AMACR; brown chromogen: p63, cytokeratin 5, and cytokeratin 14), and ERG staining are shown. Bar: 100 μm.
Table 1.
Clinicopathologic characteristics of four cases that were mapped for TMPRSS2:ERG breakpoints in adjacent G3 and G4 tumors.
| Patient | Race | Age | Fixative | RP Gleason score | RP Stage | Tumor volume (%) | G3 and G4 relationship | Gleason Pattern | Morphologic features |
|---|---|---|---|---|---|---|---|---|---|
| 1 | W | 50 | Formalin | 3+4=7 | pT3b | 35 | areas with intervening stroma | G3 | acinar |
| G4 | acinar with ductal features, cribriform and fused | ||||||||
| 2 | W | 66 | Formalin | 3+4=7 | pT2c | 5~10 | continuous with distinct G3 and G4 areas | G3 | acinar |
| G4 | acinar, cribriform and fused | ||||||||
| 3 | B | 63 | Paxgene | 3+4=7 & Tertiary 5 | pT3b | 40 | intermixed with G4-dominant nodules | G3 | acinar |
| G4 | acinar, cribriform and fused | ||||||||
| 4 | W | 52 | Formalin | 3+4=7 | pT2c | 10 | continuum with distinct G3 and G4 areas | G3 | acinar, scant mucin secretion |
| G4 | acinar, cribriform and fused, extensive mucin secretion |
Cells from adjacent ERG-positive G3 and G4 tumor foci were isolated by laser capture microdissection (Fig. 2A). Genomic DNA (gDNA) and RNA then were extracted from each, and quantitative RT-PCR was used to confirm expression of the common TMPRSS2:ERG fusion mRNA in each sample (not shown). The gDNA was amplified and then enriched for the TMPRSS2:ERG fusion by hybridization to a probe library containing oligonucleotides spanning the TMPRSS2 and ERG loci, as well as the AR and PTEN loci. The enriched DNA then was amplified, barcoded, and pooled for multiplexed paired-end Illumina sequencing. Sequencing of TMPRSS2 and ERG was achieved at an average of 700-fold and 300-fold coverage, respectively, across the entire locus at comparable efficiencies for each sample.
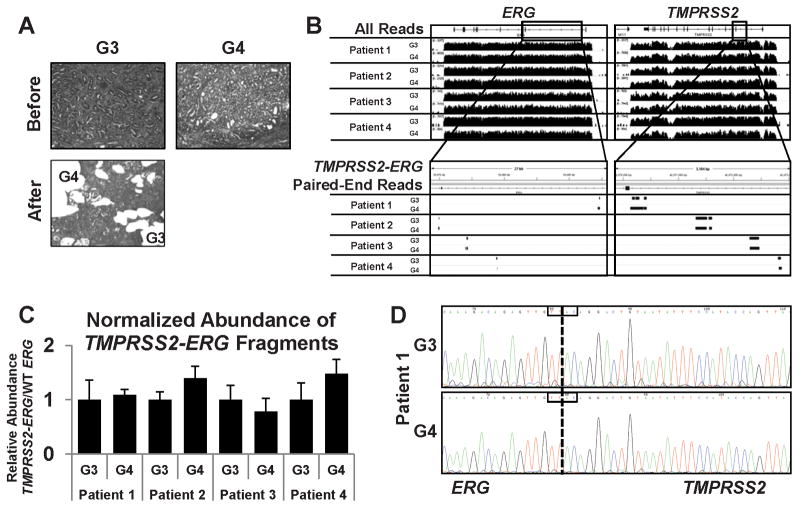
Figure 2.
Next-generation sequencing for determination of TMPRSS2:ERG breakpoints. A, before (left) and after (right) photographs of representative laser-capture microdissected regions from adjacent G3 and G4 glands from the same slide (Patient 2). B, top: coverage of ERG locus (left) and TMPRSS2 locus (right) from G3 and G4 cancer in each patient. Bottom: Zoom-in and filtering of paired-end alignments corresponding to both ERG (left) and TMPRSS2 (right). C, quantitative PCR on amplified genomic DNA prior to hybrid capture to measure relative abundance of TMPRSS2:ERG fragments. Data is relative to the abundance of wild-type ERG and normalized to G3 for each patient, using patient-specific fusion primers for TMPRSS2:ERG. Error bars: ± s.d. D, Sanger sequencing to confirm sequence of TMPRSS2:ERG breakpoint in Patient 1 from G3 glands (top) and G4 glands (bottom). Breakpoint was amplified from original genomic DNA isolated from G3 and G4 glands using breakpoint-specific primers. The boxed region reflect a four-base microhomology present in both TMPRSS2 and ERG sequences indicating that the physical break could have occurred at any of the four bases.
To identify fragments containing the breakpoints, reads with computed distances between the paired-end reads greater than 400 bases (the maximum size expected for most fragments in the library) were filtered for TMPRSS2 and ERG. Supplementary Table 2 shows the number of fragments containing breakpoints versus total reads. While there were differences in total reads (reflecting unequal library pooling), the fraction of reads containing the breakpoints was comparable in each G3 versus G4 tumor. This indicated that the fraction of cells containing the gene fusion was similar and that these reads were not derived from small numbers of cross-contaminating tumor cells. The fragments with reads in TMPRSS2 and ERG mapped to distinct sites in each patient, but mapped to the exact same sites in the adjacent G3 and G4 tumors (Fig. 2B, bottom). Quantitative PCR (qPCR) on the amplified gDNA using breakpoint-specific PCR primers for each patient confirmed that cells with the TMPRSS2:ERG fusions were present at comparable levels in the adjacent G3 and G4 tumors (Fig. 2C). This was further confirmed by qPCR of the unamplified gDNA (not shown). Finally, sequencing of the PCR amplified products from each G3 and G4 tumor confirmed that the breakpoints from each patient were distinct, but were identical in the adjacent G3 and G4 tumors (Fig. 2D, Supplementary Fig. S2). The precise coordinates for each break, the fusion mechanisms (translocation in two and deletion in two), and the inferred repair mechanisms are summarized in Supplementary Table S3.
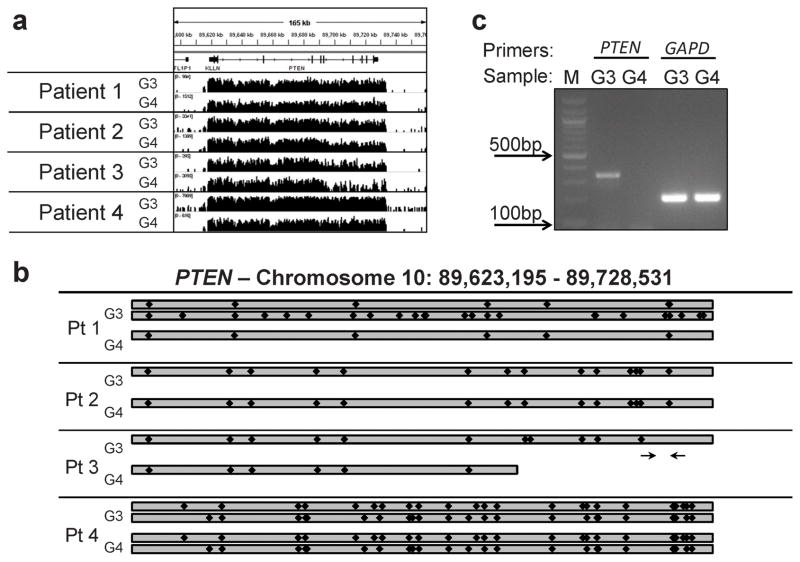
Frequent genomic alterations that occur during PCa progression include AR gene amplification and PTEN loss or mutation. The AR gene did not show evidence of amplification or mutation (not shown), consistent with these alterations occurring primarily in tumors that relapse after androgen deprivation. In contrast, raw genomic coverage for PTEN inferred a region of deletion in the G4 tumor from Patient 3 (Fig. 3A). More quantitative analysis of PTEN reads (Supplementary Table S4) and SNPs (Supplementary Table S5) revealed that one copy of PTEN was lost in both the G3 and G4 tumor from Patient 3, and that the G4 tumor had sustained a further loss of a portion of the remaining PTEN gene (Fig. 3B). The analysis in Patient 1 showed that both copies of PTEN were intact in the G3 tumor, while there was loss of heterozygosity (LOH) across the PTEN locus in the G4 tumor, indicating loss of one gene. In Patient 2 we found LOH both in the G3 and G4 tumors, indicating that each had lost one copy, but did not find further loss or mutations in the G4 tumor. There was no evidence of loss or coding region mutations in the G3 or G4 tumors from Patient 4. Finally, using RT-PCR we confirmed that the G4 tumor of Patient 3 had no detectable PTEN mRNA (Fig. 3C).
Figure 3.
PTEN status in each patient. A, coverage of the PTEN locus from G3 and G4 tumor. B, representation of the PTEN status for each patient based on the relative abundance of previously-reported SNPs (indicated by black diamonds) in the aligned sequences. C, RT-PCR of total cellular RNA isolated from G3 and G4 glands from Patient 3 with primers specific for PTEN and GAPDH. The PTEN primers are in exons 6 and 7, as depicted by arrows in (B).
Discussion
This study establishes that G3 tumors can progress to G4 or derive from a common precursor lesion. This common precursor may be PIN in some cases, although PIN lesions we found in the vicinity of the adjacent G3 and G4 tumors were not consistently concordant for ERG, and both ERG positive and negative PIN lesions were found in some cases (Supplementary Fig. S3). The frequency of clonal G3 and G4 tumors is not yet clear, but it is not uncommon to find G4 tumors adjacent to or intermixed with G3 tumors, particularly in higher volume G3 tumors, which suggests that progression of G3 tumors or their precursors to G4 is not a rare event. We suggest that these G3 tumors that can progress to G4, or evolve from a common precursor, may be molecularly distinct from isolated G3 tumors that are not associated with higher grades. One such molecular alteration may be loss of one PTEN allele, as this was found in two of the four G4-associated G3 tumors we examined, but appears to be less common in G3 tumors that are not associated with G4 (Gleason score 6 tumors) (20). However, further large-scale studies are needed to determine whether loss of one PTEN allele is more common in these G4-associated G3 tumors, and whether further PTEN loss mediates progression to G4.
While isolated G3 tumors versus G4-associated G3 tumors may be readily distinguished by thorough examination of RP specimens, prostate needle biopsies containing only G3 tumor could reflect a larger clonal G4 tumor that was not sampled, or a tumor that is likely to progress to G4 (either directly from the G3 or from a common precursor). Therefore, determining whether there are molecular alterations that distinguish G4-assocated G3 tumors, which can then be assessed on prostate needle biopsies, would greatly aid in the identification of men with Gleason score 6 on their biopsies who are likely harboring a higher grade tumor or are at increased risk of progression. Further molecular analyses of these clonally related G3 and G4 tumors should also identify molecular correlates of these Gleason patterns and mechanisms of progression to G4. Finally, comparisons with G4 in Gleason score 8 and 9 tumors may reveal that G3-associated G4 tumors are a distinct subset that contribute to the molecular and clinical heterogeneity of Gleason score 7 tumors.
Supplementary Material
Acknowledgments
Financial support: This work was supported by grants to S.P.B. from the NIH (Prostate SPORE P50 CA090381), a DF/HCC Mazzone Award, and awards from The V Foundation for Cancer Research and the Prostate Cancer Foundation. A.G.S was supported by an NIH T32 CA081156. H. Y. was supported by a Career Development Award from the Prostate and Breast Cancer Research Program at the BIDMC.
We sincerely thank Dr. Elizabeth Genega for her support, revision of the IRB protocol, guiding tissue procurement, reviewing tumor grades, and constructive suggestions, Stephen Duggan for maintaining our tissue bank, and Cesar Vazquez for transporting samples. We also express sincere gratitude for the support of the BIDMC urologists Drs. Andrew Wagner, William DeWolf, and Martin Sanda.
Footnotes
Conflicts of interest: The authors have no conflicts of interest to declare.
References
- 1.Draisma G, Boer R, Otto SJ, van der Cruijsen IW, Damhuis RA, Schroder FH, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95:868–78. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 2.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293:2095–101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 3.Eggener SE, Scardino PT, Walsh PC, Han M, Partin AW, Trock BJ, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185:869–75. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klotz L, Zhang L, Lam A, Nam R, Mamedov A, Loblaw A. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol. 2010;28:126–31. doi: 10.1200/JCO.2009.24.2180. [DOI] [PubMed] [Google Scholar]
- 5.Tosoian JJ, Trock BJ, Landis P, Feng Z, Epstein JI, Partin AW, et al. Active surveillance program for prostate cancer: an update of the Johns Hopkins experience. J Clin Oncol. 2011;29:2185–90. doi: 10.1200/JCO.2010.32.8112. [DOI] [PubMed] [Google Scholar]
- 6.Epstein JI, Walsh PC, Carter HB. Dedifferentiation of prostate cancer grade with time in men followed expectantly for stage T1c disease. J Urol. 2001;166:1688–91. [PubMed] [Google Scholar]
- 7.Sheridan TB, Carter HB, Wang W, Landis PB, Epstein JI. Change in prostate cancer grade over time in men followed expectantly for stage T1c disease. J Urol. 2008;179:901–4. doi: 10.1016/j.juro.2007.10.062. discussion 4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porten SP, Whitson JM, Cowan JE, Cooperberg MR, Shinohara K, Perez N, et al. Changes in prostate cancer grade on serial biopsy in men undergoing active surveillance. J Clin Oncol. 2011;29:2795–800. doi: 10.1200/JCO.2010.33.0134. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi M, Ishida H, Shindo T, Niwa S, Kino M, Kawamura K, et al. Molecular analysis of multifocal prostate cancer by comparative genomic hybridization. Prostate. 2008;68:1715–24. doi: 10.1002/pros.20832. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Bostwick DG, Li G, Wang Q, Hu N, Vortmeyer AO, et al. Allelic imbalance in the clonal evolution of prostate carcinoma. Cancer. 1999;85:2017–22. doi: 10.1002/(sici)1097-0142(19990501)85:9<2017::aid-cncr20>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L, Song SY, Pretlow TG, Abdul-Karim FW, Kung HJ, Dawson DV, et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J Natl Cancer Inst. 1998;90:233–7. doi: 10.1093/jnci/90.3.233. [DOI] [PubMed] [Google Scholar]
- 12.Macintosh CA, Stower M, Reid N, Maitland NJ. Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res. 1998;58:23–8. [PubMed] [Google Scholar]
- 13.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–42. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 15.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–75. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–11. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miettinen M, Wang ZF, Paetau A, Tan SH, Dobi A, Srivastava S, et al. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 2011;35:432–41. doi: 10.1097/PAS.0b013e318206b67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–3. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–5. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 20.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–73. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.