Abstract
Copy-number variants (CNVs) represent a functionally and evolutionarily important class of variation. Here we take advantage of the use of pooled sequencing to detect CNVs with large differences in allele frequency between population samples. We present a method for detecting CNVs in pooled population samples using a combination of paired-end sequences and read-depth. Highly differentiated CNVs show large differences in the number of paired-end reads supporting individual alleles and large differences in read-depth between population samples. We complement this approach with one that uses a hidden Markov model to find larger regions differing in read-depth between samples. Using novel pooled sequence data from two populations of Drosophila melanogaster along a latitudinal cline, we demonstrate the utility of our method for identifying CNVs involved in local adaptation.
1. Introduction
Technological advancements over the last two decades have given researchers the ability to efficiently and accurately search for genetic differences between individuals across entire genomes. While initial efforts identified millions of single nucleotide polymorphisms (SNPs) within human populations [1], it was soon discovered that any two individuals also differ in copy-number at many large genomic regions encompassing whole genes or parts of genes [2]. In recent years, many more such copy-number variants (CNVs) have been detected using microarray hybridization intensity or sequence read-depth [3–5], paired-end/mate-pair sequencing [6,7], or both types of evidence together [8,9]. These CNVs are ubiquitous across eukaryotes, with large numbers also present in chimpanzees [10], mice [11], Arabidopsis [4], fruit flies [12,13], yeast [14], and many other species. These polymorphisms have attracted a great deal of attention because their large size suggests that they could have a considerable functional impact [2]. This hypothesis has been borne out by the large number of CNVs found to cause or increase the risk of various diseases in humans, including autism [15], schizophrenia [16], Charcot–Marie–Tooth disease [17], Crohn’s disease [18], and Parkinson’s [19].
The availability of large population genomic data sets has also allowed for genome-wide tests of recent and ongoing adaptive natural selection on CNVs, especially in humans [20,21]. Methods that detect signatures of adaptive evolution—such as long haplotypes with reduced nucleotide diversity [22,23] or large differences in allele frequencies across selective environments [24]— have been used to provide evidence for selection on CNVs [3,5]. These and other studies have identified a large number of putatively adaptive CNVs (>100 listed in [25]), with fitness benefits ranging from improved digestion of starches [26] to reduced HIV susceptibility [27]. Thus, CNVs appear to be an important source of adaptive as well as deleterious mutations in humans, and likely in other organisms as well.
A cost-effective and powerful way to detect variants with large differences in allele frequencies among populations is to pool and sequence large numbers of individuals from each of several populations using next-generation sequencing technologies (e.g. refs. [28,29]). While this pooling approach does not provide information on individual haplotypes, it can be used to accurately estimate other important population-genetic parameters [30,31]. Such an approach is also very effective at detecting locally adapted alleles when interbreeding between the sampled populations is frequent, as allele frequencies at neutral loci (i.e. those not affected by spatially varying selection) are homogenized very rapidly and linkage disequilibrium between selected polymorphisms and nearby variants is quickly reduced [32]. This is an ideal scenario for identifying polymorphisms that facilitate adaptation to local environments because one allele is favored by natural selection in one population and another allele is favored in other populations.
Searching for CNVs with a greater-than-expected difference in allele frequencies among populations has been particularly effective at identifying candidate adaptive CNVs in humans [25]. However, previous uses of pooled population sequences to detect highly differentiated CNVs has been limited to procedures examining the tails of distributions of differences in read-depth between two populations [28]. In this paper, we describe a principled method leveraging pooled sequencing data from two geographically dispersed but interbreeding populations in order to detect differentiated CNVs with high resolution. This method combines information from paired-end reads with a hidden Markov model designed to detect large differences in read-depth. We demonstrate the utility of this method on data from Drosophila melanogaster sampled from opposite ends of a latitudinal cline along the East Coast of the United States. Our search identifies 140 CNVs with the most highly differentiated allele frequencies between the two ends of the cline. These data and functional enrichment analyses strongly suggest that many of these CNVs are experiencing spatially varying selection.
2. Method for detecting CNVs with differentiated allele frequencies
2.1. Method overview
Here we describe in detail a method that leverages pooled paired-end next-generation sequence data to detect CNVs that differ substantially in allele frequency between two populations; in the data presented below these populations are at opposite ends of an environmental cline. Briefly, this method begins by searching for read pairs suggestive of deletions or tandem duplications. These read pairs are then combined into single putative events using a simple clustering algorithm, and putative events that are supported by many more read pairs from one population than the other are retained. For each of these putative duplications (Figure 1A) or deletions (Figure 1B), read-depth across the entire CNV region is examined in both populations, and those with a significant difference in read-depth between the two ends are considered CNV candidates for spatially varying selection. Because this paired-end approach may not detect all CNVs, we also use a hidden Markov model (HMM) to detect CNVs with differentiated allele frequencies based on read-depth alone. Both of these approaches will fail to detect CNVs segregating at low frequencies, depending on the depth of coverage to which pooled samples were sequenced.
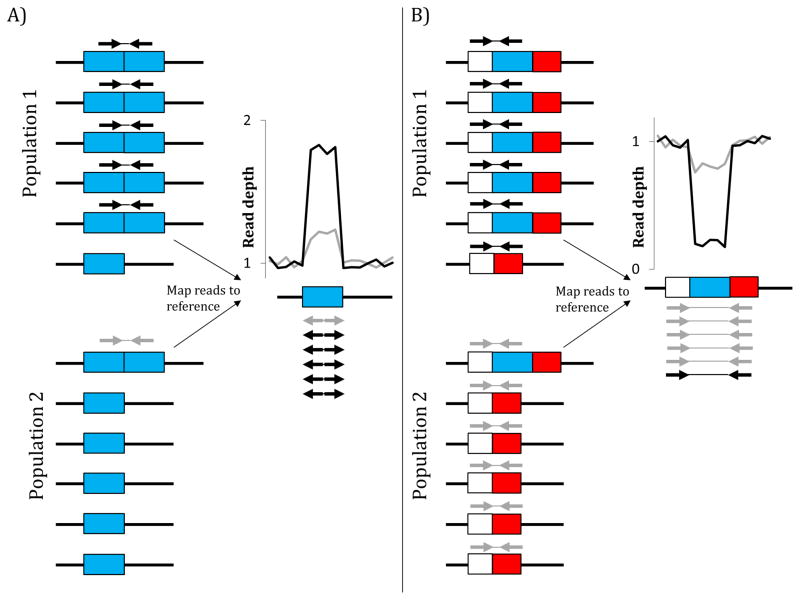
Figure 1.
Detecting CNVs differing in allele frequency between two populations each sequenced from pooled DNA. A) Paired-ends spanning a tandem duplication are mapped to the reference genome in “everted” orientation. Thus, if one population has many more such reads than the other, then the duplication likely differs in allele frequency between populations (e.g., the excess of black versus grey reads in this example). Read-depth within the duplicated locus would also be much higher in the population in which the duplication appears at higher frequency (black line versus grey line). B) When paired-ends from a region with a deletion are mapped to the reference, those from chromosomes with the deletion allele that span the breakpoints of the deletion are mapped much further apart than expected. A deletion differing in allele frequency between two populations will exhibit far more of these read-pairs and lower read-depth (black) within the deletion in one population than the other (grey).
2.2. Capturing and sequencing of D. melanogaster samples for test data
Our method was tested on sequence data from two pooled DNA samples of Drosophila melanogaster from opposite ends of a latitudinal cline in temperature and seasonality along the East Coast of the United States. 36 flies were captured from Bowdoinham, Maine and 30 were captured from Homestead, Florida. DNA was extracted from these two populations and pooled into two samples that were each sequenced using the Illumina Genome Analyzer IIx using short-insert paired-end libraries (~244 bp for Maine and ~213 bp for Florida on average) with 72 bp reads on each end. 60,730,268 read pairs were sequenced from the Maine sample and 48,771,223 were sequenced from Florida. After mapping to the euchromatic portion of the genome, the Maine data had an average read-depth of 49.39X and Florida had an average depth of 38.10X.
2.3. Detecting differentiated CNVs using paired-end sequencing
2.3.1. Mapping reads and detecting paired-ends indicative of CNVs
Our first method to detect CNVs under spatially varying selection leverages paired-end reads mapped in a manner suggestive of a duplication or deletion relative to the reference genome. In order to find such read pairs, we first map reads to the reference genome. In the case of our test data, we used BWA [33] to map reads to release 5 of the D. melanogaster genome assembly [34]. Whenever a read can be mapped equally well to multiple locations, and the mapping location(s) of the other read in the pair do not resolve this ambiguity, BWA randomly selects one of these locations to place the read. Other strategies, such as mapping the read to each location or discarding the read, are also compatible with the approach we describe here. In order to detect putative duplications, we then search for read pairs mapping to the same chromosome but in “everted” orientation, where the two reads are oriented away from one another rather than toward one another as expected. Such read pairs are expected in the case of head-to-tail tandem duplications, when read pairs crossing from the end of the first copy into the beginning of the second copy can only map to the first copy, to the left of the first read in the pair (Figure 1A; also illustrated in Figure 3A from ref. [35]). While this approach will only successfully identify tandem duplications in head-to-tail orientation, the vast majority of duplication polymorphisms in D. melanogaster meet these criteria [12,13]. For other species in which an appreciable number of duplication polymorphisms are non-tandem—such as humans [36]—this method would need to be extended to detect signatures of other types of duplications. Paired-end reads indicative of deletions relative to the reference are simply those that map in proper orientation but further apart than expected given the distribution of insert sizes [6]. We consider the most extreme 1% of read-pairs with respect to mapping distance from one another as potentially indicative of deletions. Discordantly-mapped paired-ends can also be used to detect inversions [6]: indeed, using an approach analogous to that described here we find that at least two known large inversions, In(2L)t and In(3L)P, are differentiated along the East Coast of the United States in Drosophila (data not shown).
2.3.2. Clustering discordant read-pairs
Sequenced read-pairs signifying copy-number variants (referred to as discordant read-pairs or discordant inserts) were clustered into distinct putative polymorphisms using a simple greedy clustering algorithm. Briefly, each discordant insert is initially assigned to its own cluster, and then pairs of clusters are examined and merged into a single cluster if any pair of inserts, one from each cluster, meets the following two criteria: 1) The coordinates of the reads must differ by no more than the largest expected insert-size (350 bp for our data); 2) For deletions, the inferred insert-sizes of the two inserts must differ by no more than the typical difference between insert-sizes from the sequenced library (200 bp for our data). If these criteria are met, then it is possible that the two inserts support the same mutation. This process is repeated until no more clusters can be merged. All clusters containing only a single discordant read-pair were ignored. When clustering putative deletions with our Drosophila data, there were a large number of clusters containing only a single insert. This is because we sequenced a large number of inserts, resulting in hundreds of thousands of inserts in the upper 1% tail with respect to insert-size that are likely not the result of deletions. Because a normal insert misclassified as discordant may therefore appear near a deletion supported by true discordant inserts, when examining any pair of deletion-supporting insert clusters <C1,C2> for overlap, we merged C1 and C2 only if 75% of all possible pairs of inserts (one insert from C1 and one insert from C2) met the merging criteria. Finally, after clustering was completed, any inserts within a cluster not appearing to support the putative mutation (according to the merging criteria) were removed from the cluster. Clustering is performed separately for each population.
2.3.3. Detecting highly differentiated CNVs
Once putative CNVs have been identified by clustering discordant paired-ends, we next search for polymorphisms differing in allele frequency between the two populations. This is done by first determining whether each polymorphism (i.e. each cluster of discordant inserts) present in one sample is present in the other using the merging criteria described in the previous section, and then comparing the numbers of distinct inserts supporting the event in each population; this number is zero when the event is not detected in a population. To reduce the number of false positive CNVs, we then removed all deletions supported by fewer than four paired-end reads. For each CNV, we calculate the difference between the number of inserts (after correcting for the total number of read pairs mapped to the reference genome from each sample) supporting the mutation in each cline-end, and retain all CNVs with a large difference (i.e., in either of the 5% tails of the empirical distribution of insert-number differences for deletions or 2.5% for deletions, where more stringency is required). For these events we then counted in each sample the total number of read pairs mapped in proper orientation and within the expected distance range, correcting for differences in average read-depth across the euchromatic genome.
The ratio of these corrected read-depths is then used as an independent mode of confirmation of highly differentiated CNVs detected by paired-ends. This was done in the D. melanogaster data by selecting roughly 10,000 random genomic regions of various lengths and calculating the read-depth ratios between the populations for these regions, and then calculating the 5% tails for each length. A CNV was considered confirmed as differentiated by read-depth if the depth ratio was biased in the same direction as the numbers of paired-ends in the two samples, and more extreme than 95% of read-depth ratios within random genomic regions of approximately the same length.
2.4. Detecting differentiated CNVs from read-depth using a hidden Markov model
The method described above requires both discordant paired-ends and read-depth to show evidence of differentiation in allele frequency between cline-ends. Because the number of discordant-paired ends may be small due to chance, some events with biologically significant differences in allele frequency may be missed. Dispersed duplications will also be missed because we only examine paired-ends supporting tandem duplications. We therefore complement the above approach by using a hidden Markov model (HMM) to detect differentiated CNVs based on read-depth alone. The observations for this HMM are, for small adjacent windows, the ratios of the (corrected) number of mapped paired-ends from one sample within the window over the (corrected) number of mapped paired-ends from the other sample in the same window. When testing this approach on our pooled D. melanogaster data, we binned these ratios to produce discrete observation symbols, though continuous distributions can also be used. The HMM has three states: no difference in read-depth between pooled samples, higher read-depth in sample 1, and higher read-depth in sample 2. The initial, transition, and emission probability matrices, which model the density and length of differentiated CNVs, and the distribution of read-depth ratios appearing in each of the hidden states, can be trained in a supervised manner using results from the paired-end based method described above. Once the HMM has been trained, the genome can be segmented into regions that have higher copy-number in sample 1, higher copy-number in sample 2, or similar copy-number in both samples, using the Viterbi algorithm.
2.5. Estimating allele frequencies
The methods outlined above detect strongly differentiated polymorphisms not by using allele frequency estimates directly, but by searching for differences in the numbers of mapped inserts between the two populations. However, for many applications researchers may wish to estimate actual allele frequencies at putatively differentiated CNVs. For duplications relative to the reference genome, this is given by:
| (1) |
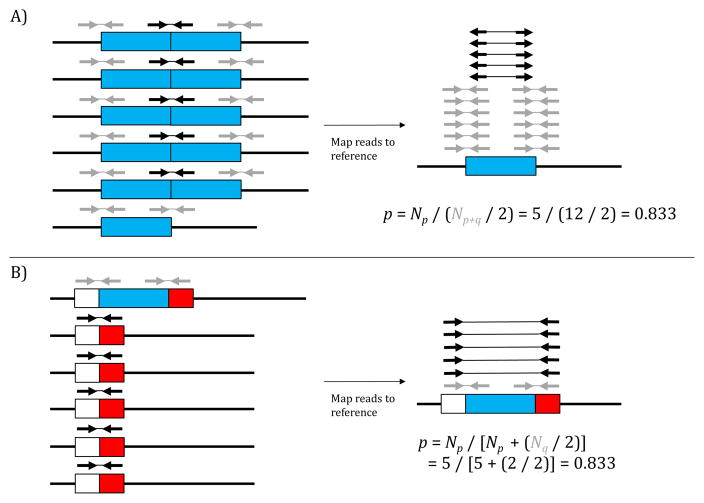
where Np is the number of “everted” inserts supporting the duplications, and Np+q is the total number of inserts mapping across either of the breakpoints and not supporting the duplication (as both chromosomes with and without the duplication will have this sequence; Figure 2A). For deletions relative to the reference genome,
Figure 2.
Estimating allele frequencies of CNVs from pooled DNA sequence data. A) For duplications, read pairs having one read flanking and one read within the duplicated locus (grey) are contributed from all chromosomes, while read-pairs mapped to the reference in “everted” orientation (black) are only sequenced from chromosomes with the duplication. B) For deletions, read pairs having one read flanking the deleted region and one read within the region (grey) are contributed only from chromosomes lacking the deletion, while read pairs spanning the entire deleted locus (black) are only derived from chromosomes with the deletion. Allele frequencies are estimated as shown in the examples using Equations 1 and 2, respectively.
| (2) |
where Np is the number of inserts mapping across the deletion breakpoints and supporting the deletion, and Nq is the total number of inserts mapping across either of the breakpoints (Figure 2B). In order to measure population differentiation, FST [37] can then be calculated using allele frequency estimates from the two samples.
Because these allele frequency estimates may be calculated from a small number of inserts they can be quite noisy, and it may be preferable to estimate allele frequencies in the two populations from read-depth across the entire CNV. This can be done by modeling expected read-depth across the genome as a response to variables such as GC composition [13,38] and density of SNPs and indels [13]. We do not describe this approach in detail here, but instead refer readers to Appendix B of ref. [13] for a more detailed discussion of modeling expected read depths in order to detect copy-number variation. Once this is done, the allele frequency of biallelic CNVs (those with only one copy polymorphic for presence/absence across individuals) is simply the percent excess or deficit of reads compared to the expectation in a given sample.
3. Assessing the utility of the method using D. melanogaster data from a latitudinal cline
3.1. Differentiated CNVs called from paired-ends and confirmed by read-depth
We assessed the utility of the method described above by searching for highly differentiated CNVs in Drosophila melanogaster along the East Coast of the United States. We pooled and sequenced DNA samples from Maine and Florida as described in Section 2.2, and then mapped paired-ends to the reference genome and clustered paired-ends indicative of deletions or tandem duplications (Sections 2.3.1 and 2.3.2), referred to as discordant paired-ends. We identified 9,428 putative deletions and 1,829 duplications relative to the reference genome. While the number of duplications is similar to those discovered in a recent examination of 37 whole-genome sequences of fruit flies captured from Raleigh, NC (2,588 duplications in [13]), we find substantially more deletions (only 3,336 in [13]), perhaps due to a large number of false positives in our set. 1,114 deletions and 129 duplications showed a large difference in the number of discordant paired-ends supporting the event in the two samples; we also calculated average read-depth across these putative CNVs in each sample. 102 deletions and 29 duplications were confirmed as differentiated CNVs by read-depth (Section 2.2.3), far more than the 5% expected by chance given our confirmation cutoffs (P=1.95×10−10 for deletions; P<2.2×10−16 for duplications; χ2 tests). In order to further assess the accuracy of this approach, we asked how many of these CNVs were found in a recent examination of 37 whole-genome sequences from Raleigh, North Carolina [13]. Because CNVs in these Raleigh genomes were detected from read-depth alone, an approach that has lower sensitivity to detect smaller variants, we examined only events >500 base pairs in length. We find that 31 of 52 (59.6%) of our differentiated CNVs >500 bp in length are also found in the Raleigh genomes (based on mutual 50% overlap with events in [13], or with paired-end sequences collected from two of these genomes indicative of CNVs). This is likely a substantial underestimate of our accuracy because some highly differentiated CNVs may have very low frequency in Raleigh, and suggests that most false CNV calls are removed by our two complementary tests for differentiated allele frequencies. Furthermore, the high correlation between the difference in number of paired-ends supporting a CNV in each sample and the ratio of read-depths between the two samples (ρ=0.786; P<2.2×10−16) suggests that both of these independent measures are estimates of allele frequency differentiation, and that we are accurately detecting differentiated CNVs. The average lengths of these deletions and duplications relative to the reference genome were 1,985 and 5,506 bp, respectively. Larger duplications than deletions have been observed previously in polymorphism data [13].
Because this method is designed to use pooled data, with the goal of finding polymorphisms differentiated across pooled samples, its performance cannot be compared to previous CNV-detection methods (e.g., [7–9,39–41]) in a straightforward manner. Indeed, to our knowledge the only existing method for detecting structural variants from pooled data is designed to detect transposable element insertion polymorphisms [42]. Thus, while many existing methods for detecting CNVs could be extended to the problem we address here, a comparison of the effectiveness of these methods with ours is beyond the scope of this paper.
3.2. Differentiated CNVs detected from read-depth using an HMM
Because the discordant paired-end approach may not detect all differentiated CNVs (Section 2.4), we supplemented this search using a hidden Markov model (HMM) examining only read-depth. Briefly, this HMM was used to segment the genome into three hidden states: differentiated CNVs with higher copy-number (and substantially higher read-depth) in Maine (State 1), regions with no differentiated CNVs (approximately equal read-depth; State 2), and differentiated CNVs with higher copy-number in Florida (State 3). It should be noted that this approach can only identify regions that differ in copy-number between the populations, and which population has higher copy-number—it does not determine whether the CNV is a duplication or deletion relative to the reference genome. It also does not detect regions with CNVs that do not differ in frequency between the two pools, unlike the paired-end method.
Observations for the HMM were ratios of read-depths of the two samples (Florida:Maine) in 100 bp windows, binned into one of the following categories of ratios: [0, 0.67), [0.67, 0.8), [0.8, 1), [1, 1.25), [1.25, 1.5), [1.5, ∞). We estimated the initial probabilities (the probability of the first genomic window lying within a differentiated CNV or not), transition probabilities modeling the average length of differentiated CNVs and the average distance between them, and emission probabilities (modeling the distribution of Florida:Maine read-depth ratios both within and outside of differentiated CNVs) from the properties of differentiated CNVs detected via the discordant paired-end method, yielding the following vectors/matrices (with minor manual adjustment based on prior expectations):
In order to infer the most likely sequence of hidden states across the genome, we ran the Viterbi and traceback algorithms on windowed Florida:Maine read-depth ratios, finding 11 highly differentiated CNVs, or stretches of genomic sequence assigned to either State 1 (elevated copy-number in Maine) or State 3 (elevated in Florida), with an average length of 5,776 bp. We found that two of these calls were also detected using paired-ends, implying that the remaining nine are either highly differentiated CNVs that the paired-end approach failed to detect or false positives. Of these nine CNVs, two were identified in previous analyses of highly differentiated genomic regions (containing Cyp12d, and Ace, respectively; Turner et al. 2008), and another two were detected in the Raleigh genomes [13]. Thus, these events may be true positives, underscoring the complementarity of the HMM approach to the paired-end approach discussed above.
3.3. Evidence of natural selection acting on differentiated CNVs
Although we have several lines of evidence that the vast majority of the putatively differentiated CNVs described in the sections above are true variants, additional evidence is required to show that these CNVs are not evolving neutrally with respect to the environmental cline. We estimated FST and found that many of these CNVs have high estimates (30 have FST > 0.2), but we cannot assess the significance of this given that the neutral expectation for our data is unknown (synonymous SNPs may be linked to nearby selected polymorphisms), and our FST estimates may have considerable variance. Thus, the best way to evaluate the impact of natural selection on these CNVs may be to search for enrichment of certain genes and functional categories. We noticed that several CNVs contain complete or partial cytochrome P450 genes, including Cyp28d2, Cyp12d1-p, Cyp12d1-d, Cyp6a17, Cyp6a22, Cyp6a23, Cyp12c1, Cyp313a4, and Cyp12a4. This is more than are expected by chance (P=0.0032; permutation test of the 140 most differentiated CNVs; P<0.0001 when testing for an excess of CNVs containing at least one Cyp gene—both of these tests control for spatial clustering of related genes). Members of this superfamily are often involved in insecticide resistance [43], and overexpression of Cyp12d1 [44] and Cyp12a4 [45] have been shown to increase insecticide resistance; this selective pressure may therefore be the cause of the extensive differentiation seen at these genes. In addition, Cyp6a17 has been shown to affect temperature preference [46], implying that insecticide resistance may not be the only geographically dependent fitness effect conferred by cytochrome P450s in Drosophila. Ace (acetylcholinesterase), another gene involved in insecticide resistance [47] and previously identified as lying partially within a CNV differentiated along this cline [48], was also found in our analysis.
We searched for overrepresented Gene Ontology (GO) terms associated with genes lying within differentiated CNVs, using the hypergeometric distribution as our null hypothesis for each term. It is important to note that GO enrichment analyses conducted on CNVs or other large regions can be biased away from the null distribution by the clustering of functionally related genes [48,49]. We therefore allowed each term to be counted at most once per CNV before calculating significance. In order to correct for multiple testing, we calculated the false discovery rate (FDR) following ref. [50]. We identified a large number of terms with FDR <0.1, including 67 biological process terms. In addition to terms involved with response to pesticides (e.g., response to organophosphorus, response to carbamate), this set of enriched terms included several related to neuronal development and activity, including regulation of short-term neuronal synaptic plasticity, synaptic transmission, and synaptic target attraction. The enrichment of these terms is not driven by the overrepresented insecticide resistance genes discussed above, suggesting that CNVs confer distinct spatially dependent fitness benefits related to nervous system development and insecticide tolerance.
The overrepresentation of the functional categories listed above lends further confidence to our assertion that a substantial fraction of the CNVs detected by the method described here are under spatially varying selection. Although the results of this type of enrichment analysis should not be taken as proof of the action of natural selection [49], they do support our assertion that natural selection is driving differentiation of CNVs in D. melanogaster along the East Coast, thereby demonstrating the utility of our method for identifying CNVs under spatially varying selection.
4. Discussion
The method presented here accurately detects differentiated copy-number variants from pooled DNA sequence data, and we show that many of the CNVs identified likely reside in regions experiencing spatially varying selection. Because of the high level of gene flow between the two Drosophila samples examined here, differentiation at neutral variants is short-lived, and regions with polymorphisms differing in allele frequency between the two samples are quite small, often on the order of 5 kb or less [48]. Thus, while it is difficult to be certain that any given CNV identified by the approach described here is indeed responsible for allele frequency differentiation, it is likely that many of the CNVs identified by our method are indeed beneficial mutations. We believe this approach has the potential to identify CNVs under spatially varying selection in other species and environmental gradients, and significantly improve our understanding of the contribution of copy-number variation to adaptive evolution.
Contributor Information
DANIEL R. SCHRIDER, Email: dschride@indiana.edu, Department of Biology and School of Informatics and Computing, Indiana University, 1001 E Third St., Bloomington, IN 47405, USA.
DAVID J BEGUN, Email: djbegun@ucdavis.edu, Department of Evolution and Ecology, University of California, 3350A Storer Hall, Davis, CA 95616, USA.
MATTHEW W HAHN, Email: mwh@indiana.edu, Department of Biology and School of Informatics and Computing, Indiana University, 1001 E Third St., Bloomington, IN 47405, USA.
References
- 1.Sachidanandam R, et al. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Conrad DF, et al. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ossowski S, et al. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redon R, et al. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuzun E, et al. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 7.Hormozdiari F, et al. Genome Res. 2011;21:2203–2212. doi: 10.1101/gr.120501.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medvedev P, et al. Genome Res. 2010;20:1613–1622. doi: 10.1101/gr.106344.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handsaker RE, et al. Nat Genet. 2011;43:269–U126. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry GH, et al. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graubert TA, et al. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson JJ, et al. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 13.Langley CH, et al. Genetics. 2012 doi: 10.1534/genetics.1112.142018. [DOI] [Google Scholar]
- 14.Carreto L, et al. BMC Genomics. 2008;9:524. doi: 10.1186/1471-2164-9-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girirajan S, et al. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-De-Luca D, et al. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupski JR. Nat Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 18.McCarroll SA, et al. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singleton AB, et al. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 20.Altshuler D, et al. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altshuler DM, et al. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabeti PC, et al. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 23.Voight BF, et al. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreiro LB, et al. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 25.Iskow RC, Gokcumen O, Lee C. Trends Genet. 2012;28:245–257. doi: 10.1016/j.tig.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry GH, et al. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez E, et al. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 28.Kolaczkowski B, et al. Genetics. 2011;187:245–260. doi: 10.1534/genetics.110.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng C, et al. Genetics. 2012;190:1417–1432. doi: 10.1534/genetics.111.137794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futschik A, Schlotterer C. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, et al. PLoS ONE. 2012;7:e41901. doi: 10.1371/journal.pone.0041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slatkin M. Genetics. 1981;99:323–335. doi: 10.1093/genetics/99.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams MD, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 35.Cooper GM, et al. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrider DR, Hahn MW. Mol Biol Evol. 2010;27:103–111. doi: 10.1093/molbev/msp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright S. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alkan C, et al. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abyzov A, et al. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, et al. Nat Methods. 2009;6:473–474. doi: 10.1038/nmeth.f.256. [DOI] [PubMed] [Google Scholar]
- 41.Sindi S, et al. Bioinformatics. 2009;25:I222–I230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kofler R, Betancourt AJ, Schlotterer C. PLoS Genet. 2012;8:e1002487. doi: 10.1371/journal.pgen.1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranson H, et al. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- 44.Daborn PJ, et al. Insect Biochem Mol Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Bogwitz MR, et al. Proc Natl Acad Sci U S A. 2005;102:12807–12812. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang J, Kim J, Choi K-W. PLoS ONE. 2011;6:e29800. doi: 10.1371/journal.pone.0029800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menozzi P, et al. BMC Evol Biol. 2004;4:4. doi: 10.1186/1471-2148-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner TL, et al. Genetics. 2008;179:455–473. doi: 10.1534/genetics.107.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavlidis P, et al. Mol Biol Evol. 2012 doi: 10.1093/molbev/mss1136. [DOI] [Google Scholar]
- 50.Storey JD. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2002;64:479–498. [Google Scholar]