Abstract
The rapid development of sequencing technologies makes thousands to millions of genetic attributes available for testing associations with various biological traits. Searching this enormous high-dimensional data space imposes a great computational challenge in genome-wide association studies. We introduce a network-based approach to supervise the search for three-locus models of disease susceptibility. Such statistical epistasis networks (SEN) are built using strong pairwise epistatic interactions and provide a global interaction map to search for higher-order interactions by prioritizing genetic attributes clustered together in the networks. Applying this approach to a population-based bladder cancer dataset, we found a high susceptibility three-way model of genetic variations in DNA repair and immune regulation pathways, which holds great potential for studying the etiology of bladder cancer with further biological validations. We demonstrate that our SEN-supervised search is able to find a small subset of three-locus models with significantly high associations at a substantially reduced computational cost.
Keywords: Epistasis, High-order genetic interactions, GWAS, Statistical epistasis networks, MDR
1. Introduction
The goal of genome-wide association studies (GWAS) is to identify and characterize susceptibility genes that can help diagnose, treat, and prevent common human diseases.1–3 However, most existing association analyses employ main-effect-centered strategies that assume a simple genetic architecture and are thus only able to find very limited single-locus effects on disease risks.4 The non-additive effect of gene-gene interactions, i.e. epistasis, has been recognized playing an important role explaining the complex relationship between the genetic and phenotypic variations.5–7 Thus, identifying and characterizing genetic interactions across multiple loci have become the focus of current association studies.8–10 However, this imposes a great computational challenge in high-dimensional data analyses. Specifically, for a genetics dataset consisting of n loci, the computational complexity of enumerating all possible two-locus combinations is O(n2), and it increases exponentially with the order of combinations considered. Given the sizes of current genome-wide data (n ~ 106) and the next-generation whole-genome sequencing11 data (n ~ 109), it requires 3 × 104 to 3 × 1013 years to enumerate and evaluate all three-locus models, using a 1000-node computer cluster where each node is assumed to be able to process 1000 models per second. Therefore, new data-mining technologies with advanced and efficient pre-screening and attribute-selection strategies are needed in large-scale genetic association studies.12–15
In this article, we propose a network-based model-prioritization approach that is able to identify high-order association models at a significantly reduced computational cost than exhaustive enumerations. The networks were built by including strong pairwise epistatic interactions as edges and their two end genetic attributes as vertices, as in the framework of statistical epistasis networks (SEN) previously developed by Hu et al.16 Following the hypothesis that strong pairwise interactions may indicate the existence of higher-order interactions, we propose to i) quantify all pairwise epistatic interactions in a given genetics dataset; ii) construct pairwise statistical epistasis networks; iii) identify attributes that are clustered together by traversing the networks; iv) evaluate the clustered attributes for higher-order interactions. This distinguishes our approach the most from many existing attribute-selection strategies and advances the detection of higher-order interactions since hypothetically it is much less likely for a higher-order interaction to exist without showing any lower-order interactions than without showing any main effects.17,18
In the present study, we consider searching for three-locus interaction models and use the multifactor dimensionality reduction (MDR) algorithm and software to evaluate the associations of the models found by our SEN-supervised search. MDR is a data-mining strategy for detecting and characterizing gene-gene interactions associated with a discrete disease status.19–22 It pools multi-locus genotypes from multiple single-nucleotide polymorphisms (SNPs) into high-risk and low-risk groups. Specifically, a multi-locus genotype combination is considered high-risk if it has subjects with a ratio of cases to controls higher than a given threshold; otherwise it is considered low-risk. The clustering of all multi-locus genotype combinations into high-risk and low-risk is then evaluated for its ability to classify and predict disease status through cross-validations. Population-based data are partitioned into a training set and a testing set. The attribute combination with the highest training accuracy is chosen as the best model and is subsequently evaluated using the testing set. The article by Moore et al22 provides a good overview of the development of MDR. MDR is model-free, i.e. no particular genetic models are assumed, and non-parametric, i.e. no parameters are estimated, and is thus an ideal independent classifier to evaluate our SEN-supervised model search.
We previously identified a pairwise interaction network by applying SEN to a large population-based bladder cancer dataset.16 Such a network showed significant topological properties compared to the null networks built from permuted data. We believe that this large connected structure captures the complex genetic architecture of bladder cancer and is a promising guide-map for searching higher-order combinations of attributes that may jointly modify the disease outcome. Here, we use this bladder cancer pairwise interaction network to supervise the search for high-association three-locus models using a fast network traversing algorithm that identifies trios clustered together.
2. Methods
2.1. Bladder cancer dataset
The dataset used in this study includes 1422 SNPs from about 500 cancer susceptibility genes for 491 bladder cancer cases and 791 healthy controls.23,24 The bladder cancer cases were collected among New Hampshire residents of ages 25 to 74 years, diagnosed from July 1, 1994 to June 30, 2001 and identified in the State Cancer Registry. Controls less than 65 years of age were selected using population lists obtained from the New Hampshire Department of Transportation, while controls aged 65 and older were chosen from data files provided by the Centers for Medicare & Medicaid Services (CMS) of New Hampshire. Most (> 95%) of the subjects were of Caucasian origin. Informed consent was obtained from each participant and all procedures and study materials were approved by the Committee for the Protection of Human Subjects at Dartmouth College. DNA was isolated from peripheral circulating blood lymphocyte specimens using Qiagen genomic DNA extraction kits (QIAGEN Inc., Valencia, CA). Genotyping was performed on all DNA samples of sufficient concentration, using the GoldenGate Assay system by Illumina’s Custom Genetic Analysis service (Illumina, Inc., San Diego, CA). Out of the submitted samples, 99.5% were successfully genotyped, and samples repeated on multiple plates yielded the same call for 99.9% of the SNPs.
2.2. Statistical epistasis networks (SEN)
We have previously developed a network approach to inferring statistical epistasis of bladder cancer.16 First, entropy-based information-theoretic measures were used to quantify pairwise interactions22,25–28 for all two-locus models in the bladder cancer dataset. Specifically, for two genetic attributes G1, G2, and the phenotypic status C, mutual information I(G1;C) and I(G2;C) measure the shared information, or dependency, between individual genotypes and the phenotype, i.e. the main effects. In addition, by joining G1 and G2 together, I(G1,G2;C) measures how much of the phenotypic status that combining G1 and G2 can explain. The epistatic interaction strength between G1 and G2 can then be defined using information gain IG(G1;G2;C) = I(G1,G2;C)−I(G1;C)−I(G2;C). As such, IG(G1;G2;C) is the gained mutual information about C from considering genetic attributes G1 and G2 together, i.e. the synergy between G1 and G2 on the phenotype C. Moreover, normalizing the main effect I(G1;C) and the interaction effect IG(G1;G2;C), by dividing them by the entropy of the phenotype H(C), provides the association of a single attribute or a pairwise interaction with the phenotype C, i.e. the percentage of the phenotypic status that a genotype can explain.
Second, we ranked all possible pairwise interactions between SNPs according to their relative strength and subsequently built a series of statistical epistasis networks by incrementally adding edges if their corresponding pairwise interaction strength was stronger than a given cutoff value. Topological properties were analyzed for the network at each cutoff value including the size of the network (the number of its vertices and the number of its edges), the connectivity of the network (the size of its largest connected component), and its vertex degree distribution. Permutation testing was used to generate a null distribution of those topological properties by building permuted-data networks through the same construction process and using the same cutoffs.
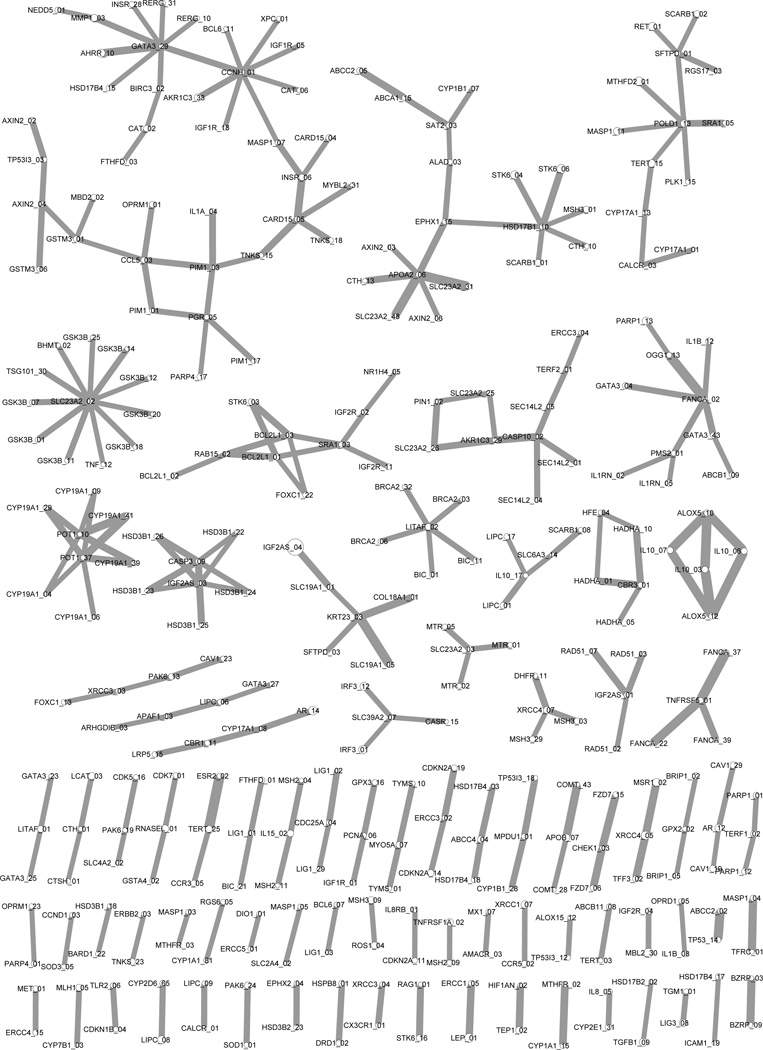
Then, a threshold of the pairwise interaction strength was determined by finding the cutoff when the topological properties of the real-data network differentiated the most from the null distribution.16 Such a systematically derived and most significant epistasis network of bladder cancer is shown in Fig. 1. This network provided a global map of strong pairwise epistatic interactions associated with bladder cancer. It was able to show not only the neighborhood structure of each attribute, but also the topology of a set of clustered attributes. Thus it serves as a very promising tool to identify higher-order genetic models.
Fig. 1.
The derived statistical epistasis network of bladder cancer. The network includes 319 SNPs (vertices) and 255 pairwise interactions (edges). The size of a vertex represents the strength of the main effect of its corresponding SNP, with the disease association ranging from 0.001% to 1.614%. The width of an edge indicates the strength of its corresponding interaction, with the disease association ranging from 1.354% to 2.052%.
2.3. SEN-supervised search for three-locus genetic models
SEN is essentially an attribute-prioritization approach. However, different from many existing main-effect-centered pruning methods, our network strategy prioritizes attribute pairs that show strong or significant interactions. In addition, organizing these strong interacting pairs in the network format provides a landscape of interaction structures. We hypothesize that the sets of attributes that are clustered together in the bladder cancer network may better explain the case-control outcome than the non-clustered sets. Therefore, we propose to use SEN to supervise the search for multi-locus association models. As the first attempt, in this study, we consider the search for three-locus models and use MDR to assess the associations of three-locus models.
The clustering of vertices, or attributes, in a network is determined based on their pairwise distances. In Graph Theory,29 the distance d(v1, v2) of a pair of vertices v1 and v2 is defined as the minimal number of edges for one vertex to reach the other. Two vertices v1 and v2 are neighbors if d(v1, v2) = 1. Given three vertices v1, v2, and v3, we define their trio distance dtrio(v1, v2, v3) as the sum of all pairwise distances, i.e. dtrio(v1, v2, v3) = d(v1, v2) + d(v1, v3) + d(v2, v3). Therefore, for trios with dtrio = 3, any two of them are directly joined by an edge, and if a trio has dtrio = 4, one vertex is directly connected to the other two but the other two are not joined by an edge. We define that a trio of attributes are clustered in a network if their dtrio ≤ 4; otherwise we say that they are not clustered together.
All three-locus models of clustered trios can be identified by traversing the SEN, represented as a graph G with |V| vertices and |E| edges, using the following algorithm. It reads G and outputs a list of trios of vertices that are connected together. The algorithm has a computational complexity O(|V| × k2), where k is the maximum number of neighbors of a vertex in G:
| vertices = G.getVertices(); |
| for each v in vertices do |
| neighbors = v.getNeighbors(); |
| for each u1 in neighbors |
| for each u2 in neighbors do |
| output {u1, v, u2}; |
Note that in our bladder cancer epistasis network (Fig. 1) k = 11 ≪ |V|, so the complexity of the above algorithm O(|V| × k2) ≈ O(|V|). Thus the SEN-supervised search significantly reduces the computational complexity compared to enumerating all three-locus combinations.
3. Results
We first applied a χ2 test of independence to identify SNPs with significant main effects. For all 1422 SNPs from the entire dataset, using a Bonferroni-corrected significance level of α = 0.05, we found only one significant main-effect attribute IGF2AS_04 (p = 1.052 × 10−6). This SNP had one interacting neighbor SLC19A1_01 captured in our SEN (Fig. 1), and this pairwise interaction was previously reported.30 Thus we removed IGF2AS_04 from our interaction analysis to avoid its dominance effect when combined with other attributes.
Next, for the other 318 SNPs identified in the bladder cancer network, we ran MDR exhaustively on all 1-way, 2-way , and 3-way combinations. We analyzed the correlation between MDR accuracies and SNP neighborhood structures in the network, in order to see whether clustered SNPs in the network have better disease status prediction accuracies than non-clustered ones.
3.1. MDR accuracy comparison of clustered and non-clustered SNP trios
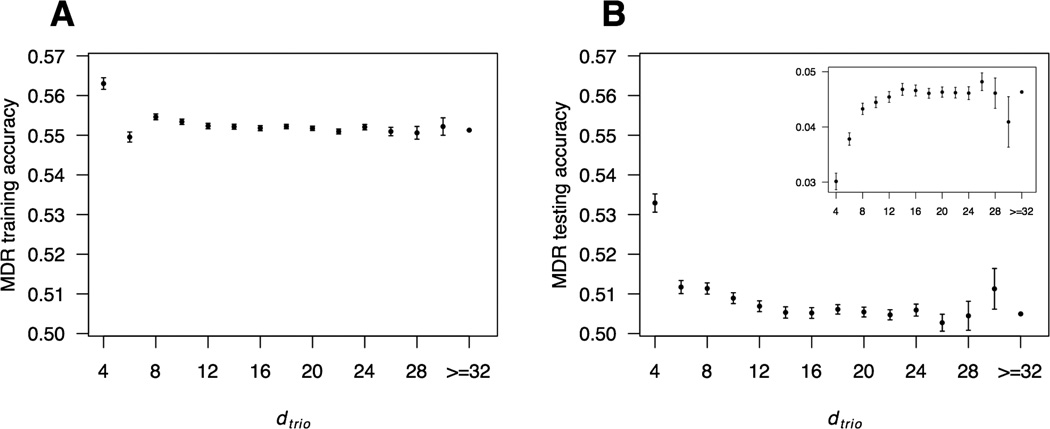
We categorized all 5,309,116 trios according to their trio distances and show the MDR accuracies in each distance category (Fig. 2). We observe that, since there are no triangles in the network, the minimal trio distance is 4. In addition, trios of distances greater than 32 are not connected in the network, i.e. at least two out of the three vertices do not have a path connecting them. The clustered trios of distance 4 have significantly higher training and testing accuracies than the trios in all other distance categories, while those other distance categories do not statistically distinguish among themselves. Moreover, the clustered trios have better consistencies between training and testing accuracies (Fig. 2B inset).
Fig. 2.
The 3-way MDR A) training accuracy and B) testing accuracy relative to the trio distance. Points are mean values and bars show the 95% confidence intervals. The inset depicts Δ = training accuracy − testing accuracy, which indicates the level of over-fitting. A lower value of Δ means a better prediction consistency for training and testing data.
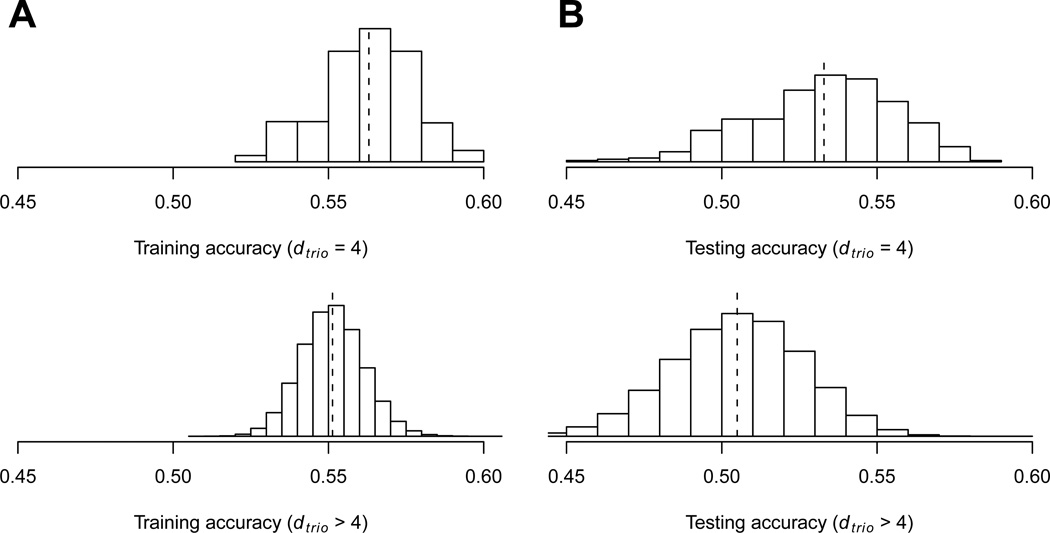
We then binned all dtrio > 4 three-locus models together as non-clustered trios, and compared their distributions of MDR training and testing accuracies to those of the clustered trios (Fig. 3). As seen from the figure, clustered trios have both better training and testing accuracies compared to non-clustered trios. Therefore, using the pairwise SEN structure was able to identify a good subset of three-locus combinations that improved the phenotypic status prediction accuracy.
Fig. 3.
Distributions of 3-way MDR A) training and B) testing accuracies for clustered (dtrio = 4) and non-clustered (dtrio > 4) trios. The mean of each distribution is shown using a vertical dashed line. There are 391 clustered trios and 5, 309, 116− 391 = 5, 308, 725 non-clustered trios.
We also performed a correlation analysis on the MDR accuracies at different combination orders. Table 1 shows that, in general, three-way accuracies had stronger correlations with two-way accuracies than those with one-way accuracies. Compared to non-clustered trios, the three-way accuracies of clustered trios were less correlated with one-way accuracies. That is, three-locus models of clustered trios were less biased towards high main-effects attributes. When correlating two-way with three-way accuracies, compared to non-clustered trios, clustered trios had a lower dependency on training data but a higher dependency on testing data.
Table 1.
Spearman’s rank correlation of MDR accuracies at different model orders
| 1-way vs. 3-way | 2-way vs. 3-way | |
|---|---|---|
| Training balanced accuracy | ||
| Clustered trios | ρ = 0.1863 (p = 1.27 × 10−10) | ρ = 0.4319 (p < 2.2 × 10−16) |
| Non-clustered trios | ρ = 0.2934 (p < 2.2 × 10−16) | ρ = 0.5897 (p < 2.2 × 10−16) |
| Testing balanced accuracy | ||
| Clustered trios | ρ = 0.1060 (p = 2.77 × 10−4) | ρ = 0.4027 (p < 2.2 × 10−16) |
| Non-clustered trios | ρ = 0.1946 (p < 2.2 × 10−16) | ρ = 0.3795 (p < 2.2 × 10−16) |
3.2. SEN-supervised MDR three-locus models
As shown previously, SEN-supervised search yielded a small subset of three-locus combinations (391 out of 5,309,116) based on their clustering structure in the network, and this small subset had significantly better three-way MDR accuracies compared to the others. In this section, we examined the results of these SEN-supervised MDR models, and tested whether the observations from such a model-selection process were statistically significant.
For these 391 SEN-filtered trios, their best and average MDR accuracies are reported in Table 2. We performed two sets of significance tests to assess the p-values for each observation. First, we randomly resampled 391 trios out of the total 5,309,116 and repeated it 1000 times. Second, on the 318 vertices identified in the network, we permuted their neighborhood structures by swapping edges. For each edge swapping, two edges, e.g. e1 = {v11, v12} and e2 = {v21, v22}, were picked randomly, and then their end vertices were swapped to form two new edges . This was a standard network randomization procedure where the total number of neighbors for each vertex was preserved but its interacting partners were randomized. For each permutation, we performed edge swapping 10 × |E| times, where |E| is the total number of edges in the network (Fig. 1). Such a permutation process provided null networks with randomized pairwise interactions. Again, we generated 1000 permuted networks and used them to identify the clustered-trio subsets. Then MDR analyses were applied to both sets of permuted data and the assessed significances of the real observations are shown in Table 2. As we can see, all observations from the subset found by SEN-supervised search were statistically significant.
Table 2.
MDR results of the clustered trios and their levels of statistical significance
| Observed-value | Significance | ||
|---|---|---|---|
| random-resample | edge-swap | ||
| Best training accuracy | 0.5992 | p = 0.005 | p = 0.002 |
| Best testing accuracy | 0.5873 | p = 0.002 | p < 0.001 |
| Average training accuracy | 0.5630 | p < 0.001 | p < 0.001 |
| Average testing accuracy | 0.5329 | p < 0.001 | p < 0.001 |
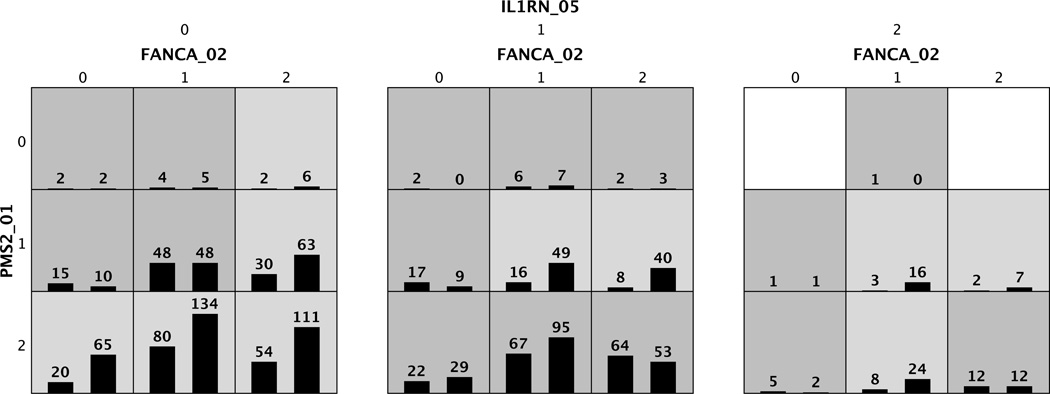
The best three-locus MDR model using SEN-supervised search was FANCA_02, PMS2_01, and IL1RN_05, with a training balanced accuracy 0.5992 and a testing balanced accuracy 0.5783 (p = 1 × 10−5 using a standard permutation test). This model included two DNA repair genes and one immune regulation gene. Fig. 4 summarizes the MDR analysis for the best model. Out of all 27 possible genotype combinations, 25 had observed samples, 15 genotypes were predicted as high-risks (dark-grey cells), and 10 genotypes were predicted as low-risks (light-grey cells).
Fig. 4.
Summary of the best MDR model using SEN-supervised search. A three-locus model has 27 multi-factorial cells, each of which is filled with the distribution of cases (left bars) and controls (right bars) for the corresponding genotypes. A cell is left blank if there are no samples falling into its genotype. Each non-empty cell is labeled either “high-risk” (dark grey) or “low-risk” (light grey) based on its case-control ratio.
3.3. Comparing SEN-supervised search to other common MDR filters
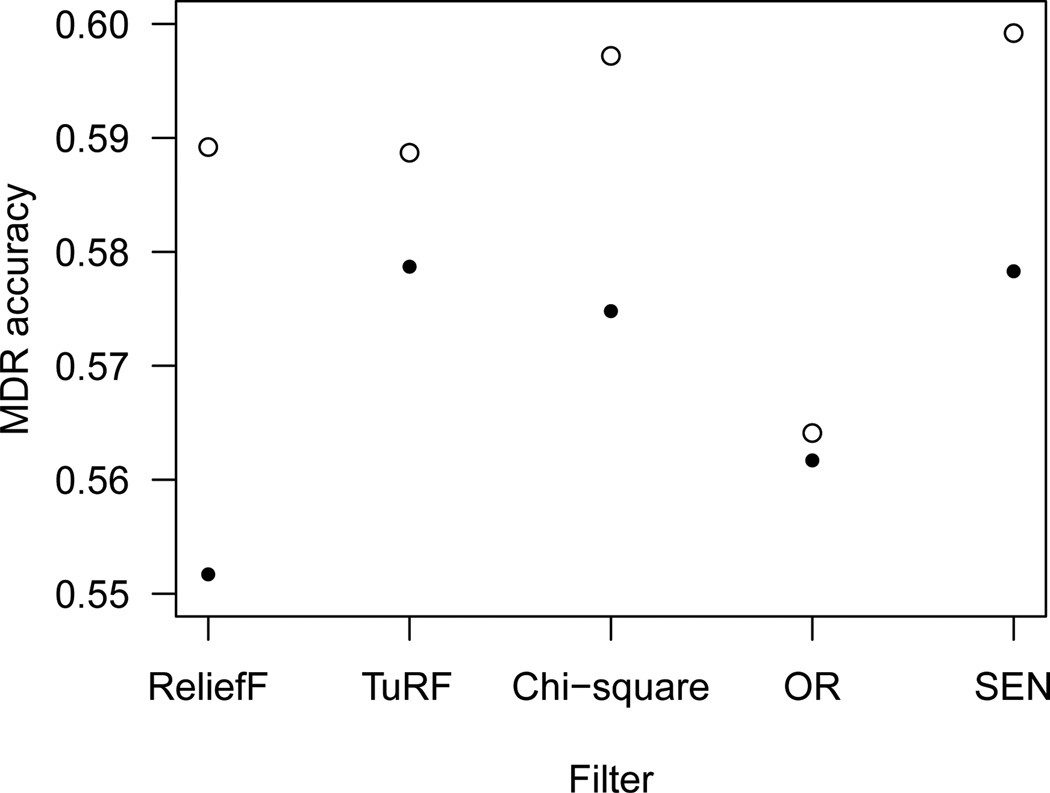
Due to the exhaustive enumeration nature of MDR, attribute-selection is usually used for large genome-wide data. We implemented four most commonly used filters, ReliefF,31 TuRF,32 Chi-square, and Odds Ratio (OR), on the bladder cancer data (1422 SNPs), and compared the best models they found to our best model using SEN-supervised search (Fig. 5). For each of the four other filters, we chose its top 15 most important attributes and ran MDR on all three-locus combinations of them. This also provided a comparable number of models for MDR to evaluate since SEN-supervised search yielded 391 three-locus combinations. As seen in the figure, our SEN-supervised search found the best three-locus model compared to all the other common attribute-selection strategies.
Fig. 5.
Results of the best three-locus MDR models using five different attribute-selection or model-prioritization techniques. Circles represent training balanced accuracies and solid points are testing balanced accuracies.
4. Discussion
Epistasis has been recognized playing an important role in understanding the mapping between genetic and phenotypic variations.8–10 Detecting and characterizing epistasis is a very challenging data-mining task due to the fact that the epistatic interactions could involve multiple genetic attributes from a pair to a large set, and this undetermined order of interactions imposes enormous computational complexities for enumerating all possible combinations of genetic attributes for varying orders in genome-wide data.15 Various pre-screening techniques have been proposed to filter potentially important attributes for further higher-order combination analyses. However, most of them adopt main-effect-centered strategies and may overlook attributes that are important in interactions but only show weak main effects.17
In this article, we proposed a network-guided approach to searching three-locus genetic models for association studies. The network was built by including strong pairwise epistatic interactions, and we were able to show that trios clustered together in this network have higher associations than those non-clustered ones. Traversing the pairwise statistical epistasis networks (SEN) to search clustered three-locus models significantly reduces the computational complexity of enumerating all possible three-locus combinations. Thus our SEN-supervised model search can serve a very promising prioritization method and can be combined with many existing association-mining techniques, such as MDR used in this study.
We had previously developed a network approach to characterizing statistical epistasis interactions in genetic association studies.16 In this framework, all pairwise interactions in a genetic dataset were quantified using information gain, an information-theoretic measure based on Shannon entropy.33 Then networks were built by including pairs of attributes, as edges and two end vertices, if their pairwise interaction strengths were greater than a theoretically-derived threshold. This threshold was determined systematically by analyzing network topo-logical properties and comparing them to null networks built using permuted data through the same construction process. This SEN approach advanced many existing genetic association methods by focusing on interactions rather than individual genetic factors. Moreover, by organizing interactions in the form of networks, SEN provided a global connection map and suggested clustering of multiple attributes that might have joint effects on the phenotype.
The present study explored the clustering structure captured in our previous SEN application to a bladder cancer dataset (Fig. 1). Using a fast network-traversing algorithm, the three-locus models of clustered trios were identified and further evaluated using MDR. These models were shown having both significantly higher training and testing MDR accuracies than the three-locus models of non-clustered trios (Fig. 2 and Fig. 3). Moreover, the clustered models had less over-fitting (Fig. 2B inset). These results show that the SEN-supervised search was able to identify a small subset of three-locus models with significantly high associations at a very moderate computational cost. Note that even if the computational complexity of building a pairwise interaction network (O(|V|2)) is considered together with the SEN-supervised search (O(|V| × k2) ≈ O(|V|)), where |V| is the total number of attributes and k is the maximum number of neighbors of an attribute in the network, the computational cost is still far less than enumerating all possible three-locus combinations (O(|V|3)). This reduction of computational complexity is even more encouraging in the era of genome-wide and whole-genome studies where thousands to millions of genetic attributes are considered.
The best three-locus MDR model identified using the SEN-supervised search includes FANCA_02 (rs2239359), PMS2_01 (rs3735295), and IL1RN_05 (rs419598). All three SNPs had very limited main effects with one-way MDR testing accuracies 0.4929, 0.5110, and 0.5276, respectively. The falcon anemia complementation group A (FANCA) gene produces DNA repair protein that may operate in a post replication repair or a cell cycle checkpoint function. Postmeiotic segregation increased 2 (PMS2) is a component of the post-replicative DNA mismatch repair system. Interleukin 1 receptor antagonist (IL1RN) encodes the protein that inhibits the activities of interleukin 1 alpha (IL1A) and interleukin 1 beta (IL1B), and modulates a variety of interleukin 1 related immune and inflammatory responses. The three genes have moderate biological relationships,34 all have been found associated with various cancers, and both DNA repair and immune regulation are considered major biological processes involved in bladder carcinogenesis.35–37 However, the interaction effect among the three genes associated with bladder cancer has never been reported previously. One could speculate, nevertheless, that defects in the protective cell cycle checkpoint and DNA repair functions could lead to attempts to replicate damaged DNA. Immune surveillance would be the remaining protective mechanism to eliminate potential tumor cells. Thus, this trio of genetic variations could increase the probability of tumor cell expansion. We expect that with further biological validations, our findings could help explain the etiology and the complex genetic architecture of bladder cancer.
With the fast development of sequencing technologies, more and more large-scale biomedical data are becoming available. Although this presents exciting opportunities for genetic association studies to explain many common human diseases, mining these high-dimensional data to identify important genetic factors with non-linear interaction effects is a daunting endeavor. In this article, we proposed a network-guided search approach that is able to efficiently identify high-association three-locus genetic models. Our approach prioritizes genetic attributes that have strong pairwise interaction effects. This differentiates our method from most existing pre-screening strategies that focus on individual attributes with significant main effects. The effectiveness of our approach was validated using MDR. In future research, we expect to extend our SEN-supervised approach to the search for higher-order models and to expand its applications to more data-mining and classification techniques.
Acknowledgements
This work was supported by the National Institute of Health (NIH) grants R01-LM009012, R01-LM010098, and R01-AI59694 to JHM, and P42-ES007373 and R01-CA5749367 to MRK.
Contributor Information
Ting Hu, Department of Genetics, Geisel School of Medicine, Dartmouth College, Hanover, NH 03755, USA.
Angeline S. Andrew, Department of Community and Family Medicine, Geisel School of Medicine, Dartmouth College, Hanover, NH 03755, USA
Margaret R. Karagas, Department of Community and Family Medicine, Geisel School of Medicine, Dartmouth College, Hanover, NH 03755, USA
Jason H. Moore, Institute for Quantitative Biomedical Sciences, Departments of Genetics and Community and Family Medicine, Geisel School of Medicine, Dartmouth College, Hanover, NH 03755, USA.
References
- 1.Hirschhorn JN, Daly MJ. Nature Review Genetics. 2005;6:95. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 2.Wang WYS, Barratt BJ, Clayton DG, Todd JA. Nature Review Genetics. 2005;6:109. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, Singleton A. New England Journal of Medicine. 2009;360:1759. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, Collin FS, Cox NJ, Goldstein DB, Hindorff LA. Nature. 2009;461:747. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore JH. Human Heredity. 2003;56:73. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 6.Carlborg O, Haley CS. Nature Review Genetics. 2004;5:618. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- 7.Moore JH, Williams SM. BioEssays. 2005;27:637. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- 8.Cordell HJ. Human Molecular Genetics. 2002;11:2463. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 9.Cordell HJ. Nature Review Genetics. 2009;10:392. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore JH, Williams SM. The American Journal of Human Genetics. 2009;85:309. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shendure J, Ji H. Nature Biotechnology. 2008;26:1135. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 12.Moore JH, Ritchie MD. Journal of the Amarican Medical Association. 2004;291:1642. [Google Scholar]
- 13.Moore JH, White BC. Genetic Programming Theory and Practice IV. 2005:969. [Google Scholar]
- 14.Nunkesser R, Bernholt T, Schwender H, Ickstadt K, Wegener I. Bioinformatics. 2007;23:3280. doi: 10.1093/bioinformatics/btm522. [DOI] [PubMed] [Google Scholar]
- 15.Moore JH, Asselbergs FW, Williams SM. Bioinformatics. 2010;26:445. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu T, Sinnott-Armstrong NA, Kiralis JW, Andrew AS, Karagas MR, Moore JH. BMC Bioinformatics. 2011;12:364. doi: 10.1186/1471-2105-12-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Culverhouse R, Suarez BK, Lin J, Reich T. American Journal of Human Genetics. 2002;70:461. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wongseree W, Assawamakin A, Piroonratana T, Sinsomros S, Limwongse C, Chaiyaratana N. BMC Bioinformatics. 2009;10:294. doi: 10.1186/1471-2105-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. The American Journal of Human Genetics. 2001;69:138. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn LW, Ritchie MD, Moore JH. Bioinformatics. 2003;19:376. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie MD, Hahn LW, Moore JH. Genetic Epidemiology. 2003;24:150. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 22.Moore JH, Gilbert JC, Tsai C-T, Chiang F-T, Holden T, Barney N, White BC. Journal of Theoretical Biology. 2006;241:252. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 23.Karagas MR, Tosteson TD, Blum J, Morris JS, Baron JA, Klaue B. Environmental Health Perspectives. 1998;106:1047. doi: 10.1289/ehp.98106s41047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrew AS, Nelson HH, Kelsey KT, Moore JH, Meng AC, Casella DP, Tosteson TD, Schned AR, Karagas MR. Carcinogenesis. 2006;27:1030. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 25.Jakulin A, Bratko I. Lecture Notes in Artificial Intelligence. 2003;2838:229. [Google Scholar]
- 26.Anastassiou D. Molecular Systems Biology. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore JH, Barney N, Tsai C-T, Chiang F-T, Gui J, White BC. Human Heredity. 2007;63:120. doi: 10.1159/000099184. [DOI] [PubMed] [Google Scholar]
- 28.McKinney BA, Crowe JE, Guo J, Tian D. PLoS Genetics. 2009;5:e1000432. doi: 10.1371/journal.pgen.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West DB. Introduction to Graph Theory: Second edition. Prentice Hall; 2001. [Google Scholar]
- 30.Andrew AS, Gui J, Sanderson AC, Mason RA, Morlock EV, Schned AR, Kelsey KT, Marsit CJ, Moore JH, Karagas MR. Human Genetics. 2009;125:527. doi: 10.1007/s00439-009-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kononenko I. Lecture Notes in Computer Science. 1994;784:171. [Google Scholar]
- 32.Moore JH, White BC. Lecture Notes in Computer Sceicne. 2007;4447:166. [Google Scholar]
- 33.Cover TM, Thomas JA. Elements of Information Theory: Second Edition. Wiley; 2006. [Google Scholar]
- 34.Wong AK, Park CY, Greene CS, Bongo LA, Guan Y, Troyanskaya OG. Nucleic Acids Research. 2012;40:W484. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Omar EM, Carrington M, Chow W-H, McCol KEL, Bream JH, Young HA, Herrera J, Lissowska J, Yuan C-C, Rothman N, Lanyon G, Martin M, Fraumeni JF, Jr, Rabkin CS. Nature. 2000;404:398. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 36.Southey MC, Jenkins MA, Mead L, Whitty J, Trivett M, et al. Journal Of Clinical Oncology. 2005;23:6524. doi: 10.1200/JCO.2005.04.671. [DOI] [PubMed] [Google Scholar]
- 37.Michiels S, Laplanche A, Boulet T, Dessen P, Guillonneau B, Mejean A, Desgrandchamps F, Lathrop M, Sarasin A, Benhamou S. Carcinogenesis. 2009;30:763. doi: 10.1093/carcin/bgp046. [DOI] [PubMed] [Google Scholar]