Summary
Although human induced pluripotent stem cells (hiPSCs) have enormous potential in regenerative medicine, their epigenetic variability suggests that some lines may not be suitable for human therapy. There are currently few benchmarks for assessing quality. Here we show that X-inactivation markers can be used to separate hiPSC lines into distinct epigenetic classes and that the classes are phenotypically distinct. Loss of XIST expression is strongly correlated with upregulation of X-linked oncogenes, accelerated growth rate in vitro, and poorer differentiation in vivo. Whereas differences in X-inactivation potential result in epigenetic variability of female hiPSC lines, male hiPSC lines generally resemble each other and do not overexpress the oncogenes. Neither physiological oxygen levels nor HDAC inhibitors offer advantages to culturing female hiPSC lines. We conclude that female hiPSCs may be epigenetically less stable in culture and caution that loss of XIST may result in qualitatively less desirable stem cell lines.
Introduction
With the potential to differentiate into cells of three germ lineages ex vivo, human embryonic stem cells (hESCs) hold immense promise for the field of regenerative medicine. Their derivation from the early human embryos has, however, limited the extent to which hESCs can be generated to meet the needs of an immunologically diverse population. A major breakthrough, therefore, has been creation of patient-specific hESC-like cells from somatic cells by reprogramming through defined pluripotency factors OCT4, SOX2, KLF4, and c-MYC (OSKM) (Takahashi and Yamanaka, 2006; Yu et al., 2007). These “human induced pluripotent stem cells” (hiPSCs) share similar gene expression profiles, morphologies, and differentiation potential with hESCs (Wernig et al., 2007; Maherali et al., 2008), and mouse-derived iPSCs can be passaged through the germline (Okita et al., 2007). Although hiPSCs solve major ethical issues, recent studies have revealed that they may be as genetically and epigenetically fluid as hESCs (Kim et al., 2010; Bock et al., 2011; Gore et al., 2011). There may also be greater expression anomalies in hiPSCs (Rugg-Gunn et al., 2005, 2007; Adewumi et al., 2007; Pick et al., 2009). Because mutation and epigenetic change can lead to cancer and other diseases, these observations imply that some hiPSC lines may not be suitable in a clinical setting. However, apart from karyotype and a limited panel of differentiation markers, there are currently few established benchmarks for assessing hiPSC quality and suitability.
Interestingly, unlike mouse embryonic stem cells (mESCs), hESCs vary tremendously in their potential to undergo X chromosome inactivation (XCI) (Hoffman et al., 2005; Adewumi et al., 2007; Hall et al., 2008; Shen et al., 2008; Silva et al., 2008; Dvash et al., 2010; Lengner et al., 2010), an epigenetic event that is tightly coupled to cell differentiation both in vivo during epiblast differentiation and ex vivo in cultured embryonic stem cells (Payer and Lee, 2008). During XCI, one of two female X chromosomes is transcriptionally repressed to achieve similar X-linked gene dosage as males. This process depends on expression of the long noncoding Xist RNA, which is upregulated just prior to the initiation of chromosome-wide silencing (Marahrens et al., 1997; Wutz and Jaenisch, 2000), and results in recruitment of repressive chromatin to the X. Whereas all XX female mESCs faithfully recapitulate XCI in culture, female hESCs have been grouped into three classes based on differences in their ability to do so. Class I lines initially carry two active Xs (XaXa) but can upregulate XIST and undergo XCI during cell differentiation, suggesting that they most closely approximate the mESC ideal. Class II lines already possess one inactive X (XaXi) and may therefore be partially differentiated. Class III lines largely already underwent XCI but subsequently lost XIST expression, raising questions about their epigenetic stability (Silva et al., 2008; Dvash et al., 2010). Whether these epigenetic classes themselves have practical implications remains unclear. However, because the XCI phenotype may correlate with differentiation potential, XIST has been proposed as a benchmark for assessing hESC quality (Silva et al., 2008).
Indeed, use of the XIST marker led to identification of more XaXa hESCs at early passage (Dvash et al., 2010), discovery that physiological oxygen concentrations are preferable for deriving class I cell lines, and demonstration that stressful ex vivo conditions are associated with conversion to the class III epigenotype (Lengner et al., 2010). These observations have lately generated much interest in the X chromosome status of hiPSCs and raised the question of whether XIST could also be used as a benchmark for hiPSC quality. In the mouse system, reprogramming of somatic cells to iPSCs is accompanied by X reactivation (Maherali et al., 2007). By contrast, recent studies in the human system have reported varying results, with some demonstrating that reprogramming does not reactivate the Xi of parental fibroblasts (Tchieu et al., 2010; Cheung et al., 2011), and others observing that some hiPSC lines have reactivated Xi (Lagarkova et al., 2010; Marchetto et al., 2010). Unresolved, therefore, is whether hiPSCs ever attain the XaXa state associated with pluripotency in the mouse system and in class I hESCs. Also unclear is whether X chromosome states can be used as a readout for female hiPSC quality. Below, we investigate the XCI status and implications of XCI differences in female hiPSCs. We report genome-wide signatures associated with loss of XIST expression and demonstrate sex-specific differences, the combination of which caution that female hiPSCs may be inherently more difficult to maintain by existing protocols.
Results
X Reactivation and Inactivation in Female hiPSCs
Given contrasting reports on X reactivation during establishment of hiPSC lines, we revisited this question by following the X-transcriptional status of new female hiPSC lines derived from IMR-90, a diploid human fetal fibroblast line that has been used extensively to generate hiPSCs (Yu et al., 2007). Cells were reprogrammed using virally expressed OSKM (Park et al., 2008a) and colonies picked between days 28 and 32 (Figure S1A available online). Immunostaining (Figure S1B) and qRT-PCR (Figure S1C) showed expression of pluripotency markers; bisulfite sequencing showed appropriate demethylation of endogenous OCT4 and NANOG promoters (Figure S1D); and qRT-PCR demonstrated silencing of viral factors (Figure S1E). The hiPSCs could differentiate into three germ lineages (Figures S2A and S2B) and form teratomas in NOD-SCID mice (Figure S2A). Furthermore, differentiation induced expression of lineage-specific markers (Figure S2C) and karyotypes confirmed a 46,XX constitution (Figure S3). These data demonstrate successful generation of new female hiPSC lines. For XIST analysis, we used earliest possible passages (p.0–7) to circumvent potential problems associated with long-term culture.
To examine XCI status, we performed RNA FISH and observed one XIST cloud in 58%–84% of nuclei immediately after reprogramming (p.0) (Figures 1A and 1B, Table 1). These 14 distinct clones were derived from five different individuals, including IMR-90 (46,XX), a 47,XXY cell line, a 46,XX “MM” line, and two lines from MM's twin daughters (“TA” and “TB”). An H3K27me3 domain indicative of XIST-mediated Polycomb recruitment was also present (Figure 1C). These findings demonstrate XCI in a majority of cells in each line. Allele-specific analyses of gene expression via single-nucleotide polymorphism (SNP) arrays showed monoallelic expression of X-linked genes for two hiPSC lines (Figure S4), consistent with the absence of X reactivation after reprogramming (Tchieu et al., 2010; Cheung et al., 2011).
Figure 1. Partial X Reactivation and High-Frequency Class III Conversion in Female hiPSCs.
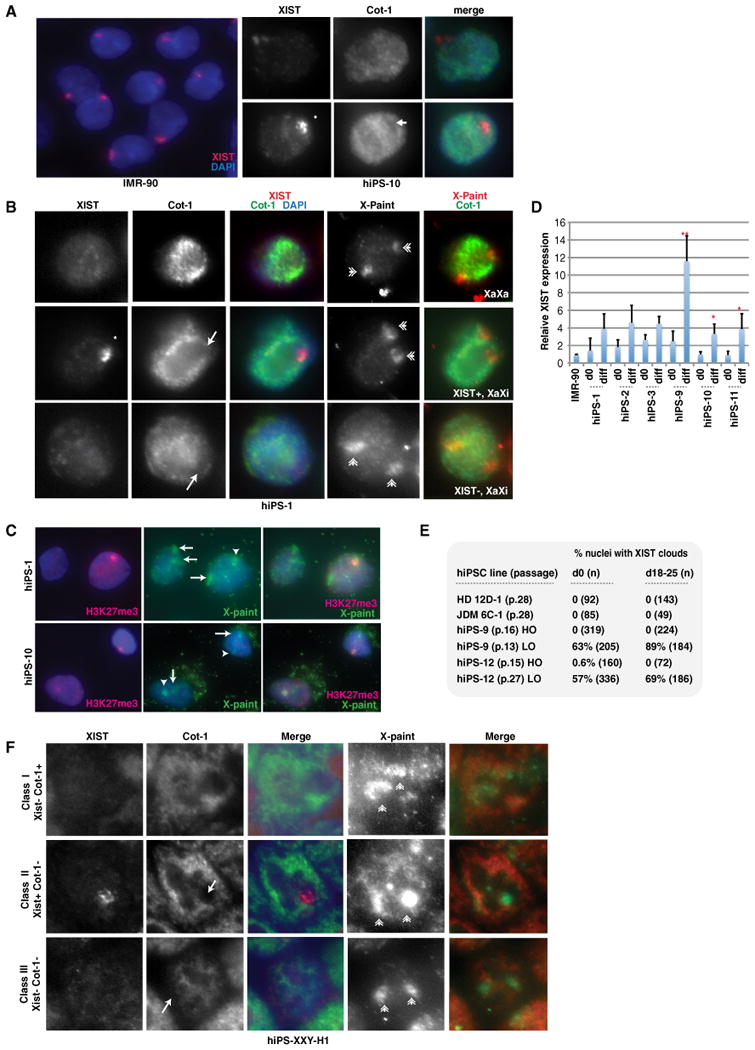
(A) RNA FISH of IMR-90 and undifferentiated hiPS-10. XIST RNA, red; Cot-1 RNA, green; asterisk, XIST cloud; arrow, COT-1 hole.
(B) RNA FISH for XIST and Cot-1, followed by X-paint DNA FISH. Arrows, Cot-1 holes; asterisk, XIST cloud; double arrowheads, X chromosomes. Shown is hiPS-1 p.6.
(C) Immunostaining for H3K27me3 (red) followed by DNA FISH (green) for X chromosomes in differentiated (d16) hiPSCs.
(D) Real-time PCR of XIST expression. Ct values were normalized to IMR-90 cells (set to 1) and GAPDH, and values represent averages of triplicates. Error bars indicate standard deviations (SD) of the mean. p values were calculated with one-tailed Student's t test assuming equal variance; *p = 0.04; **p = 0.004. See also Figure S4.
(E) Summary of XIST RNA FISH. n, sample size. LO, 4% oxygen; HO, 20% oxygen.
(F) Three classes of XXY hiPSCs (d0, p.4). Arrows, Cot-1 holes; asterisk, XIST cloud; double arrowheads, X chromosomes. See also Figure S5.
Table 1.
X Chromosome States in hiPSC Lines
| Undifferentiated d0 | Differentiated d14–d20 | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Sample Size (n) | XIST+ Cells | XIST− Cells | XIST− Cells | Sample size (n) | XIST+ Cells | Class Designation | |
| hiPSC line (passage) | XaXi | XaXa | XaXi* (Xi* in Cot-1 hole) | ||||
| hiPS-1 (p.6) | 78 | 60% | 6.4% | 33% | 95 | 55% | class II predominant |
| hiPS-2 (p.32) | 100 | 68% | 8% | 24% | 153 | 84% | class II predominant |
| hiPS-3 (p.14) | 117 | 58% | 7% | 35% | 273 | 54% | class II predominant |
| hiPS-9 (p.7) | 111 | 78% | 4.5% | 17% | 161 | 91% | class II predominant |
| hiPS-10 (p.29) | 154 | 78% | 3.2% | 20% | 139 | 80% | class II predominant |
| hiPS-11 (p.6) | 106 | 67% | 8% | 24% | 287 | 80% | class II predominant |
| hiPS-12 (p.6) | 79 | 65% | 14% | 22% | 71 | 72% | class II predominant |
|
| |||||||
| hiPS-2 class III (p.50) | 145 | 0 | ND | ND | 80 | 0% | class III |
| hiPS-9 class III (p.15) | 113 | 0 | 0 | 100% | 224 | 0% | class III |
| hiPS-12 class III (p.27) | 105 | 0 | 0 | 100% | 72 | 0% | class III |
|
| |||||||
| hiPS-XXY-L1 (p.7) | 127 | 75% | 7% | 18% | 149 | 91% | class II predominant |
| hiPS-XXY-L3 (p.7) | 279 | 71% | 232 | 90% | |||
| hiPS-XXY-L4 (p.7) | 357 | 66% | 61 | 84% | |||
| hiPS-XXY-H1 (p.7) | 109 | 81% | 2.80% | 17% | 148 | 91% | class II predominant |
| hiPS-XXY-H2 (p.7) | 194 | 75% | 285 | 94% | |||
| hiPS-XXY-H3 (p.7) | 192 | 89% | 205 | 93% | |||
| hiPS-XXY-H5 (p.7) | 332 | 72% | 134 | 93% | |||
|
| |||||||
| hiPS-MM 11 (p.0) | 121 | 84% | 4% | 11% | ND | ||
| hiPS-MM 13 (p.0) | 112 | 76% | 8% | 18% | ND | ||
| hiPS-TA 11 (p.0) | 116 | 71% | 6% | 23% | ND | ||
| hiPS-TA 12 (p.0) | 138 | 76% | 4% | 20% | ND | ||
| hiPS-TA 13 (p.0) | 77 | 68% | 8% | 25% | ND | ||
| hiPS-TB 10 (p.0) | 83 | 74% | 8% | 18% | ND | ||
| hiPS-TB 12 (p.0) | 137 | 80% | 5% | 15% | ND | ||
| hiPS-TB 13 (p.0) | 85 | 73% | 5% | 22% | ND | ||
However, we also observed absence of XIST in 15%–40% of cells, raising several possibilities with respect to XCI. First, XIST− cells could represent cells that underwent X reactivation and attained the XaXa state of pluripotent stem cells. In this scenario, reprogramming would be accompanied by X reactivation, and XaXi cells might represent those that spontaneously reinactivated one X, as is often observed for hESCs (Higgins et al., 2007; Dvash et al., 2010; Lengner et al., 2010). Alternatively, the large number of XIST+ cells might indicate that X reactivation never occurred during reprogramming and resulting hiPSCs merely retained the Xi of parental cells, as proposed by two previous studies (Tchieu et al., 2010; Cheung et al., 2011). In this scenario, XIST− cells would represent spontaneous loss of XIST expression characteristic of class III hESCs (Silva et al., 2008; Dvash et al., 2010).
To distinguish these possibilities, we performed serial RNA-DNA FISH. We first carried out two-color RNA FISH on undenatured nuclei to visualize XIST and Cot-1 expression. The Cot-1 staining pattern provides an overview of nascent transcription from a nuclear domain (Hall et al., 2002; Clemson et al., 2006; Namekawa et al., 2010). After RNA FISH, we denatured the samples and performed DNA FISH by using X-painting probes to locate the X chromosomes (Figure 1B). Two types of XIST− cells were observed in all hiPSCs, irrespective of reprogramming method (e.g., with or without VPA), passage number, and genetic background (Table 1).
One type of XIST− cells showed two Cot-1+ X chromosomes, implying active transcription of both Xs (class I). This inferred XaXa state suggests occurrence of X reactivation during reprogramming. To determine whether percentages of XIST+ cells must increased during cell differentiation, we placed hiPSCs in differentiation conditions for 14–50 days. Indeed, XIST expression increased (Figure 1D, Table 1; e.g., hiPS-2, hiPS-11, hiPS-12), as would be expected of differentiating XaXa cells. We suggest that hiPS-2, hiPS-9, hiPS-10, and hiPS-11 contain a small fraction of class I cells mixed with class II and III cells. Because class I cells accounted for only 2%–14% of cells (Table 1), biallelic expression from this subpopulation would not have been discernible by allele-specific SNP analysis (Figure S4).
The second type of XIST− cells displayed one Cot-1− X chromosome, indicating X chromosome repression in spite of being XIST− (class III state) (Figure 1B). This class III phenotype resembled spontaneous conversion to a class III phenotype in hESCs (Silva et al., 2008; Dvash et al., 2010). Initially, class III cells comprised less than one-third of each hiPSC line (Table 1). During routine culture, three of the class II-predominant hiPSC lines evolved to 100% class III (Table 1, hiPS-2, hiPS-9, and hiPS-12). In the three sublines, XIST expression was absent before and after differentiation, and one X chromosome lay within a Cot-1 hole (Figures 1B and 1F). Two previously published female hiPSCs lines, HD 12D-1 and JDM 6C-1, derived from patients with Huntington's disease and type I diabetes mellitus, respectively (Park et al., 2008a), were also class III, with 0% XIST expression before and after differentiation (Figure 1E). Examination of nascent transcription from X-linked PGK1 indicated that, of XIST− cells with detectable PGK1 signal, approximately half showed biallelic PGK1 and half showed monoallelic expression in hiPS-9, hiPS-10, and hiPS-11 lines on day 0 (data not shown), consistent with the idea that the XIST− subpopulation is a class I-III mixture.
Thus, hiPSCs and hESCs share the tendency to lose XIST expression in culture. Once lost, XIST expression was never regained (data not shown), though a Cot-1 hole indicative of repetitive element silencing persisted. These data show that our female hiPSC lines consist of a mixture of class I, II, and III cells. The presence of XaXa cells (class I) argues that X reactivation takes place in a fraction of cells during reprogramming. The XaXi cells (class II) indicates either that XaXa cells spontaneously undergo reinactivation of one X or that a fraction of hiPSCs never underwent X reactivation. Although the hiPSC lines have a mixed population, class II cells dominate at early passage. The tendency to become class III during culture demonstrates a level of epigenetic fluidity characteristic of female hESCs.
Effects of Oxygen and HDAC Inhibitors
Previous work showed that physiological (4%) oxygen instead of ambient (20%) levels preserves the class I state of hESCs (Lengner et al., 2010) and enhances reprogramming to iPSCs (Utikal et al., 2009; Yoshida et al., 2009). We investigated whether physiological oxygen might be beneficial for hiPSCs. Here we used a fibroblast line, derived from an 18-week-old 47,XXY fetus. It yielded 39 colonies at 20% oxygen, in contrast to IMR-90, which typically yielded 1–6 colonies from 50,000 starting cells. Furthermore, when reprogrammed at 4% oxygen, the XXY line produced twice as many colonies (∼80) (Utikal et al., 2009; Yoshida et al., 2009). The XXY fibroblasts behaved similarly to 46,XX cells with respect to XCI, as XIST RNA was expressed from a single X (Poplinski et al., 2010). We expanded four clones reprogrammed in ambient oxygen (hiPS-XXY-H1, -H2, -H3, -H5) and three in physiological oxygen (hiPS-XXY-L1, -L3, -L4). Immunostaining and qRT-PCR confirmed expression of pluripotency markers in all seven; each also demonstrated EB formation and outgrowth during differentiation (Figure S5A and data not shown). In general, hiPSCs maintained in 4% oxygen better preserved their morphology and showed less oxidative stress (Figure S5B). XIST RNA FISH showed that reprogramming at 4% oxygen had no effect on XIST expression (Table 1), as the XXY lines remained predominantly class II, with 66%–89% expressing XIST on day 0 and 84%–94% after differentiation (Table 1, Figure 1F). The class I subpopulation was invariably low (2.8%–7%). Class III cells were also present in each isolate. Thus, oxygen levels have no major effect on XIST in hiPSCs.
Previous work also showed that HDAC inhibitors promote a more favorable epigenetic state for hESCs. Specifically, treating H9 containing a mixture of XaXa/XaXi cells resulted in a homogeneous XaXa population capable of upregulating XIST upon differentiation (Ware et al., 2009). To determine whether the effects extended to hiPSCs, we treated hiPSCs with HDAC inhibitors, sodium butyrate, and vorinostat for 5–8 passages and examined XIST during differentiation. HDAC inhibition did not change XIST profiles from day 0 to day 18 in any of six lines (Table S1). All class II-predominant lines continued to show XIST clouds in 40%–70% of cells, and three class III lines from three distinct individuals showed no rescue of XIST expression. Therefore, HDAC inhibition has no obvious beneficial effect for XIST in hiPSCs.
To determine whether the effect might be specific to hESCs, we treated two class I-predominant hESCs lines, HUES-9 and H9 (Silva et al., 2008; Ware et al., 2009), for five passages and examined XIST (Table S1). Consistent with previous analysis (Ware et al., 2009), HDAC inhibition increased the number of XaXa cells on day 0 and yielded cells with XIST clouds after differentiation. Treatment of HUES-9 resulted in a modest increase of XIST+ cells during differentiation but did not increase the number of XaXa cells on day 0, as observed with H9. In our hands, recovery after cryopreservation, general growth, and morphology of both hESCs and hiPSCs were enhanced, consistent with the previous report (Ware et al., 2009). We conclude that HDAC inhibitors do not improve XIST profiles for hiPSCs but may better rescue XIST in female hESCs (Diaz Perez et al., 2012).
Genome-wide Transcription Profiling Reveals Class-Specific Differences
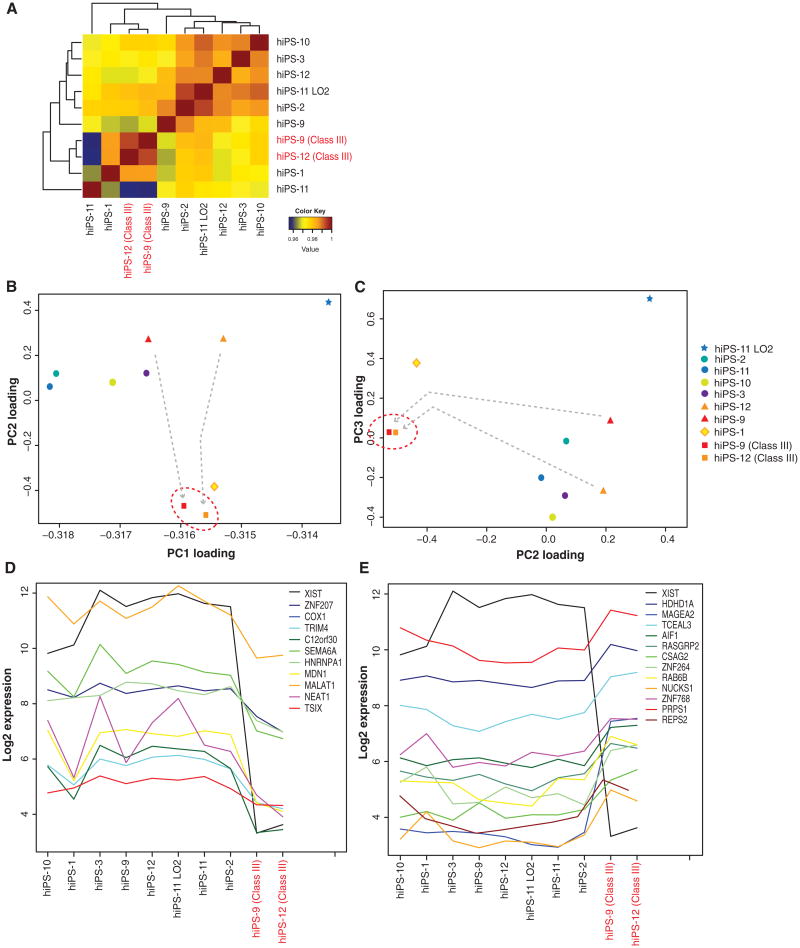
Whether the class III state has significant biological consequences is currently unknown. To address this question, we compared genome-wide expression profiles by microarray analysis of ten hiPSCs lines and sublines, all of which were derived from IMR-90. Hierarchical clustering revealed that all class II-predominant cell lines showed highly correlated expression patterns among each other (Figure 2A). Furthermore, class III lines were strongly correlated with each other. Intriguingly, class III sublines of hiPS-9 and hiPS-12 resembled each other more than they resembled class II parents and other class II lines. Departures from their parental lines were more dramatic than differences for hiPSCs grown in 20% versus 4% oxygen (hiPS-11 versus hiPS-11LO2).
Figure 2. Class III Female hiPSCs Have Unique Global Gene Expression Patterns.
(A) Pearson correlation coefficients between whole sets of gene expression levels (RMA normalization) in the ten female hiPSC samples. hiPS-2 p.9, hiPS-9 p.7, hiPS-10 p.24, hiPS-3 p.14, hiPS-12 p.23, hiPS-11 p.16: high O2; hiPS-11 p.16: low O2; hiPS-9 p.19 c.III; hiPS-12 p.30 c.III.
(B and C) PCA of gene expression patterns in indicated samples. Plot of component loadings shows relations of each microarray sample (RMA normalization) in PC1 versus PC2 (B) and of PC2 versus PC3 (C). Class II to III conversion indicated by arrows.
(D and E) Expression levels for genes downregulated (D) and upregulated (E) in class III samples. Shown are top genes with highest correlation (D) or anti-correlation (E) to XIST expression, among those that are differentially expressed in at least six out of eight class II versus class III lines. See also Figure S6.
Principal component analysis (PCA) supported these observations (Figures 2B and 2C). In multiple dimensions, class III hiPS-9 and hiPS-12 sublines were significantly closer to each other than to parental class II hiPS-9 and hiPS-12 counterparts and to all other class II lines. Interestingly, the hiPS-1 profile was closer to class III than to other class II. This correlated with the larger subpopulation of class III cells within hiPS-1 (33%, Table 1). hiPS-1 may be in transition to class III. Thus, loss of XIST expression is associated with significant shifts in global expression profiles, suggesting that the class III state is a distinct epigenotype that develops during culture.
Class III Association with Upregulation of Cancer-Related Genes
To determine what genes were affected, we looked for class-specific differences in gene expression. We used ANOVA-based estimates of statistical significance with conservative modeling of gene-specific intersample variance implemented in NIA Array Analysis webtool (Sharov et al., 2005). Among genes showing significant differential expression (FDR < 0.05), only ten coding genes were consistently upregulated more than 2-fold in class III compared to all class II lines (Table 2; Figure S6). Interestingly, among the genes upregulated in class III hiPSC lines, X-linked genes were significantly overrepresented (four out of ten genes, p = 7 × 10−5). This caught our attention, given that loss of Xist has been shown to result in partial X reactivation in murine cells (Csankovszki et al., 2001; Zhang et al., 2007) and overexpression of X genes has been correlated with cancer (Richardson et al., 2006; Pageau et al., 2007).
Table 2.
Genes with Greatest Expression Change in Class III hiPSCs
| Gene Name | Description | Chr. | FDR |
|---|---|---|---|
| Genes Showing Greatest Upregulation | |||
|
| |||
| MAGEA6 | melanoma antigen family A, 6 | X | 0 |
| MAGEA2 /// MAGEA2B | melanoma antigen family A, 2 /// melanoma antigen family A, 2B | X | 0 |
| ACP5 | acid phosphatase 5, tartrate resistant | 19 | 0 |
| TCEAL3 | transcription elongation factor A (SII)-like 3 | X | 0.0001 |
| HDHD1A | haloacid dehalogenase-like hydrolase domain containing 1A | X | 0.0007 |
| RAB6B | RAB6B, member RAS oncogene family | 3 | 0.0017 |
| AIF1 | allograft inflammatory factor 1 | 6 | 0.0082 |
| CHP2 | calcineurin B homologous protein 2 | 16 | 0.0111 |
| LOC285965 | hypothetical protein LOC285965 | 7 | 0.0231 |
| LOC100131199 | LOC100131199 transmembrane protein 178-like (Homo sapiens) | 7 | 0.0475 |
|
| |||
| Genes Showing Greatest Downregulation | |||
|
| |||
| MALAT1 | metastasis associated lung adenocarcinoma transcript 1 (nonprotein coding) | 11 | 0 |
| XIST | X (inactive)-specific transcript (nonprotein coding) | X | 0 |
| SEMA6A | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 5 | 0 |
| AI380514.1 | tg01e02.x1 NCI_CGAP_CLL1 Homo sapienscDNA clone IMAGE:2107514 3-, mRNA sequence | 2 | 0 |
| NAA25 | N(alpha)-acetyltransferase 25, NatB auxiliary subunit | 12 | 0 |
| MED17 | mediator complex subunit 17 | 11 | 0 |
| FARSB | phenylalanyl-tRNA synthetase, beta subunit | 2 | 0 |
| AA524609.1 | nh34c11.s1 NCI_CGAP_Pr3 Homo sapiens cDNA clone IMAGE:954260 similar to contains Alu repetitive element;, mRNA sequence | / | 0 |
| BC020935.1 | similar to otoconin 90, clone IMAGE:4277593 | 13 | 0.0001 |
| BF223214.1 | 7q30f03.x1 NCI_CGAP_GC6 Homo sapiens cDNA clone IMAGE:3699965 3-, mRNA sequence | 6 | 0.0003 |
| AI684643.1 | wa84h10.x1 Soares_NFL_T_GBC_S1 Homo sapiens cDNA clone IMAGE:2302915 3-, mRNA sequence | 12 | 0.0005 |
| FTX | FTX, NCRNA00182 nonprotein coding RNA 182 (Homo sapiens) | X | 0.0006 |
| FN1 | fibronectin 1 | 2 | 0.0007 |
| LOC649305 | hypothetical LOC649305 | 8 | 0.0008 |
| AW135003.1 | UI-H-BI1-abt-c-08-0-UI.s1 NCI_CGAP_Sub3 Homo sapiens cDNA clone IMAGE:2712951 3-, mRNA sequence | 11 | 0.0028 |
| FST | follistatin | 5 | 0.0033 |
| NPPB | natriuretic peptide precursor B | 1 | 0.0058 |
| AV699781.1 | AV699781 GKC Homo sapiens cDNA clone GKCEKC01 3-, mRNA sequence | / | 0.0061 |
| FNBP4 | formin binding protein 4 | 11 | 0.0063 |
| AW197431.1 | xm39b03.x1 NCI_CGAP_GC6 Homo sapiens cDNA clone IMAGE:2686541 3- similar to contains element KER repetitive element; mRNA sequence | 12 | 0.0064 |
| IGFBP5 | insulin-like growth factor binding protein 5 | 2 | 0.0167 |
| NEAT1 | NEAT1 nuclear paraspeckle assembly transcript 1 (nonprotein coding) (Homo sapiens) | 11 | 0.0211 |
| CER1 | cerberus 1, cysteine knot superfamily, homolog (Xenopus laevis) | 9 | 0.0265 |
| GAD1 | glutamate decarboxylase 1 (brain, 67 kDa) | 2 | 0.0484 |
Two of the upregulated X-linked genes, MAGEA2 and MAGEA6, are highly expressed in cancers (Rogner et al., 1995). Overexpression of five others has also been implicated in cancer and metastasis, including RAB6B, a member of the RAS oncogene family; CHP2 in ovarian tumors; ACP5 in various cancers; and AIF1 in breast tumor growth. TCEAL3, LOC100131199, and LOC285965 have no known function. Thus, at least six of ten upregulated coding genes specific to class III lines are previously identified cancer genes.
We then asked which genes were consistently downregulated by at least 2-fold in class III cells compared to all class II samples (Table 2; Figure S6). X-linked genes were not overrepresented in this list, as might be expected because XIST is an X silencer. Apart from XIST, the only other X-linked locus in the top hits list was FTX, a noncoding gene near XIST with undefined function (Chureau et al., 2002). Other downregulated genes of interest were known tumor suppressors, including FN1, a fibronectin involved in cell adhesion. Noncoding RNAs MALAT1 and NEAT1 (of nuclear speckles and paraspeckles associated with cancers) were also downregulated (Ji et al., 2003; Sunwoo et al., 2009).
Taken together, the genome-wide profiles argue for class-specific associations with cancer genes and raises the question of how many of the class III changes could be attributed to or strongly correlated with loss of XIST. To address this, we identified genes whose expression levels had highest Pearson correlation coefficients with XIST levels across all samples. We included genes that were upregulated in all class III lines compared to at least six of eight class II lines. Several hundred met these criteria (see Table S2 for complete list), of which 30 with greatest correlation are shown in Table 3. Intersecting the list of XIST-correlated genes with known cancer genes from MSKCC CancerGenes resource (Higgins et al., 2007) revealed nine tumor suppressors (CDC14B, CDK6, CNOT7, IDH1, IGFBP5, PCDH10, PLXNC1, RBBP4, STK4) and seven oncogenes (BCL11A, CHD1L, FGFR1, FUS, FYN, RAB12, SOS1) that were differentially expressed between class II and class III. Genes with highest correlation with XIST include SEMA6A, MALAT1, and FTX and genes for oxidative stress response, COX1 (R = 0.94) and PRDX2 (R = 0.838) (Figure 2D, Table 3). Also highly correlated were members of the Mediator complex (MED6, MED17), a transcriptional coactivator complex found at promoters of active genes in pluripotent cells, and MLL2, a histone 3 lysine-4 (H3K4) methyltransferase responsible for bulk methylation of H3K4me3 associated with transcriptional activation. Using DAVID Bioinformatics Resource (Huang et al., 2009), we observed significant enrichment for genes involved in transcription (FDR = 0.001), transcriptional repression (FDR = 0.0017), and transcriptional regulation (FDR = 6.02 × 10−4). DAVID analysis of Table S2 also confirmed enrichment for many genes involved in RNA processing (FDR = 0.006), splicing (FDR = 0.054), binding (FDR = 0.034) stability, and export (e.g., FUS, HNRNPA1, SFPQ, HNRNPD, SFRS15, SFRS4).
Table 3.
Genes with Greatest Expression Correlation with XIST
| Gene | Description | Chr. | FDR | Corr. | # Chip |
|---|---|---|---|---|---|
| Positive Correlation | |||||
|
| |||||
| XIST | X (inactive)-specific transcript (nonprotein coding) | X | 0 | 1 | 8 |
| ZNF207 | zinc finger protein 207 | 17 | 0.0034 | 0.9446 | 8 |
| COX1 | cytochrome coxidase I | MT | 0 | 0.9444 | 8 |
| NAA25 | N(alpha)-acetyltransferase 25, NatB auxiliary subunit | 12 | 0 | 0.9423 | 8 |
| TRIM4 | tripartite motif-containing 4 | 7 | 0.0001 | 0.9416 | 7 |
| SEMA6A | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 5 | 0 | 0.9415 | 8 |
| AV699781.1 | AV738585 CB Homo sapiens cDNA clone CBFAWD05 5-, mRNA sequence | / | 0.0258 | 0.9396 | 8 |
| AA524609.1 | nh34c11.s1 NCI_CGAP_Pr3 Homo sapiens cDNA clone IMAGE:954260 similar to contains Alu repetitive element; mRNA sequence | / | 0 | 0.9384 | 8 |
| HNRNPA1 /// LOC728844 | heterogeneous nuclear ribonucleoprotein A1 /// hypothetical LOC728844 | 12 | 0.0044 | 0.9370 | 8 |
| N72610 | za46h03.s1 Soares fetal liver spleen 1NFLS Homo sapiens cDNA clone IMAGE:295637 3-, mRNA sequence | / | 0.0287 | 0.9270 | 6 |
| AI247478 | qh56c08.x1 Soares_fetal_liver_spleen_1NFLS_S1 Homo sapiens cDNA clone IMAGE:1848686 3- similar to contains Alu repetitive element; contains element PTR5 repetitive element; mRNA sequence | 8 | 0.0007 | 0.9261 | 4 |
| BE503070 | hz83b02.x1 NCI_CGAP_Lu24 Homo sapiens cDNA clone IMAGE:3214539 3-, mRNA sequence | 5 | 0.0001 | 0.9204 | 8 |
| MDN1 | MDN1, midasin homolog (yeast) | 6 | 0 | 0.9120 | 8 |
| AI806781 | wf15b12.x1 Soares_NFL_T_GBC_S1 Homo sapiens cDNA clone IMAGE:2350655 3-, mRNA sequence | 17 | 0 | 0.9092 | 8 |
| BG281679 | 602402364F1 NIH_MGC_20 Homo sapiens cDNA clone IMAGE:4544871 5-, mRNA sequence | / | 0.0056 | 0.9090 | 4 |
| BF223214 | 7q30f03.x1 NCI_CGAP_GC6 Homo sapiens cDNA clone IMAGE:3699965 3-, mRNA sequence | 6 | 0.0003 | 0.9081 | 8 |
| AI539426 | te46d04.x1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:2089735 3-, mRNA sequence | 12 | 0.0039 | 0.9035 | 8 |
| AI380514 | tg01e02.x1 NCI_CGAP_CLL1 Homo sapiens cDNA clone IMAGE:2107514 3-, mRNA sequence | 2 | 0 | 0.9027 | 8 |
| W86781 | zh64a03.s1 Soares_fetal_liver_spleen_1NFLS_S1 Homo sapiens cDNA clone IMAGE:416812 3-, mRNA sequence | 20 | 0.0003 | 0.9020 | 8 |
| AI056872 | oz03e12.x1 Soares_fetal_liver_spleen_1NFLS_S1 Homo sapiens cDNA clone IMAGE:1674286 3-, mRNA sequence | 6 | 0.0007 | 0.9018 | 1 |
| MLL2 | myeloid/lymphoid or mixed-lineage leukemia 2 | 12 | 0.0035 | 0.9016 | 8 |
| FUS | fusion (involved in t(12;16) in malignant liposarcoma) | 16 | 0.0012 | 0.9016 | 8 |
| AA398740 | zt75f06.s1 Soares_testis_NHT Homo sapiens cDNA clone IMAGE:728195 3-, mRNA sequence | 1 | 0.0003 | 0.9016 | 7 |
| CHD1L | chromodomain helicase DNA binding protein 1-like | 1 | 0.0006 | 0.9006 | 8 |
| FTX | NCRNA00182 non-protein coding RNA 182 (Homo sapiens) | X | 0.0006 | 0.9005 | 8 |
| CHPT1 | choline phosphotransferase 1 | 12 | 0.0023 | 0.8984 | 3 |
| AI367034.1 | qq40f01.x1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:1935001 3- similar to contains Alu repetitive element; mRNA sequence | 2 | 0.0081 | 0.8958 | 8 |
| MALAT1 | metastasis associated lung adenocarcinoma transcript 1 (nonprotein coding) | 11 | 0 | 0.8948 | 8 |
| AA436194.1 | zv22f03.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:754397 3- similar to contains Alu repetitive element;contains element PTR7 repetitive element; mRNA sequence | 4 | 0.0434 | 0.8948 | 8 |
| MED17 | mediator complex subunit 17 | 11 | 0 | 0.8939 | 8 |
|
| |||||
| Negative Correlation | |||||
|
| |||||
| HDHD1A | haloacid dehalogenase-like hydrolase domain containing 1A | X | 0.0007 | −0.9824 | 8 |
| MAGEA2 /// MAGEA2B | melanoma antigen family A, 2 /// melanoma antigen family A, 2B | X | 0 | −0.9801 | 3 |
| TCEAL3 | transcription elongation factor A (SII)-like 3 | X | 0.0001 | −0.9560 | 8 |
| AIF1 | allograft inflammatory factor 1 | 6 | 0.0082 | −0.9557 | 0 |
| RASGRP2 | RAS guanyl releasing protein 2 (calcium and DAG-regulated) | 11 | 0.0211 | −0.9533 | 7 |
| ACP5 | acid phosphatase 5, tartrate resistant | 19 | 0 | −0.9490 | 0 |
| SDCCAG8 | serologically defined colon cancer antigen 8 | 1 | 0.0017 | −0.9474 | 1 |
| NXT2 | nuclear transport factor 2-like export factor 2 | X | 0.0014 | −0.9461 | 0 |
| MAGEA6 | melanoma antigen family A, 6 | X | 0 | −0.9437 | 0 |
| CHP2 | calcineurin B homologous protein 2 | 16 | 0.0111 | −0.9284 | 3 |
| CSAG2 /// CSAG3 | CSAG family, member 2 /// CSAG family, member 3 | X | 0 | −0.9256 | 0 |
| C20orf94 | chromosome 20 open reading frame 94 | 20 | 0.0178 | −0.9185 | 3 |
| ZNF264 | zinc finger protein 264 | 19 | 0.0078 | −0.9169 | 7 |
| RAB6B | RAB6B, member RAS oncogene family | 3 | 0.0017 | −0.9138 | 2 |
| C1orf77 | chromosome 1 open reading frame 77 | 1 | 0.0035 | −0.9127 | 8 |
| NUCKS1 | nuclear casein kinase and cyclin-dependent kinase substrate 1 | 1 | 0.003 | −0.9112 | 3 |
| MBNL3 | muscleblind-like 3 (Drosophila)(Homo sapiens) | X | 0.0035 | −0.9096 | 2 |
| ZNF768 | zinc finger protein 768 | 16 | 0.0064 | −0.9038 | 7 |
| LOC284242 | hypothetical protein LOC284242 | 18 | 0.0191 | −0.9036 | 7 |
| PRPS1 | phosphoribosyl pyrophosphate synthetase 1 | X | 0.0173 | −0.9019 | 8 |
| REPS2 | RALBP1 associated Eps domain containing 2 | X | 0.0197 | −0.9009 | 6 |
The same analysis was applied to genes whose expression levels had the greatest anticorrelation (negative Pearson correlation coefficients) with XIST levels across all samples (Table 3; complete gene list in Table S3), of which 12 with highest negative Pearson correlation coefficients are plotted in Figure 2E. There was considerable overlap between Tables S2A and S3B, and X-linked genes were significantly overrepresented (p = 2 × 10−8, for 9 of 21 genes being X-linked in Table 3). Again, cancer genes were also highly represented. In addition to those in Table 2, CSAG2, NUCKS1, REPS2, MTA2, RAB6B, RAP2C, and VAV1 showed anticorrelation with XIST. Oncogenes MAGE2A (R = −0.980) and MAGE6A (R = −0.944) showed especially high correlation. DAVID analysis of Table S3 yielded no significant enrichment for any group of genes. Notably, oncogenes as a general class were not significantly enriched. Taken together, these argue for enriched expression only of oncogenes residing on the X, resulting from loss of XIST-mediated suppression in cis. We conclude that loss of XIST expression is strongly correlated with X gene overexpression, hyperexpression of select X-linked oncogenes, and repression of select tumor suppressors.
Male and Female Differences in hiPSC Quality
Because male cells do not undergo XCI, male hiPSCs cannot be subclassified by XIST expression. However, the strong genome-wide positive and negative correlations identified above for female hiPSCs might be used in lieu of XIST to address male hiPSC quality. Could male hiPSCs be subcategorized on the basis of genome-wide expression profiles? Do some male hiPSCs exhibit aberrant expression of cancer genes? To address these questions, we analyzed gene expression profiles of published male and female hiPSCs derived from normal fibro-blasts by reprogramming with either virally introduced factors, modified RNA, or direct protein delivery (Maherali et al., 2008; Park et al., 2008b; Kim et al., 2009; Jia et al., 2010; Mayshar et al., 2010; Tchieu et al., 2010; Warren et al., 2010). We also queried whether variability occurred in hESC lines (Westfall et al., 2008; Chen et al., 2011) and compared male hiPSC and female hESC profiles to our female hiPSCs, including a low-oxygen line (hiPS-2 cIII LO) and two disease-model hiPSC lines (12D-1, 6C-1) created elsewhere but passaged in our laboratory (Park et al., 2008a). Our diverse sampling therefore tested cell lines of distinct provenance, with fibroblasts derived from multiple individuals and hiPSCs created in 12 different labs.
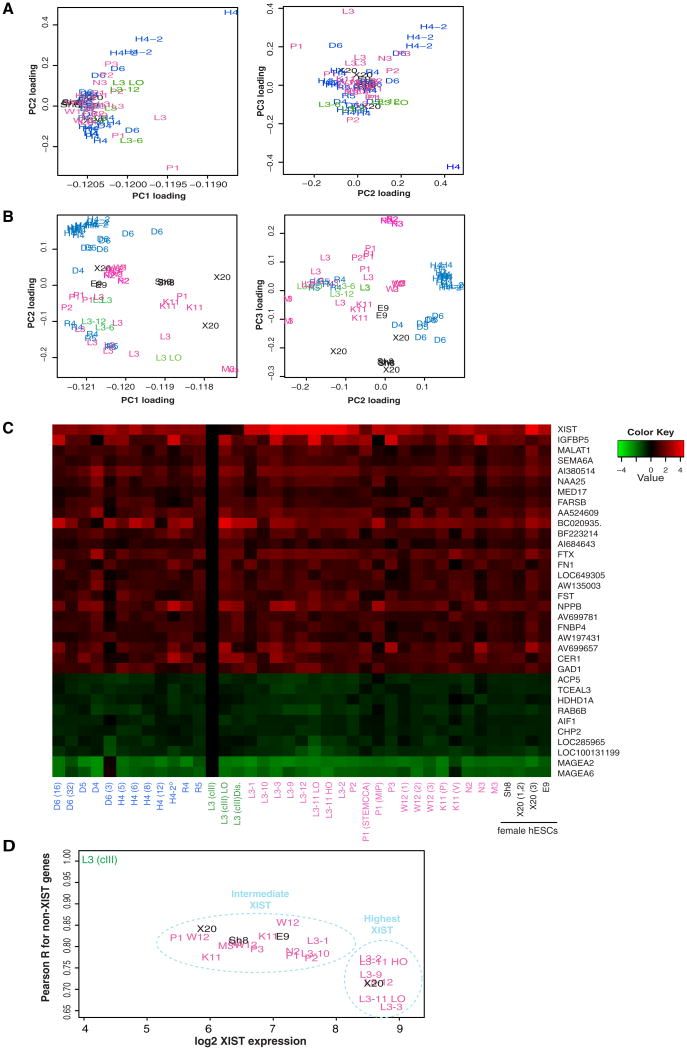
We first performed hierarchical clustering and PCA loading analyses. Because hiPSC lines are known to have a tendency to cluster by laboratory of origin (Guenther et al., 2010; Laurent et al., 2011) and because variations could arise from biases between microarray batches (“batch effects”), we analyzed the data without in silico correction of batch effects or with correction via the ComBat method (Figures 3A and 3B; Johnson et al., 2007). Several patterns emerged with either method. First, there is a tendency for each type of cell line to cluster together, irrespective of lab origin. For example, female hESCs clustered together (black), as did female hiPSCs (pink) and male hiPSCs (blue). Furthermore, class III female lines (green) grouped together but away from female hiPSCs and hESCs. In general, female hiPSC lines showed greater variation among one another than did male hiPSC lines among themselves (blue male lines versus pink female lines; Figures 3A and 3B). Interestingly, while male hiPSCs tend to group together apart from female hiPSCs, RNA-reprogrammed male lines (R4, R5) (Warren et al., 2010) appeared to better resemble female hESCs and hiPSCs. The secondarily reprogrammed male hiPSC line (H4-2) was also set apart from other male hiPSCs (most evident with ComBat correction; Figure 3A). These differences pertained to cell lines derived not only in different laboratories but also within any given laboratory, as evidenced by loose groupings observed in multiple PCA dimensions (Figures 3A and 3B).
Figure 3. Microarray Analyses of Male versus Female hiPSCs and hESCs.
(A and B) PCA shown in two dimensions for ComBat-corrected (A) and uncorrected (B) samples. See Table S4 for list of samples, GEO numbers, PubMed ID, and abbreviations. Class III hiPSCs from this study (L3) in green; L3 LO, hiPS-2 c.III in low oxygen p.50; L3-6, hiPS-6C-1 c.III p. 28; L3-12, hiPS-12D-1 c.III p.28. Blue, male hiPSCs; pink, female hiPSCs; black, female hESCs.
(C) Expression heatmaps normalized to hiPS-9 and hiPS-12 c.III average (expression set as 0). Shown are genes up- and downregulated in class III hiPSCs (Table 2). L3 (cIII), hiPS-9, hiPS-12 c.III; L3 (cIII) LO, hiPS-2 c.III; L3 (cIII) Dis., hiPS 6C-1, 12D-1 c.III. Averages shown for duplicate and triplicate samples.
(D) XIST expression in indicated lines plotted against correlation of expression pattern across differentially expressed genes.
Given a strong positive association between XIST loss and overexpression of select X-linked oncogenes (Tables 2, 3, and S4), we next asked whether molecular signatures of male hiPSCs could be compared against those of female hiPSCs to infer stem cell quality. For the genes differentially expressed in class III versus II (Table 2), we evaluated expression profiles in female hiPSCs, male hiPSCs, and female hESCs and compared them to the average of class III lines, hiPS-9 and hiPS-12 (Figure 3C, L3 [cIII]), as the basis for comparison. As expected, all three class III lines (green) showed low XIST expression, whereas class II lines of various provenance (pink) showed significantly more XIST expression. The dark red values for male hiPSCs (blue) were consistent with low-level XIST expression known to occur in male mouse embryonic stem cells (Beard et al., 1995), consistent with their successful reprogramming.
In general, male hiPSC expression profiles resembled those of class II female hiPSCs (except XIST levels were lower than in class II lines but higher than in class III lines, consistent with pinpoint XIST expression in undifferentiated ESCs of mice) (Figure 3C). The male profiles, however, significantly deviated from those of the hiPS-9/hiPS-12 class III average. For example, male hiPSCs lines generally did not show increased expression of the oncogenes upregulated in class III lines (e.g., MAGEA2, MAGEA6, RAB6B, TCEAL3, and ACP5). Main exceptions were male D6(3) and D6(32), which displayed increased MAGEA2 and MAGEA6 expression, and the secondarily reprogrammed male lines (shown as an average, H4-2°), which showed increased expression of many genes upregulated in class III lines (e.g., TCEAL3, ACP5, CHP2, RAB6B;Table 3). By contrast, the additional class III female lines exhibited a trend toward greater expression of the most correlated marker genes from Table 2. For example, L3(cIII)LO (hiPS-2 cIII grown in low oxygen) and L3(cIII)Dis (disease lines, hiPS 6C-1 and 12D-1) had similarly increased expression of TCEAL3, RAB6B, LOC285965, and CHP2.
Even among class II female lines, casual examination hinted at a correlation between degree of XIST expression and likeness to the class III profile. For instance, hiPS-1 (L3-1, Figure 3C), shown above to be a class II-III intermediate (Figures 2B and 2C), resembled the class III profile (Figure 3C). This suspicion was confirmed by direct quantitative analysis of profile similarities calculated as Pearson correlation coefficients of expression values on the set of genes differentially expressed in class III (Table 2), excluding XIST itself. This analysis revealed a trend of monotonic decrease with increasing level of XIST expression (Figure 3D). Two loose groupings of female hiPSCs were apparent. Cell lines with highest XIST expression occupied the bottom right region of the plot, demonstrating the highest dissimilarity to class III. Those with intermediate XIST expression were located in the center (e.g., hiPS-1 [a.k.a. L3-1]), demonstrating a drift toward the class III reference in the top-left corner.
Taken together, these results argue that upregulation of X-linked oncogenes and other loci revealed in Tables 2 and 3 is a property of female hiPSCs when they lose XIST expression. Though not generally a feature of male hiPSCs, secondarily reprogrammed male lines may more closely resemble class III female lines. We believe that expression differences in class III lines are due to epigenetic change rather than to genomic alterations, as microarray-based comparative genomic hybridization (CGH) on paired sets of class II and III lines revealed no gross copy number changes (Figure S7). No deletions within the XIST locus were observed in each case. Thus, we demonstrate that hiPSCs of both male and female origin could be evaluated by comparison to the deviant class III profiles.
Loss of XIST Results in Accelerated Growth In Vitro and Poor Differentiation In Vivo
Here we tested whether resemblance to class III has functional consequences. In light of increased oncogene expression, we asked whether class III lines grow faster in culture. We measured growth rates of multiple undifferentiated class II and III cells in four independent experiments over 20–35 days and plotted numbers of cells (Figures 4A and 4B) and colonies (Figure 4C). In multiple replicates, class III hiPSC lines generally exhibited a shorter doubling time than their class II parents and other class II lines (Figures 4A and 4B). They also grew more quickly than male hiPSCs. This was the case inhigh and physiological oxygen conditions. Interestingly, hiPS-1—the class II-III transitional cell line—exhibited a growth rate more similar to class III cells (Figure 4A), thus correlating with its class III-like expression profiles (Figures 3A and 3C). Other transitional lines (identified by fewer XIST+ clouds) also displayed faster growth rates than their class II parents (compare hiPS-12 p.28 to hiPS-12 p.32). Faster growth rates did not appear to be a consequence of adaptation to culture, as we tested class II cell lines (hiPS-2, hiPS-11, hiPS-12) at early (p.14) and later (p.32) passages and found no consistent significant change in growth rates (Figure 4B). Interestingly, we also observed that class III lines recovered faster after routine passaging, as these lines typically yielded greater colony numbers and larger colony sizes 1 day after passaging when compared to class II lines and male hiPSCs.
Figure 4. Comparative In Vitro Growth Rates and In Vivo Differentiation.
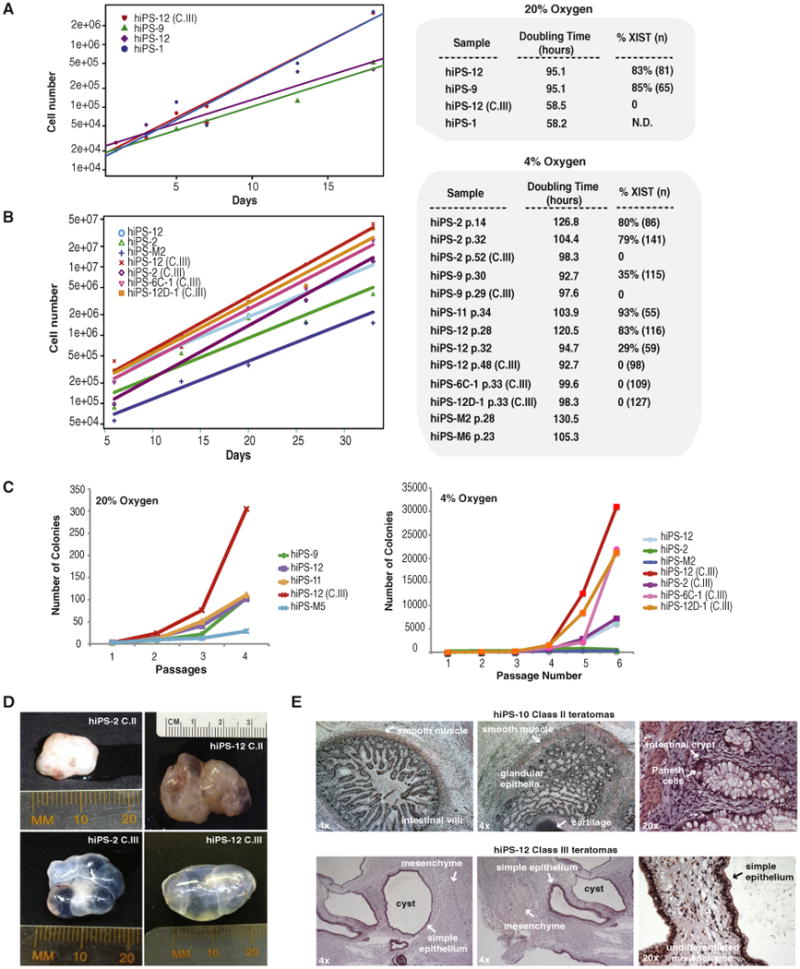
(A) Growth profiles for indicated hiPSC lines in ambient oxygen. Doubling times calculated from line equations. One colony for each line was mechanically passaged and plated in ten replicate, MEF-coated wells. Cells were trypsinized and counted. Averages shown. Percentages of XIST+ nuclei at the end of the experiment shown. n, sample size; N.D., not determined. Two biological replicates performed; similar results; one shown.
(B) Growth profiles for indicated lines in physiological oxygen. Three colonies for each line were plated in quadruplicate on MEF-coated plates, then processed as in (A).
(C) Growth differences as a function of passage number at ambient or physiological oxygen. Average values shown.
(D) Teratomas from matched class II-III sublines of hiPS-2 and hiPS-12. (E) Representative histologic sections of class II and III teratomas.
We next investigated the ability of class III cells to form teratomas. In general, hiPSCs are known to form teratomas when injected into immunocompromised mice. Although both class II and III lines could do so, their in vivo differentiation capacities were markedly different (Figures 4D and 4E). Class II teratomas showed prominent differentiation into structures recapitulating adult organs and tissues, such as cartilage and small intestine, including a range of cell types found in mature intestine including mucin-producing epithelial cells and Paneth cells, secondary organization into villi, and investing layers of circular and longitudinal smooth muscle (Figures 4D, 4E, and S2A). Intriguingly, all teratomas derived from four representative class II sublines (hiPS-2, hiPS-10, hiPS-11, and hiPS-12) formed solid masses (5 of 5); by contrast, teratomas derived from the matched class III sublines (hiPS-2 and hiPS-12) and disease model line (6C-1) were all cystic (11 of 11), with the cysts lined by simple epithelia and undifferentiated mesenchymal tissue, with little to no differentiated cell types (Figures 4D and 4E). Two of the 11 cystic teratomas had small solid masses with a low degree of differentiation into all three germ layers. Notably, one prior report found that male hESCs formed solid teratomas but one female hESCs line with unknown XIST expression status produced cystic teratomas (Mikkola et al., 2006). Our observed differences between matched class II-III lines argue that class III cells may generally form poorly differentiated teratomas of cystic nature. The poor differentiation is consistent with a cancer-like state. On the basis of these observations, we suggest that XCI class designations of female hiPSCs may have practical implications for stem cell therapy.
Discussion
Here, we have studied genome-wide expression profiles of multiple new and existing hiPSC lines and shown that the XCI marker XIST RNA can be used as a readout to assess one aspect of female hiPSC quality. The gene expression profiles have identified molecular signatures that distinguish XIST+ (class II) and XIST− (class III) female hiPSC lines. Loss of XIST expression in class III cells is associated with upregulation of oncogenes, several of which are X linked, and downregulation of several tumor suppressors. We do not know whether loss of XIST is directly responsible for these expression differences. An alternative possibility is that conditions that lead to loss of XIST expression cause other changes genome-wide. In either case, we presume that these changes are generally undesirable and can therefore be used as additional benchmarks of hiPSC quality. Indeed, the class III changes correlate with faster growth in culture. Notably, these changes are not generally observed for male hiPSC lines. We also observed differences in differentiation in vivo, as shown by formation of predominantly cystic, poorly differentiated teratomas in immunocompromised mice.
These data argue for class- and sex-specific differences in epigenetic stability of hiPSCs that depend in large part on the ability to maintain XCI. One major implication is that the epigenetic state of female hiPSCs may be more difficult to maintain in culture, at least by current protocols. Neither physiological oxygen nor HDAC inhibitors offered any advantage nor more efficient X reactivation. Several recent works suggest that hiPSCs are not equivalent to the more extensively reprogrammed “naive” female hiPSCs, which apparently contain two Xa and may therefore represent the best model for X reactivation in hiPSCs (Hanna et al., 2010; Pomp et al., 2011; Wang et al., 2011). However, it is not clear whether these naive hiPSCs contain a pure population of class I cells or rather a mixture of class I-III cells. The epigenetic stability of XIST after extended culture is also uncertain. Better protocols are needed in order to avoid the potentially unfavorable genome-wide changes seen in many female hiPSC lines.
Another major implication may be that class III female hiPSC lines are best avoided for in vivo human therapy because of (1) the upregulation of some X-linked cancer genes, (2) faster-than-normal growth rates ex vivo, and (3) poor differentiation potential in vivo. Some hiPSC lines may evolve into the class III state more readily than others, perhaps because of underlying genetic and copy number variation between parental cell lines or the number of viral OSKM integrations. Although we do not know whether upregulation of X-linked and other oncogenes is a direct consequence of XIST repression, we surmise that the absence of XIST in class III lines may promote reactivation of undesirable X-linked genes, given recent work showing that conditionally deleting Xist on Xi of mouse somatic cells and loss of XIST in hESCs can result in piecemeal X reactivation (Csankovszki et al., 2001; Shen et al., 2008; Diaz Perez et al., 2012). If X reactivation occurs in these lines, they do not occur on all X genes at once (one X still resides in a Cot-1 hole). Nevertheless, the possibility of general X reactivation over time should present significant concern and urge caution in using some female hiPSC lines in cell regeneration programs. We therefore encourage the use of XCI markers as a benchmark to assess quality of all female hiPSCs and, by inference, hESC lines. Going forward, we suggest that XIST expression in combination with differentiation potential be used to assess stem cell quality.
Experimental Procedures
hiPSC Culture and Derivation
IMR-90 fibroblast line (ATCC; CCL-186) was cultured in EMEM medium with 10% FBS and XXY line (Corielle GM03102) with 15% FBS. Human iPSCs were maintained on irradiated MEFs with hESC medium (DMEM/F12, 10% knockout serum replacement [Invitrogen], L-glutamine, nonessential amino acids, 2-mercaptoethanol, penicillin/streptomycin, and 10 μg bFGF). For hiPSC derivation, 105 fibroblasts were infected with retrovirus (pEYK cassette with 4F) (Park et al., 2008a) at ambient oxygen, then 48 hr later transferred to MEF-coated plates at either ambient or 4% oxygen. MM, TA, and TB fibro-blasts were reprogrammed with the tet-inducible lentiviral STEMCCA and rtTA (without MEFs, no VPA during reprogramming or ROCK inihibitor). For hiPSC-1, hiPSC-2, hiPSC-3, hiPSC-9, hiPSC-10, hiPSC-11, and hiPSC-12, cells were treated with 1 mM VPA for 7 days (10 days for XXY lines), and colonies were picked 1 month after infection. Use of VPA did not impact occurrence of class I, II, or III cells, as MM, TB, and TA lines were reprogrammed without it. ROCK inhibitor Y-27632 (Calbiochem) was used for the first 2 days during the first two passages and after thawing cells. All hiPSCs were passaged manually. For HDACi treatment, sodium butyrate (0.1 mM; Sigma) and vorinostat (400 nM; Cayman) were freshly diluted and added daily. For cell growth experiments, colonies were split by cell rollers (Invitrogen) and one colony (about ten clumps) or three colonies for each line were transferred to one well of a96-well plate (one well of 12-well plate for three colonies). Cells were plated in triplicate. After day 7, duplicate wells containing three to five colonies were transferred to two wells (12-well plate). Cells were harvested weekly. Half the culture was counted and the other half passaged onto MEF-coated wells for four to six passages. Colony number was determined by counting undifferentiated colonies at each passage. The derivation of hiPSCs from fibroblasts obtained from ATCC and Corielle (lacking patient identification) does not require a human subjects oversight committee.
In Vitro and In Vivo Differentiation of hiPSCs
Human iPSC colonies were dislodged with cell scraper and transferred to low attachment 6-well plates containing hESC differentiation media (hiPSC media without b-FGF with 20% FBS). EBs were transferred to gelatin-coated plates (day 7) and cultured for additional 8–14 days. For EB germ lineage testing, hiPSCs were dispersed then grown in ultra-low attachment 6-well plates (Nunc) in hESC media without b-FGF supplemented with 1% FBS for 19 days. EBs were fixed in PBS with 4% paraformaldehyde (PFA), pelleted in low-melt agarose, paraffin embedded, sectioned (5 μm), and then stained with H&E. For teratoma injections, one to two 10 cm plates of confluent hiPSCs (no MEFs) were pelleted and mixed with equal volume of 2 × Matrigel (200 μl/injection). Tumors appeared 6–12 weeks after injection and were dissected and fixed overnight with 4% PFA, then sectioned and stained with H&E. These injections were performed with oversight of the Institutional Animal Care and Use Committee (IACUC) at Massachusetts General Hospital.
Microarray Experiments
Total RNA isolated with Trizol and converted to cDNA via NuGEN Ovation V2 Amplification system. cDNA was hybridized to Affymetrix Human Genome U133 Plus 2.0 Arrays (Microarray Core Facility, Dana Farber Cancer Institute). Samples: hiPS-1 p.31, hiPS-2 p.9, hiPS-3 p.14, hiPS-9 p.7, hiPS-10 p.24, hiPS-11 p.16, hiPS-11 p.16 (4% O2), hiPS-12 p.23 (4% O2), hiPS-9 p.17 (XIST−), and hiPS-12 p.30 (XIST−).
Microarray expression data were normalized by RMA (Irizarry et al., 2003). Hierarchical clustering and PCA were performed on the total sets of RMA expression values. MAS5 (Liu et al., 2002) and MBEI (Li and Hung Wong, 2001) normalization produced similar results. For the analysis of differential expression, we used the estimates of false discovery rate (FDR; ANOVA-based) with variance adjustment, implemented in NIA Array Analysis (Sharov et al., 2005). The tool was used with default parameters, except for z score threshold for outliers set to 10,000. The definition of differentially expressed genes was based on the combination of high statistical significance (FDR cutoff 0.05) and the magnitude of expression change. When overall expression was consistent between samples and the two compared groups corresponded to tight clusters, differentially expressed transcripts were defined with FDR < 0.05 and ≥2-fold change between group expression means. In the absence of tight clustering (class II female samples), differentially expressed transcripts were defined based on FDR cutoffs (FDR < 0.05) and n (individual samples that consistently showed at least 2-fold expression change compared to other group). For the eight samples of class II, we used the strict cutoff of n = 8 (all samples) and a more relaxed cutoff of n = 6. Correlation with XIST expression was measured by Pearson correlation coefficient calculated for the expression values. ComBat method for the compensation of microarray batch effects (Johnson et al., 2007) was run with default parameters on RMA expression values in the set of Affymetrix microarrays for different cell lines. DAVID functional annotation tool (Huang et al., 2009) was run online on the extended sets of differentially expressed genes.
Supplementary Material
Acknowledgments
We thank members of the J.T.L. laboratory for critical insight and discussion; G. Daley for pEYK,HD12D-1, and JDM 6C-1; G. Mostoslovasky for STEMCCA lentiviral cassette; H. Willard and J. Lawrence for PGK1 probe; L. Daheron, T. Ahfeldt, and R. Alagappan for technical assistance; and C. Ware for advice on HDACi. This work was funded by NIH RO1-GM58839 and ARRA supplement to J.T.L. and NIH R21MH087896 and a Stanley Medical Research Institute award to S.J.H. J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures (including RNA/DNA FISH, immunostaining, real-time PCR bisulfate sequencing, allele-specific expression analyses, and CGH analysis), seven figures, and four tables and can be found with this article online at doi:10.1016/j.stem.2012.03.008.
Accession Numbers: This microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession number GSE36233.
References
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. International Stem Cell Initiative Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BZ, Yu SL, Singh S, Kao LP, Tsai ZY, Yang PC, Chen BH, Shoei-Lung Li S. Identification of microRNAs expressed highly in pancreatic islet-like cell clusters differentiated from human embryonic stem cells. Cell Biol Int. 2011;35:29–37. doi: 10.1042/CBI20090081. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Perez SV, Kim R, Li Z, Marquez VE, Patel S, Plath K, Clark AT. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Hum Mol Genet. 2012;21:751–764. doi: 10.1093/hmg/ddr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoSONE. 2010;5:e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C, Lawrence JB. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35(Database issue):D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarkova MA, Shutova MV, Bogomazova AN, Vassina EM, Glazov EA, Zhang P, Rizvanov AA, Chestkov IV, Kiselev SL. Induction of pluripotency in human endothelial cells resets epigenetic profile on genome scale. Cell Cycle. 2010;9:937–946. doi: 10.4161/cc.9.5.10869. [DOI] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, Smeekens SP. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Mikkola M, Olsson C, Palgi J, Ustinov J, Palomaki T, Horelli-Kuitunen N, Knuutila S, Lundin K, Otonkoski T, Tuuri T. Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Dev Biol. 2006;6:40. doi: 10.1186/1471-213X-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30:3187–3205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pageau GJ, Hall LL, Ganesan S, Livingston DM, Lawrence JB. The disappearing Barr body in breast and ovarian cancers. Nat Rev Cancer. 2007;7:628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008b;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- Pomp O, Dreesen O, Leong DF, Meller-Pomp O, Tan TT, Zhou F, Colman A. Unexpected X chromosome skewing during culture and reprogramming of human somatic cells can be alleviated by exogenous telo-merase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Poplinski A, Wieacker P, Kliesch S, Gromoll J. Severe XIST hypomethylation clearly distinguishes (SRY+) 46,XX-maleness from Klinefelter syndrome. Eur J Endocrinol. 2010;162:169–175. doi: 10.1530/EJE-09-0768. [DOI] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Rogner UC, Wilke K, Steck E, Korn B, Poustka A. The melanoma antigen gene (MAGE) family is clustered in the chromosomal band Xq28. Genomics. 1995;29:725–731. doi: 10.1006/geno.1995.9945. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16:R243–R251. doi: 10.1093/hmg/ddm245. Spec No 2. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci USA. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci USA. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibro-blasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Westfall SD, Sachdev S, Das P, Hearne LB, Hannink M, Roberts RM, Ezashi T. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. Stem Cells Dev. 2008;17:869–881. doi: 10.1089/scd.2007.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.