Abstract
The cellular and molecular mechanisms of neurodevelopmental conditions such as autism spectrum disorders have been studied intensively for decades. The unavailability of live patient neurons for research, however, has represented a major obstacle in the elucidation of the disease etiologies. Recently, the development of induced pluripotent stem cell (iPSC) technology allows for the generation of human neurons from somatic cells of patients. We review ongoing studies using iPSCs as an approach to model neurodevelopmental disorders, the promise and caveats of this technique and its potential for drug screening. The reproducible findings of relevant phenotypes in Rett syndrome iPSC-derived neurons suggest that iPSC technology offers a novel and unique opportunity for the understanding of and the development of therapeutics for other autism spectrum disorders.
Introduction
The limited potential of neuronal samples from post-mortem brains and the inability to isolate populations of neurons from living subjects has blocked progress toward understanding the cellular and molecular mechanisms behind several neurodevelopmental disorders. Studies of postmortem tissue are problematic in developmental disorders as disease onset usually precedes death by decades. Moreover, frozen tissue sections are of limited use for studying cellular physiology and neural networks. Peripheral tissues, such as blood, are not suitable for relevant biological experiments since they are not the target tissue. Mathematical or computational models are also restricted by nature. Brain imaging allows you to study circuitries at a low magnification and does not reveal details of short circuitries in the brain. Finally, animal models often do not recapitulate complex human diseases, and have been particularly problematic in the case of human neurodevelopmental disease such as autism. Thus, the field lacks a human model that could provide unlimited supplies of neurons so experiments can be performed in controlled situations.
Genetic reprogramming provides a complementary model as it allows the genomes of human individuals afflicted with neurodevelopmental diseases to be captured in a pluripotent stem cell line. Reprogramming of somatic cells to a pluripotent state by overexpression of specific genes has been accomplished using mouse and human cells [1,2••]. These reprogrammed cell types, named induced pluripotent stem cells (iPSCs) can be derived from cells isolated from peripheral tissues of normal individuals or people affected from several conditions [3]. Isogenic pluripotent cells are attractive not only for their potential therapeutic use with lower risk of immune rejection, but also for their prospects to further understanding of complex diseases with heritable and sporadic conditions [4,5]. iPSCs can then be differentiated to human neurons to evaluate whether the captured genome alters cellular phenotypes in a similar manner as predicted by the clinical data or other mechanistic models. Although iPSCs have been generated for several neurological diseases the demonstration of disease-specific pathogenesis and phenotypic rescue in relevant cell types is a current challenge in the field, with only a handful proof-of-principle examples to date [6]. Nonetheless, the examples reflect the potential that this new model brings to disease modeling.
Considerations about iPSCs as a model for neurodevelopmental diseases
As with other models, the iPSC system also has important limitations. Cells in culture represent a research artifact. Thus, it is possible that important signaling information is missing or overstimulated in the system, masking potential cellular phenotypes or creating artificial ones. The discrimination between what is real and truly important in vivo will probably depend on validations coming from other models. Another challenge is the derivation of relevant neuronal subtypes. Specific protocols for subtypes of neurons are currently being developed and need further optimization. Alternatively, the relevant neuronal subtype needs to be sorted out or visualized using specific reporter genes. Unfortunately, the characterization of human neuronal subtypes is modest owing to the lack of knowledge on the temporal expression of specific genes and respective promoter activation. Recent efforts on human brain expression maps will certainly help [7,8].
Another important consideration is the use of appropriate controls. Intuitively, the ideal controls are the ones that differ from the patient by only the specific genetic defect. The targeted manipulation of the iPSCs to introduce genetic mutations in control cell lines or to restore the mutation from a patient cell line is a promising tool [9,10]. Another strategy to generate ‘isogenic’ cell lines is to take advantage of X-inactivation in iPSC clones coming from female cell lines. Owing to the fast X inactivation process during reprogramming, it is possible to generate iPSC cell clones carrying the mutant or the wild-type version of a X-linked affected gene. That strategy was used to model Rett syndrome, using female patients with mutations in the X-linked MeCP2 gene [11••,12]. For non-monogenetic diseases, or when the mutations are not known, such as sporadic autism, the challenge is greater. Behavioral variation between cell lines and iPSC clones from the same individual can influence phenotypic readouts. Unfortunately, the generation of individual iPSC clones is also expensive and time-consuming, restricting the number of cell lines that an individual laboratory can handle. A possible useful strategy for these types of disease is the coordination of consortium initiatives, where multiple sites would contribute to the pool of different cell types and phenotypic assays. Nonetheless, methods for generating neurons at a large scale and automated phenotypic analyses will become essential to realize such an endeavor.
Modeling neurodevelopmental diseases in a dish
A few years after the success of somatic cell reprogramming was reported in 2006, iPSC technology has been extensively used to model several neurodevelopmental diseases including a monogenic form of autism spectrum disorders (ASDs). Rett syndrome (RTT) [11••,12,13,15], sporadic form of Schizophrenia (SCZD) [16•], fragile X syndrome (FXS) [17], and Timothy syndrome (TS) [18••]. These studies were able to demonstrate that such disease-specific iPSC-derived neurons elegantly recapitulated relevant cellular and/or molecular phenotypes previously reported using different approaches, for example, postmortem human brain, and mouse model. In addition to those disorders, somatic cell reprogramming has recently been conducted for modeling another early onset neurological diseases including CDKL5-related disorder (a RTT variant) [19], and genomic imprinting disorders such as Angelman syndrome (AS) [20] and Prader-Willi syndrome (PWS) [20,21]. Successful neuronal differentiation was shown in these studies but the phenotypes of such disease-specific iPSC-derived neurons have not yet been examined. The neurodevelopmental diseases that have been modeling using human iPSC thus far and their significant findings are summarized in Table 1.
Table 1.
Examples of neurodevelopmental diseases that have been modeling using human iPSC
| Disease | Incidence | Age of neuropathological onset |
Key gene (chromosome) |
Genetic mutation in fibroblasts used for reprogramming |
Reprogramming method |
Neuronal differentiation and validation |
Relevant neuronal phenotype |
Candidate drug |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| RTT | 1:10,000 (female) [22] | ½–1½ years old [22] | MeCP2 (X) | Nonsense (Q244X) Missense (T158M, R306C) | Retrovirus (4 factors) | Yes; TuJ1+, MAP2+, GABA+, Synapsin+, VGLUT1+ Electrophysiologically active | Reduced soma size, dendritic spine density and synapses Altered Ca2+ signaling electrophysiological defect | IGF1 | Marchetto [11••] |
| Null (Δexon3–4) Missense (T158M, R306C) | Retrovirus (4 factors) | Yes; MAP2+ | Reduced soma size | No | Cheung [12] | ||||
| Nonsense (Q244X) Missense (T158M, R306C) | Retrovirus (4 factors) | Yes; TuJ1+, SCN1A/B+ | Lower expression of mature neuron markers | No | Kim [15] | ||||
| Nonsense (V247X, R294X) Missense (T158M, R306C) | Retrovirus and Lentivirus (4 factors) | Yes; TuJ1+ | Reduced nuclear size | No | Ananiev [13] | ||||
| FXS | 1:4000–1:6000 [36] | <3 years old [36] | FMR1 (X) | >200 CGG repeats in 5′UTR | Retrovirus (4 factors) | No | No | No | Urbach [37••] |
| >700 CGG repeats in 5′UTR | Retrovirus (4 factors) | Yes; TuJ1+ | Fewer and shorter neurites | No | Sheridan [17] | ||||
| SCZD | 1:100 [38] | Typically 15–25 years old In rare case, <10 years old [39] | DISC1 (1) | Δexon-intron12 region (4bp) | Integration-free episomes (4 factors) | No | No | No | Chiang [40] |
| Sporadic | Unknown | Tetracyclin-inducible lentivirus (5 factors) | Yes; TuJ1+, MAP2+, PSD95+, VGLUT1+, GAD65/67+ Electrophysiologically active | Reduced neuronal connectivity and neurite number, decreased PSD-95 expression | Loxapine | Brennand [16•] | |||
| TS | Unknown (20 cases reported worldwide [41]) | >2.5 years old [41,42] | CACNA1C (12) | Missense (G406R) | Retrovirus (4 factors) | Yes; MAP2+, VGLUT1/2+, TH+, GAD65/67+, CTIP2+, FOXP1+, SATB2+, Electrophysiologically active | Defect in Ca2+ signaling and electrophysiology, decreased SATB2 expression §Higher expression of TH and catecholamines | Roscovitine | Pasca [18••] |
| CDKL5-related disorder | Unknown (80 cases reported worldwide [43]) | 2–3 months old [44] | CDKL5 (X) | Nonsense (Q347X) Missense (T288I) | Retrovirus (4 factors) | Yes; TuJ1+, MAP2+, VGLUT1/2+, GAD65/67+ | No | No | Amenduni [19] |
| AS | 1:12,000 [45] | 2–3 years old [46] | Maternal UBE3A (15) | Maternal Δ15q11-q13 (including UBE3A) | Retrovirus (5 factors) | Yes; TuJ1+, MAP2+, SynapsinI+, PanNav+ Electrophysiologically active | No | No | Chamberlain [20] |
| PWS | 1:15,000 [47] | 2–6 years old [48] | Unknown in paternal | Paternal Δ15q11-q13 | Retrovirus (5 factors) | No | No | No | Chamberlain [20] |
| 15q11-q13 (15) | t(15;4)(q11.2;q27) | Retrovirus (4 factors) | Yes; TuJ1+, MAP2+ | No | No | Yang [21] |
4 factors, OCT4, SOX2, KLF4 and c-MYC; 5 factors, OCT4, SOX2, KLF4, c-MYC and LIN28;
not observed in animal.
Modeling Rett syndrome with iPSCs
We recently demonstrated the utility of induced pluripotent stem cells to investigate the functional consequences of mutations in the gene encoding the Methyl CpG binding protein-2 (MeCP2) on neurons from RTT patients, a syndromic form of ASD [11]. RTT patients appear to develop normally for up to 6–18 months, after which they enter a period of regression characterized by deceleration of head growth and loss of acquired motor and language skills. Patients often develop autistic behaviors, stereotypic hand wringing, abnormal breathing and seizures [22]. Postmortem analyses of affected individuals have revealed phenotypes at the cellular level, including decreased soma size, reduced dendritic branching, and altered dendritic spine density and morphology [23,24].
MeCP2 is an abundant nuclear protein implicated in a number of molecular functions, but was first identified as an epigenetic regulator of target genes by binding to methylated CpG dinucleotides [25]. Long established as a repressor of transcription, MeCP2 has recently been shown to bind active genes as well as to influence RNA splicing [22,26]. MeCP2 is found in a wide variety of tissues but appears to be most abundant in the brain [27]. Accordingly, MeCP2 appears to be critical for normal CNS development and function, and its dysfunction results in abnormal neurological phenotypes (Figure 1).
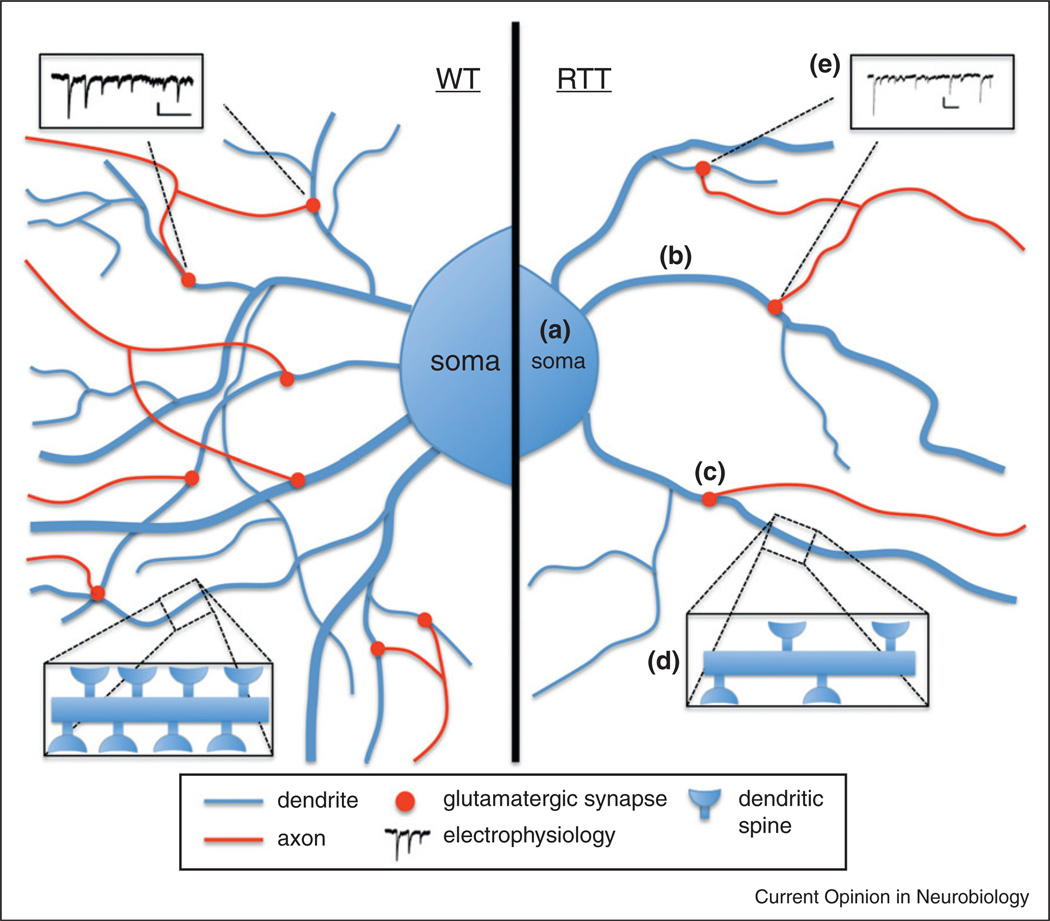
Figure 1.
Phenotypes demonstrated by human iPSC-derived RTT neurons. We generated a human cellular model for RTT by reprogramming the fibroblasts of human patients to iPSCs. Neurons differentiated from these iPSCs exhibited cellular phenotypes such as (a) smaller soma size, (b) reduced dendritic branching, and (c) fewer glutamatergic synapses and (d) dendritic spines. These morphological alterations contributed to the functional phenotype of (e) altered electrophysiology, as RTT-neurons demonstrated a decreased frequency of spontaneous postsynaptic currents.
Neurons derived from RTT-iPSCs carrying four different MeCP2 mutations showed several alterations compared to five healthy non-affected individuals, such as decreased soma size, altered dendritic spine density and reduced excitatory synapses. These phenotypes were validated using wild-type MeCP2 cDNA and specific shRNAs against MeCP2 in gain-and-loss of function experiments to demonstrate causality. Importantly, some of these cellular defects were immediately validated by independent groups, revealing the robustness and reproducibility of the system [12,13,15]. We were able to rescue the defects in the number of glutamatergic synapses using two candidate drugs, insulin-like growth factor 1 (IGF1) and gentamicin. IGF1 is considered to be a candidate for pharmacological treatment of RTT and potentially other CNS disorders in ongoing clinical trials [28]. Gentamicin, a read-through drug, was also used to rescue neurons carrying a nonsense MeCP2 mutation, by elevating the amount of MeCP2 protein.
Moreover, we took advantage of the RTT-iPSCs to demonstrate that neural progenitor cells carrying MeCP2 mutations have increased susceptibility for L1 retrotransposition. Long interspersed nuclear elements-1 (LINE-1 or L1s) are abundant retrotransposons that comprise approximately 20% of mammalian genomes [29–31] and are highly active in the nervous system [32•,33]. Our data demonstrate that L1 retrotransposition can be controlled in a tissue-specific manner and that disease-related genetic mutations can influence the frequency of neuronal L1 retrotransposition [34]. The work revealed an unexpected and novel phenomenon, adding a new layer of complexity to the understanding of genomic plasticity. These observations bring valued information for RTT and, potentially, other ASD patients, since they suggest that pre-symptomatic defects may represent novel biomarkers to be exploited as diagnostic tools. The data also suggest that early intervention may be beneficial.
Using human neurons as a drug-screening platform
Our studies performed in RTT highlighted the potential of iPSC models in toxicology and drug screening. Even better, the IGF1 overcorrection observed in some RTT neurons [11••] indicate that the iPSC technology not only can recapitulate some aspects of a genetic disease but also can be used to better design and anticipate results from translational medicine. This cellular model has the potential to lead to the discovery of new compounds to treat different neurodevelopmental diseases.
Drug-screening platforms require ‘screenable’ robust phenotypes in target cell types, such as iPSC-derived neurons. The neuronal differentiation strategies reported to date are not current capable of providing vast numbers of homogenous subtypes of neurons in a reliable, reproducible, and cost-effective fashion. However, with better markers and gene reporters, it will be possible to isolate pure populations of desired cell types in a large scale. Cellular morphology, such as soma size or dendritic spine density, can be captured using high-content imaging software. Early biochemical and gene expression read outs can be useful alternatives. However, late read outs, such as electrophysiological records, may not be ideal owing to the time in culture necessary to reach neuronal maturation. It is certainly possible to use stressors or other environmental agents to enhance the differences between control and patient groups. An alternative solution may emerge from the direct conversion of neurons from peripheral cells, skipping the pluripotent state [35]. This technology is currently inefficient in humans, difficult to scale up and has the disadvantage to skip neuronal development stages. Finally, read outs needed to be suitable for the instrumentation demanded for high-throughput screening for drug discovery. More scalable assays will allow characterization of increased numbers of control and patient neurons.
Conclusion
The iPSC strategy is a novel and complementary approach to model neurodevelopmental diseases. Although this technology is still in its early stage, it potentially demonstrated the ability to recapitulate relevant neuronal defects of those diseases. This model has the capacity to unify data generated from brain imaging, animal work, and genetics, generating downstream hypotheses that could be tested in well-controlled experiments in the relevant cell types. As several neurodevelopmental disorders share similar neurological symptoms while arising from distinct genetic variations, morphological and functional comparison of patient-specific iPSC-derived neurons would provide insight into common trends and unique phenotypes of each disease. This patient-gene-phenotype cellular analysis can ultimately contribute to the establishment of the links between genes and behavior. In the future, the iPSC approach may also be used as diagnostic tools. Finally, as the technology evolves, it will reach the point of personalized medicine, making predictions about the efficiency of certain drugs and doses in determined individuals.
Acknowledgements
The work was supported by grants from the California Institute for Regenerative Medicine (CIRM) TR2-01814, the National Institutes of Health through the NIH Director’s New Innovator Award Program, 1-DP2-OD006495-01, 1R21MH093954 from NIMH, P01 HD33113 and NS22343 from NIH, the Royal Thai Government Scholarship to T.C., the NIH predoctoral training grant T32 GM008666 to A.A and the Emerald Foundation. We would like to thank members of the Muotri laboratory for critical comments on the manuscript. This is a brief review on the fast growing field of disease modeling, we apologize for the omission of important work from colleagues that could not be described or cited here.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. The first report of cellular reprogramming using ectopic expression of pluripotent factors was elegantly described in this paper.
- 3.Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muotri AR. Modeling epilepsy with pluripotent human cells. Epilepsy Behav. 2009;14(Suppl 1):81–85. doi: 10.1016/j.yebeh.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 5.Marchetto MC, Winner B, Gage FH. Pluripotent stem cells in neurodegenerative and neurodevelopmental diseases. Hum Mol Genet. 2010;19:R71–R76. doi: 10.1093/hmg/ddq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchetto MC, Brennand KJ, Boyer LF, Gage FH. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet. 2011;20:R109–R115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu GH, Suzuki K, Qu J, Sancho-Martinez I, Yi F, Li M, Kumar S, Nivet E, Kim J, Soligalla RD, et al. Targeted gene correction of laminopathy-associated lmna mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. This work describes a comprehensive comparison of iPSC-derived neurons from Rett syndrome patients and controls using an array of techniques. It also shows rescue of a phenotype demonstrating the application of the technology for disease modeling, pathogenesis studies and a future drug screening approach.
- 12.Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of mecp2-null rett syndrome patient hips cells and isogenic controls through x-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ananiev G, Williams EC, Li H, Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from rett syndrome patients as in vitro disease model. PLoS ONE. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with rett syndrome. Proc Natl Acad Sci USA. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. This work described neuronal phenotypes associated with idiopathic schizophrenia.
- 17.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the fmr1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile x syndrome. PLoS ONE. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. This work describes defects in neurons derived from iPSC obtained from Timothy syndrome. Some of the defects could not be validated in knockout mice, suggesting intrinsic differences in human and mouse models.
- 19.Amenduni M, De Filippis R, Cheung AY, Disciglio V, Epistolato MC, Ariani F, Mari F, Mencarelli MA, Hayek Y, Renieri A, et al. Ips cells to model cdkl5-related disorders. Eur J Hum Genet. 2011;19:1246–1255. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders angelman and prader-willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Cai J, Zhang Y, Wang X, Li W, Xu J, Li F, Guo X, Deng K, Zhong M, et al. Induced pluripotent stem cells can be used to model the genomic imprinting disorder prader-willi syndrome. J Biol Chem. 2010;285:40303–40311. doi: 10.1074/jbc.M110.183392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chahrour M, Zoghbi HY. The story of rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong DD. Neuropathology of rett syndrome. J Child Neurol. 2005;20:747–753. doi: 10.1177/08830738050200090901. [DOI] [PubMed] [Google Scholar]
- 24.Schule B, Armstrong DD, Vogel H, Oviedo A, Francke U. Severe congenital encephalopathy caused by mecp2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clin Genet. 2008;74:116–126. doi: 10.1111/j.1399-0004.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- 25.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-cpg-binding protein mecp2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 26.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of rna splicing by the methylation-dependent transcriptional repressor methyl-cpg binding protein 2. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into rett syndrome: Mecp2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 28.Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, Flannery R, Jaenisch R, Sur M. Partial reversal of rett syndromelike symptoms in mecp2 mutant mice. Proc Natl Acad Sci USA. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the brown norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 31.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 32. Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. This work reveals that neural progenitor cells can support de novo somatic mutations from L1 retrotransposons.
- 33.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by mecp2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford DC, Acuna JM, Sherman SL. Fmr1 and the fragile X syndrome: human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. This work reveals intrinsic differences between an iPSC model of fragile X syndrome when compared to human ES cells carrying the mutation.
- 38.Regier DA, Narrow WE, Rae DS, Manderscheid RW, Locke BZ, Goodwin FK. The de facto us mental and addictive disorders service system. Epidemiologic catchment area prospective 1-year prevalence rates of disorders and services. Arch Gen Psychiatry. 1993;50:85–94. doi: 10.1001/archpsyc.1993.01820140007001. [DOI] [PubMed] [Google Scholar]
- 39.Sham PC, MacLean CJ, Kendler KS. A typological model of schizophrenia based on age at onset, sex and familial morbidity. Acta Psychiatr Scand. 1994;89:135–141. doi: 10.1111/j.1600-0447.1994.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 40.Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a disc1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, et al. Ca(v)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Krause U, Gravenhorst V, Kriebel T, Ruschewski W, Paul T. A rare association of long qt syndrome and syndactyly: Timothy syndrome (lqt 8) Clin Res Cardiol. 2011;100:1123–1127. doi: 10.1007/s00392-011-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rademacher N, Hambrock M, Fischer U, Moser B, Ceulemans B, Lieb W, Boor R, Stefanova I, Gillessen-Kaesbach G, Runge C, et al. Identification of a novel cdkl5 exon and pathogenic mutations in patients with severe mental retardation, early-onset seizures and rett-like features. Neurogenetics. 2011;12:165–167. doi: 10.1007/s10048-011-0277-6. [DOI] [PubMed] [Google Scholar]
- 44.Evans JC, Archer HL, Colley JP, Ravn K, Nielsen JB, Kerr A, Williams E, Christodoulou J, Gecz J, Jardine PE, et al. Early onset seizures and rett-like features associated with mutations in cdkl5. Eur J Hum Genet. 2005;13:1113–1120. doi: 10.1038/sj.ejhg.5201451. [DOI] [PubMed] [Google Scholar]
- 45.Steffenburg S, Gillberg CL, Steffenburg U, Kyllerman M. Autism in angelman syndrome: a population-based study. Pediatr Neurol. 1996;14:131–136. doi: 10.1016/0887-8994(96)00011-2. [DOI] [PubMed] [Google Scholar]
- 46.Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of angelman syndrome. Genet Med. 2010;12:385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- 47.Wattendorf DJ, Muenke M. Prader-willi syndrome. Am Fam Physician. 2005;72:827–830. [PubMed] [Google Scholar]
- 48.Gunay-Aygun M, Schwartz S, Heeger S, O’Riordan MA, Cassidy SB. The changing purpose of prader-willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics. 2001;108:E92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]