Abstract
Chromosome segregation requires the generation of force at the kinetochore—the multiprotein structure that facilitates attachment of chromosomes to spindle microtubules. This force is required both to move chromosomes and to signal the formation of proper bioriented attachments. To understand the role of force in these processes, it is critical to define how force is generated at kinetochores, the contributions of this force to chromosome movement, and how the kinetochore is structured and organized to withstand and respond to force. Classical studies and recent work provide a framework to dissect the mechanisms, functions, and consequences of force at kinetochores.
Force plays key roles in many different cellular processes by influencing objects in a way that causes them to change their speed or direction of movement. Force can take multiple forms in a cell and have very different consequences, depending on the circumstances of its action. When force pulls on an object, it creates “tension.” In contrast, a pushing force exerted upon an object is termed “compression.” To understand the contribution of force to cellular processes, it is important to determine the molecular mechanisms by which force is generated or produced at a subcellular structure, how these structures withstand the force, and how they detect and signal the presence of force. The process of mitotic chromosome segregation provides a particularly intriguing example of the importance of cellular force. During mitosis, force plays a critical role in directing the physical segregation of chromosomes and modulating the signals that sense and promote their proper attachment to the spindle. The central player in chromosome segregation is a macromolecular structure termed the kinetochore that establishes and maintains the attachment of each set of paired sister chromatids to microtubule polymers from opposing spindle poles and directs the segregation of chromosomes to the daughter cells (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009). The kinetochore plays key roles throughout mitosis, both to mediate direct attachments between microtubules and centromeric DNA (Fig. 1) and as a hub for the signaling molecules required to monitor and control faithful chromosome segregation and cell cycle progression. Because the kinetochore is the contact point between chromosomes and microtubules, the forces derived from microtubules are exerted directly on the proteins within the kinetochore. A key challenge is to understand how this force is generated and accommodated and to define the specific contributions of this force to kinetochore function.
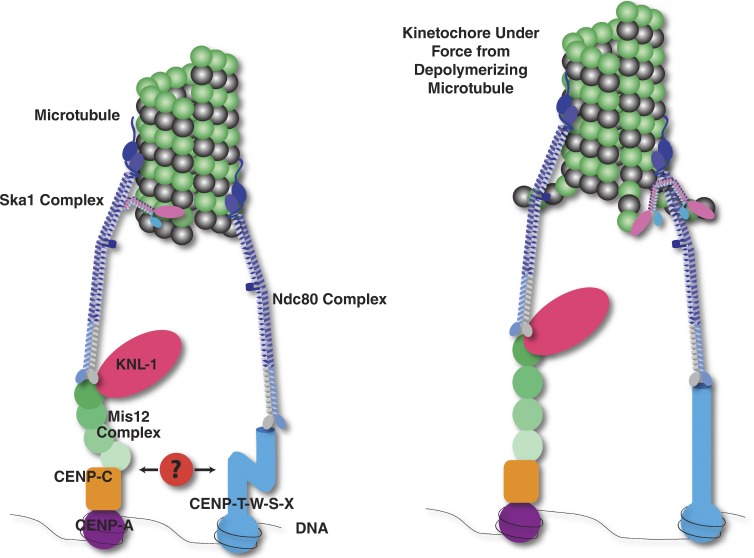
Figure 1.
Simplified diagram of the kinetochore showing the major proteins involved in the DNA–microtubule attachment. (Left) The Ndc80 complex (dark blue) binds to microtubules and forms two separate connections to kinetochores. First, the Ndc80 complex binds to the Mis12 complex (green) and KNL-1 (magenta). The Mis12 complex in turn binds to CENP-C (orange), which binds to nucleosomes containing the histone H3 variant CENP-A (purple). Second, the Ndc80 complex binds to CENP-T (light blue). CENP-T interacts with DNA as a part of a heterotetrameric nucleosome-like CENP-T–W–S–X complex. In humans, the Ndc80 complex attachment to microtubules is enhanced by an interaction with the Ska1 complex (pink and blue; Schmidt et al., 2012). Additional components may form interactions between the two connective pathways (red). (Right) Upon microtubule depolymerization, the flexible protein components of the kinetochore may rearrange. For example, recent evidence has suggested that the N and C termini of CENP-T separate under tension (Suzuki et al., 2011) and that the subunits of the Mis12 complex redistribute (Wan et al., 2009).
How much force is generated at a kinetochore?
The nature of the forces involved in partitioning chromosomes has been an active area of research for more than 50 years. Edwin Taylor and Bruce Nicklas were among the first to consider the forces that resist chromosome movement. Separate theoretical analyses predicted that ∼0.1 pN would be required to move a chromosome at 1 µm/min when resisted only by viscous cytoplasmic drag (Nicklas, 1965; Taylor, 1965). Almost 20 years after publishing his theoretical work, Nicklas was able to test the force on a single chromosome during anaphase of meiosis I (Nicklas, 1983). Using a microneedle to measure the stall force on chromosomes in grasshopper spermatocytes, Nicklas found that 700 pN could act on a chromosome (Nicklas, 1983). He estimated that the kinetochores tested in these studies were bound by ∼15 microtubules (Nicklas, 1983), suggesting that each microtubule may be capable of generating up to ∼45 pN of force. In a later study, Nicklas determined that ∼50 pN of force was produced on a chromosome during prometaphase (Nicklas, 1988). This calculation was based on observations of chromosome congression and correlations with his previous work. By Nicklas’ own admission, the microneedle assays to measure the force exerted on anaphase chromosomes had a high associated error, and it is unknown whether forces in the hundreds of piconormals would ever be produced at a kinetochore in the absence of a perturbation. Regardless, no other work since has provided a more exact measurement, and 700 pN remains the standard reference value for the force that can act at a metazoan kinetochore.
As Nicklas’ work suggested, it is likely that the force felt by kinetochores varies throughout the cell cycle and under different types of attachments (discussed later in this paper). In particular, the arrangement of paired sister chromatids attached to opposite spindle poles during metaphase would allow for the greatest tension to be applied to kinetochores. Recent work visualizing sister chromatid oscillations during metaphase has observed that at time points immediately before the switch from poleward to antipoleward motion, the poleward-moving kinetochore experiences the highest forces, at least as judged by changes in intra- and interkinetochore distances (Dumont et al., 2012; Wan et al., 2012). In addition, the antipoleward-moving kinetochore may experience passive forces (Inoué and Salmon, 1995; Maddox et al., 2003) that can also alter inter- and intrakinetochore stretch (Dumont et al., 2012; Wan et al., 2012). However, the magnitude of force during these directional switches and how this force is accommodated continues to be a subject of debate. As the higher order organization of kinetochores remains unknown, it is unclear how the forces from the multiple microtubule interactions at a single kinetochore are combined or what force is experienced by an individual protein within the kinetochore structure.
The mechanisms of force production
With the discovery of the potentially large forces produced at kinetochores (Nicklas, 1983), a major challenge has been to define the mechanisms by which this force is generated. Many initial studies focused on the contributions of the microtubule-based motors, dynein and kinesin, that were found to localize to kinetochores (Inoué and Salmon, 1995). The ability of these motors to transport cargoes along microtubules suggested that they might function similarly to move a chromosome within a cell. Individual kinesin and dynein motors have been shown to stall under ∼5–7 pN of opposing force, termed a load (Visscher et al., 1999; Gennerich et al., 2007), and the combined action of multiple motors could generate the forces that Nicklas observed. However, subsequent studies have found that chromosome movement can still largely occur in the absence of these motors in fungi (Cottingham et al., 1999; Grishchuk and McIntosh, 2006). In metazoans, motors, including the kinesin CENP-E and dynein, contribute to chromosome segregation (Sharp et al., 2000; Kapoor et al., 2006; Yang et al., 2007), although their relative importance remains unclear. An alternative hypothesis was that the microtubules themselves generated the force to move chromosomes (Inoué and Salmon, 1995). Several early studies provided evidence that microtubules could direct the movement of isolated chromosomes under conditions that would not permit motor protein function (Koshland et al., 1988; Coue et al., 1991; Hunt and McIntosh, 1998). This microtubule-derived movement could be caused by forces generated either at the kinetochore by microtubule depolymerization (Grishchuk et al., 2005) or at the spindle poles as a result of poleward flux and microtubule disassembly at the minus end (LaFountain et al., 2001, 2004; Chen and Zhang, 2004). In fact, subsequent work suggested that the stall forces measured by Nicklas were a result of minus end microtubule disassembly in equilibrium with the plus end microtubule polymerization caused by the application of tension via the microneedle (LaFountain et al., 2001, 2004; Chen and Zhang, 2004). Although it is now generally accepted that microtubules generate the primary forces responsible for chromosome movement, kinetochore-localized motors may generate some force, act as a “back-up” system when kinetochore capture by microtubules fails (Kapoor et al., 2006), generate tension via the production of the polar ejection forces (Mazumdar and Misteli, 2005), function to distribute force over additional linkages, and regulate microtubule dynamics (Bader and Vaughan, 2010; Al-Bassam and Chang, 2011). In addition to forces generated either directly or indirectly by the microtubules, a third model proposes that the chromosomes themselves may contribute to the segregation process because of entropic forces that act on the DNA (Jun and Wright, 2010; Finan et al., 2011). Although such forces would likely be very small, they may assist chromosome distribution, particularly in smaller cells.
In support of a primary role for microtubules in generating force at kinetochores, microtubules have been shown to generate pulling force during their depolymerization in vitro (Grishchuk et al., 2005; Powers et al., 2009; Akiyoshi et al., 2010; Tien et al., 2010). During microtubule polymerization, GTP-bound tubulin dimers are added to the growing microtubule plus end (Desai and Mitchison, 1997). After these dimers are incorporated into the microtubule lattice, GTP is hydrolyzed. The resulting GDP-bound tubulin dimers associate with each other along an individual protofilament and between neighboring protofilaments within the microtubule lattice to maintain a straight microtubule (Nogales, 2000; Nogales and Wang, 2006). However, when a microtubule switches to depolymerization, a process termed catastrophe, GDP-bound dimers exposed at the microtubule end lose these stabilizing interactions, causing the protofilaments to peel backward. According to measurements and calculations by Grishchuk et al. (2005), the conformational change that occurs for an individual depolymerizing protofilament can generate a power stroke of ≤5 pN, suggesting that a depolymerizing microtubule composed of 13 protofilaments could generate as much as 65 pN of force. Importantly, to harness this force and ensure proper chromosome movement, it is critical to control microtubule polymerization and depolymerization at kinetochores. The formation of kinetochore–microtubule attachments as well as the resulting tension may directly modulate microtubule dynamics by slowing microtubule depolymerization and decreasing the rate of catastrophe (Franck et al., 2007; Akiyoshi et al., 2010; Umbreit et al., 2012). In addition, microtubule polymerization factors, such as the TOG (tumor overexpressed gene) domain proteins XMAP215 and CLASP, and depolymerases, such as kinesin-13 proteins, which are present both at the kinetochore and on the spindle, also modulate microtubule behavior (Bader and Vaughan, 2010; Al-Bassam and Chang, 2011).
Although microtubule depolymerization has the capacity to generate force, a key question is how chromosome movement is coupled to microtubule depolymerization. Thus far, two models have dominated the literature to explain how kinetochores harness the force from microtubule depolymerization, although these models are not mutually exclusive. The first model, termed the “Hill sleeve” model or “biased diffusion” (Hill, 1985), postulates that the association of the kinetochore with a microtubule is formed by multiple weak interactions that can diffuse equally in either direction. However, because of a large free energy barrier that disfavors the loss of an interaction, this diffusion is biased toward the microtubule minus end as binding sites disappear from the plus end. The second model, termed the “forced walk” model (Molodtsov et al., 2005), proposes that the kinetochore is coupled to microtubules in such a way that, as the protofilaments peel backward during depolymerization, the coupling protein is pushed along the microtubule. The way in which the microtubule is connected to the kinetochore has important implications for understanding how the force manifests at the kinetochore and remains an important focus for future work.
Recent studies have focused on how kinetochores and kinetochore proteins harness the energy from microtubule depolymerization. These studies have tested key players at the kinetochore–microtubule interface, such as the Ndc80, Dam1, and Ska1 complexes (McIntosh et al., 2008; Powers et al., 2009; Welburn et al., 2009; Lampert et al., 2010; Tien et al., 2010; Schmidt et al., 2012) for their abilities to track on depolymerizing microtubules, and have attempted to analyze the kinetochore as a whole using partial purifications of kinetochores from Saccharomyces cerevisiae (Akiyoshi et al., 2010). Although individual protein complexes and isolated yeast kinetochores are able to move with depolymerizing microtubules, studies performed using optical tweezers have found that the tested proteins and complexes are able to withstand less than 10 pN of pulling force before a rupture event is observed (Powers et al., 2009; Akiyoshi et al., 2010; Tien et al., 2010). This is in contrast to the theoretical maximum of 65 pN that a microtubule has been proposed to produce during depolymerization (Grishchuk et al., 2005). It is likely that in the context of a kinetochore assembled on a chromosome, the complex architecture of the kinetochore has the capacity to harness and withstand larger forces. Thus, the in vivo load-bearing properties of the kinetochore likely depend on a combination of the properties of both the individual protein components and the organization of the entire complex.
Signaling the biorientated state of chromosomes
During mitosis, it is critical that paired sister chromatids attach to opposite spindle poles. When this biorientation fails, this error must be detected and corrected, and a signal to delay cell cycle progression must be produced to prevent chromosome missegregation. Work performed by Li and Nicklas (1995) and Nicklas et al. (1995) demonstrated that the external application of force to a chromosome using a microneedle could overcome the checkpoint signal generated by an unattached kinetochore. This and other work have supported the model that the tension produced on bioriented sister kinetochores can alter the signaling state of the kinetochore. This tension results in two apparent physical alterations to mitotic chromosome structure: an increase in the distance between paired sister kinetochores and an increase in the distance between the inner and outer kinetochore regions of a single kinetochore. Under some conditions, this inter- and intrakinetochore stretch can be uncoupled (Maresca and Salmon, 2009), and recent research has focused on the importance of intrakinetochore stretch in modulating the signals that monitor attachment state. By measuring the relative spatial positions of the different kinetochore proteins, work from several groups has found that kinetochore structure is altered when chromosomes are bioriented relative to conditions of reduced tension (Maresca and Salmon, 2009; Uchida et al., 2009; Wan et al., 2009; Suzuki et al., 2011; Dumont et al., 2012). Biorientation results in the separation of inner kinetochore components (such as CENP-A and CENP-C) from outer kinetochore components (such as Ndc80 and Mis12) as well as changes in the spatial distribution of other proteins within the kinetochore and possibly conformational changes within the proteins themselves.
Because the generation of tension is dependent on the presence of opposing forces, changes in kinetochore structure correlate with the successful bioriented arrangement of chromosomes on the metaphase plate. In contrast, when one sister kinetochore lacks an attachment to the spindle (monotelic), or if both kinetochores attach to the same pole (syntelic), it is not possible to generate similar opposing forces. However, even in these cases, some force may still be present because of the viscosity of the cytoplasm resisting chromosome movement (Nicklas, 1965; Taylor, 1965) or the action of chromokinesins that generate polar ejection forces (Mazumdar and Misteli, 2005). It remains unclear how force is exerted on a single kinetochore that simultaneously attaches to opposing spindle poles (merotelic) or how these incorrect attachments are resolved (Gregan et al., 2011; Matos and Maiato, 2011). The observed structural changes at kinetochores have been assumed to correlate with the presence of tension, but thus far, such studies have not made direct measurements of force or tension. Nevertheless, careful quantitative analysis of the dynamic changes in the distances between CENP-C and Hec1 or Cdc20 during sister chromatid oscillations has supported the model that changes in intrakinetochore distance are force dependent (Dumont et al., 2012). However, these structural alterations may also be the result of changes in the conformation, organization, or localization of proteins within the kinetochore.
Ultimately, it is important to translate the mechanical signals produced by force at kinetochores into a chemical signal that regulates the activities of kinetochore proteins. A key player in correcting errors in microtubule attachment state is the Aurora B kinase. Substrates for Aurora B show tension-sensitive phosphorylation; they are highly phosphorylated in the absence of tension and become dephosphorylated upon biorientation (Liu et al., 2009; Welburn et al., 2010). The forces generated at kinetochores have been implicated in controlling Aurora B signaling by altering the spatial separation between the kinase and its substrates (Tanaka, 2002; Liu et al., 2009), although other models for tension-sensitive Aurora B phosphorylation have also been proposed (Sandall et al., 2006). The key substrates of Aurora B are located at the outer kinetochore and can be >100 nm away from the majority of Aurora B, which is localized at the inner centromere, depending on whether the sister kinetochores are under tension (Wan et al., 2009). Therefore, structural changes caused by opposing force at kinetochores separate the kinase and its substrates. The increased separation under tension makes Aurora B less likely to phosphorylate its now distant substrates (Liu et al., 2009; Welburn et al., 2010). One effect of Aurora B phosphorylation on outer kinetochore proteins is to reduce their microtubule binding affinity (Cheeseman et al., 2006; Welburn et al., 2010; Schmidt et al., 2012). Thus, it has been proposed that the presence of tension can ultimately stabilize microtubule attachments through changes in kinetochore conformation that cause a decrease in Aurora B phosphorylation, which in turn increases the microtubule binding activities of various kinetochore components.
In addition to altering the signaling state of kinetochores, changes in force at kinetochores may also have a direct effect on microtubule binding. One recent study suggested that outer kinetochore proteins are force sensitive and show catch–slip properties (Akiyoshi et al., 2010), resulting in less frequent detachment under increasing force. This is analogous to a “Chinese finger trap” and would allow the attachment to become stabilized as the microtubule pulls on the kinetochore. Whether tension affects kinetochore–microtubule attachments directly or indirectly, force appears to play an essential role in establishing and signaling biorientation in addition to driving chromosome movement.
Theoretical considerations for force resistance
Force is a vector quantity that, when applied to a bond, decreases bond energy barriers, increasing the likelihood of bond breakage. Although the kinetochore must function under force to perform its roles properly, this force also represents a challenge with the potential for deleterious consequences to kinetochore function. Force could result in protein unfolding or the breakage of protein–protein interactions (Fig. 2). If a core kinetochore protein unfolded or if protein interactions within the kinetochore were disrupted, the connectivity between centromeric DNA and the microtubules would be compromised. The typical force required to unfold a protein or break interactions is in the range of 10–100 pN (Weisel et al., 2003; Lin et al., 2005; Kumar and Li, 2010). Nicklas did not observe an immediate rupture of chromosome–spindle attachments even while applying 700 pN on chromosomes, suggesting that the kinetochore is constructed in a way that can withstand high loads.
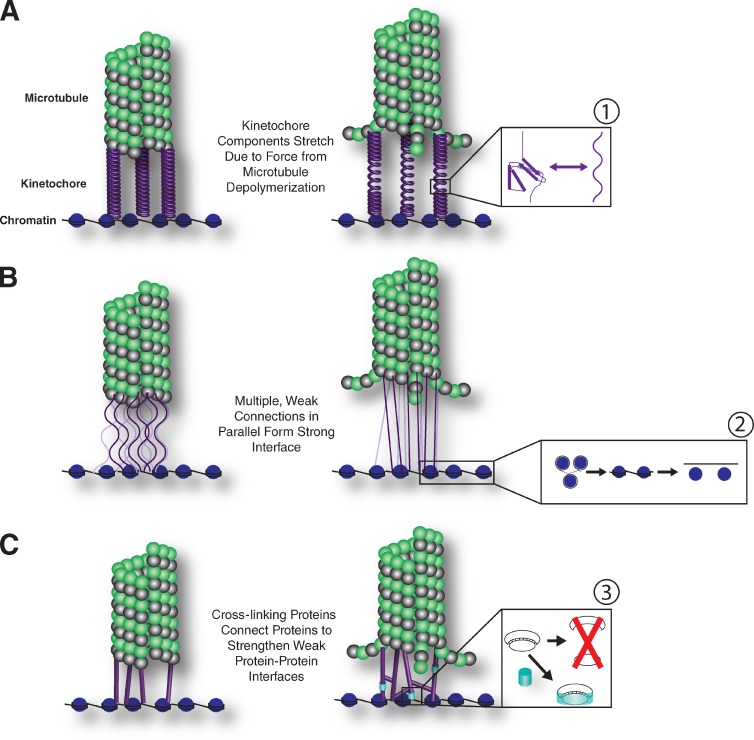
Figure 2.
Models for force response at kinetochores at both the individual protein level and global scale. (A–C) We propose three nonexclusive models for how kinetochores respond to the application of force: kinetochore proteins with elastic properties could serve to absorb some of the force produced by depolymerizing microtubules (A), multiple weak interfaces could form parallel attachments between the depolymerizing microtubule and chromosome such that the force produced by the microtubule would be diffused across multiple connections (B), and additional kinetochore components could serve as dynamic cross-linkers to diffuse force and add interactions between pairs of proteins to strengthen the protein–protein interface (C). The kinetochore protein components themselves could have multiple responses at a molecular level including that (1) under pulling forces, the bonds holding together the tertiary and secondary structure of a protein can break, causing the protein to unfold. If reversible, this would provide elastic properties, but if permanent, could lead to loss of functional kinetochore components. (2) The force generated by kinetochores is directed toward the limited number of protein–DNA interactions formed between the kinetochore proteins and the chromosome. Some tension may be relieved as the DNA wrapped around adjacent nucleosomes is pulled. This first results in the straightening out of the compact “beads on a string” structure, but with sufficient pulling force, the nucleosomes would be removed from the DNA. (3) Protein–protein interfaces held together by noncovalent bonds can break under pulling force, but the presence of additional proteins to strengthen interactions could prevent the loss of important interfaces.
At kinetochores, rupture events caused by force-dependent protein unfolding or the loss of protein–protein interfaces are likely avoided at least in part through the architecture and organization of the kinetochore. Previous theoretical work on the effects of force on protein structure and protein–protein interactions has highlighted organization and arrangement as key features for facilitating force resistance (Leckband, 2000; Evans, 2001). In a “series” arrangement, bonds are organized linearly such that the full force is felt by each component. However, in a “parallel” arrangement, the force is divided over multiple attachments arranged in parallel so that the force felt by each attachment is greatly reduced. The higher order organization of the kinetochore could diffuse the microtubule-generated force over multiple attachments, significantly decreasing the force that is felt by an individual kinetochore protein molecule.
Although the kinetochore clearly has evolved mechanisms to accommodate potentially large cellular forces, our understanding of the architecture and organization of a kinetochore remains limited. At the level of the minimal molecular path between a microtubule and centromeric DNA, the proteins involved appear to be connected linearly (Fig. 1; Gascoigne and Cheeseman, 2011; Gascoigne et al., 2011; Bock et al., 2012; Schleiffer et al., 2012). However, there are multiple connections formed between the centromere and a single microtubule. For example, as many as 10–20 kinetochore-localized Ndc80 complexes have been measured as associating with each microtubule in both fungi and vertebrate cells (Joglekar et al., 2006, 2008; Johnston et al., 2010; Lawrimore et al., 2011), supporting a parallel model. The complexities of these connections have proven a hurdle to devising methods to measure the force produced by microtubules on specific kinetochore components or the total force exerted on the kinetochore during normal mitotic processes.
In addition to defining the forces that kinetochore proteins experience, the amount of force necessary to break a bond depends on both the loading rate (force/time) and the duration of the applied force (Merkel et al., 1999). For the kinetochore, the extended periods of force experienced during metaphase (in which sister chromatids move under force in one direction for 1–2 min; Mitchison and Salmon, 1992), as well as the rapid changes in force that occur during sister chromatid oscillations, have the potential to result in a high loading rate and extended durations of applied force. As such, it will be important to account for the way that these challenges are accommodated at kinetochores. Several calculations have estimated the power output of the grasshopper and yeast spindles (Nicklas, 1988; Bloom, 2008) and provided indirect measures for the spring constant of the kinetochore based on analysis of the chromatin spring constant during anaphase (Fisher et al., 2009). However, as a result of experimental limitations, it has not been possible to precisely determine the force constant and other key force parameters at kinetochores. Without knowledge of the force constant, it is not possible to calculate the loading rate experienced by a kinetochore. Thus, defining these parameters for kinetochores is an important area for future work.
Force at the kinetochore–DNA interface
Force also has the potential to disrupt protein–DNA interactions (Fig. 2). The kinetochore is assembled on centromeric DNA, but if the kinetochore–chromatin interface were disrupted, kinetochore function would be lost. One way in which this force could be accommodated is that the force applied through the kinetochore displaces nucleosomes in pericentric regions, alleviating the mechanical stress experienced by the kinetochore itself (Bouck and Bloom, 2007; Verdaasdonk et al., 2012). Studies of the chromatin force response in S. cerevisiae have shown that a deformation of chromatin structure occurs in the regions immediately surrounding the centromere during mitosis (Pearson et al., 2001; Bouck and Bloom, 2007) and that there is an increased turnover of nucleosomes in these surrounding regions (Verdaasdonk et al., 2012). Directed analyses have measured the force required to displace nucleosomes from DNA. These studies have obtained values of between 4 and 20 pN to irreversibly remove a nucleosome from DNA, depending on the specific approach and source of nucleosomes that was used (Cui and Bustamante, 2000; Bennink et al., 2001; Brower-Toland et al., 2002; Yan et al., 2007). For these studies, force was applied to the ends of the DNA rather than perpendicular to the DNA strand as would occur at kinetochores. This difference in the directionality of force may alter the amount of force necessary to remove a nucleosome from chromatin under mitotically applied forces.
Nucleosome displacement and chromatin stretching in pericentric regions could allow the chromosome to absorb some force. However, nucleosome–DNA interactions must be maintained at the kinetochore–centromere interface. At centromeres, there are two key connections between kinetochore proteins and the underlying DNA (Gascoigne and Cheeseman, 2011; Gascoigne et al., 2011). The first occurs through the histone H3 variant, CENP-A, which epigenetically defines the centromere and forms the main site of attachment for CENP-C (Fig. 1). The other occurs via the recently identified CENP-T–W–S–X histone fold complex, which forms a heterotetrameric nucleosome-like structure (Nishino et al., 2012). Although adjacent nucleosomes surrounding the centromere could be displaced in the presence of force without severe consequences, the loss of the interaction of CENP-A or CENP-T with DNA would eliminate kinetochore function. Both the CENP-A nucleosome and the CENP-T–W–S–X complex are structurally distinct from canonical nucleosomes (Sekulic et al., 2010; Nishino et al., 2012), raising the possibility that they may have different force resistance properties. Future work characterizing the behavior of these specialized nucleosomes and the other kinetochore components will be important to understand how intrakinetochore– and kinetochore–DNA attachments are maintained in the presence of force.
How is force accommodated at kinetochores?
Although the roles of force at kinetochores have been a focus of recent work, less is known about how kinetochores are able to accommodate the forces generated at these sites. Recent work has isolated kinetochore particles from budding yeast (Akiyoshi et al., 2009) and partially reconstituted kinetochores from Xenopus laevis extract on defined templates (Guse et al., 2011). Although it is not clear how accurately these assemblies represent functional kinetochores, the reconstitution of kinetochore-like structures in vitro should allow for the analysis of its force resistance properties. At present, it remains unclear which proteins at kinetochores contribute to force resistance and how kinetochores are organized to achieve this. Current data suggest that there are two separate connections between centromeric DNA and microtubules. The first path involves an attachment of CENP-A to CENP-C followed by the Mis12 complex, which contacts KNL1 and the Ndc80 complex, with Ndc80 completing the connection to the microtubule (Fig. 1). The second connection is anchored to centromeric DNA by the CENP-T–W–S–X complex, which makes its own direct connection to the Ndc80 complex. The available biochemical data suggest that these two connections in their most minimal forms are constructed linearly and that there are two separate pools of Ndc80 that make connections to the microtubules from the Mis12 complex and CENP-T (Bock et al., 2012; Schleiffer et al., 2012; Nishino et al., 2013; Malvezzi et al., 2013). This suggests that some parts of the kinetochore might be held together by only a single protein–protein interface. However, it is possible that there are interactions between these pathways, either directly or via other protein components (Fig. 1; Gascoigne et al., 2011). If the individual protein–protein interactions within each pathway cannot withstand the force produced by the depolymerizing microtubule, the current architectural models of the kinetochore may be incomplete.
Based on the currently available structural details for the kinetochore, several different models could explain how kinetochores withstand cellular forces (Fig. 2). First, kinetochore proteins may have evolved special properties that allow them to withstand force. It is possible that a subset of kinetochore proteins have elastic properties, such as those suggested by the elongation of CENP-T (Suzuki et al., 2011). Elasticity of a protein within a series arrangement would allow it to absorb some energy, thereby decreasing the force passed through the subsequent protein–protein interfaces for at least some time, much in the same way that nucleosome displacement in pericentric chromatin could diffuse the force generated at kinetochores (Verdaasdonk et al., 2012). In this model, energy is absorbed by breaking or rearranging bonds within kinetochore proteins rather than between proteins, thereby protecting the key interfaces within the kinetochore. Second, the connections between the microtubule and centromere are likely to be arranged in a parallel manner such that they sum to a strong interface. The multiple copies of each core kinetochore protein that are present per microtubule (Joglekar et al., 2006, 2008; Johnston et al., 2010; Lawrimore et al., 2011) support at least a partial contribution from this parallel model. Third, there may be additional kinetochore proteins that are not part of the linear connectivity between the centromere and microtubule but that strengthen connections between kinetochore components that would otherwise be too weak. For example, the Tetrahymena thermophila cilia protein Bld10 has recently been proposed to structurally stabilize the basal body under the force generated during cilia beating (Bayless et al., 2012). At kinetochores, proteins could serve a similar role either by serving as dynamic cross-linkers, connecting separate linear pathways, or by reinforcing existing connections by adding contacts between proteins. It is likely that the actual force resistance properties of the kinetochore complex require a combination of all three models.
Work spanning the last 60 years has shown that the mitotic spindle can generate force that acts on kinetochores. The work we have summarized here provides a preliminary foundation for understanding the consequences of force at kinetochores, but the proposed models will change as more is discovered about kinetochore structure and organization. Defining the force resistance properties of the kinetochore will provide a better understanding of how it is able to function in the presence of force and the mechanisms by which it acts during chromosome segregation. As we look toward the future prospects of the field, the advances in the biophysical understanding of focal adhesions (Roca-Cusachs et al., 2012) provide an excellent blueprint for generating a detailed molecular picture of a large protein complex that functions under force. For focal adhesions, researchers have defined the pathway between the extracellular matrix and the cytoskeleton, analyzed the force response of each component along this pathway, and defined how cells use mechanosensors to signal to the cell. Achieving a similar understanding for the kinetochore will provide key insights into the function of this central cell division structure.
Acknowledgments
We apologize to the many people whose work we were unable to cite as a result of space constraints. We thank Marie Aubin-Tam, William Hesse, Jeff Gore, and the members of the Cheeseman laboratory for helpful discussions and critical reading of the manuscript.
This work was supported by an award to I.M. Cheeseman from the Leukemia & Lymphoma Society and a grant from the National Institutes of Health/National Institute of General Medical Sciences (GM088313). F. Rago is supported by a National Science Foundation graduate research fellowship.
References
- Akiyoshi B., Nelson C.R., Ranish J.A., Biggins S. 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23:2887–2899 10.1101/gad.1865909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Chang F. 2011. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21:604–614 10.1016/j.tcb.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader J.R., Vaughan K.T. 2010. Dynein at the kinetochore: Timing, interactions and functions. Semin. Cell Dev. Biol. 21:269–275 10.1016/j.semcdb.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B.A., Giddings T.H., Jr, Winey M., Pearson C.G. 2012. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell. 23:4820–4832 10.1091/mbc.E12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennink M.L., Leuba S.H., Leno G.H., Zlatanova J., de Grooth B.G., Greve J. 2001. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 8:606–610 10.1038/89646 [DOI] [PubMed] [Google Scholar]
- Bloom K.S. 2008. Beyond the code: the mechanical properties of DNA as they relate to mitosis. Chromosoma. 117:103–110 10.1007/s00412-007-0138-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock L.J., Pagliuca C., Kobayashi N., Grove R.A., Oku Y., Shrestha K., Alfieri C., Golfieri C., Oldani A., Dal Maschio M., et al. 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 14:614–624 10.1038/ncb2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D.C., Bloom K. 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17:741–748 10.1016/j.cub.2007.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B.D., Smith C.L., Yeh R.C., Lis J.T., Peterson C.L., Wang M.D. 2002. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 99:1960–1965 10.1073/pnas.022638399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Chen W., Zhang D. 2004. Kinetochore fibre dynamics outside the context of the spindle during anaphase. Nat. Cell Biol. 6:227–231 [DOI] [PubMed] [Google Scholar]
- Cottingham F.R., Gheber L., Miller D.L., Hoyt M.A. 1999. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147:335–350 10.1083/jcb.147.2.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M., Lombillo V.A., McIntosh J.R. 1991. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 112:1165–1175 10.1083/jcb.112.6.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Bustamante C. 2000. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 97:127–132 10.1073/pnas.97.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- Dumont S., Salmon E.D., Mitchison T.J. 2012. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 337:355–358 10.1126/science.1221886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. 2001. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128 10.1146/annurev.biophys.30.1.105 [DOI] [PubMed] [Google Scholar]
- Finan K., Cook P.R., Marenduzzo D. 2011. Non-specific (entropic) forces as major determinants of the structure of mammalian chromosomes. Chromosome Res. 19:53–61 10.1007/s10577-010-9150-y [DOI] [PubMed] [Google Scholar]
- Fisher J.K., Ballenger M., O’Brien E.T., Haase J., Superfine R., Bloom K. 2009. DNA relaxation dynamics as a probe for the intracellular environment. Proc. Natl. Acad. Sci. USA. 106:9250–9255 10.1073/pnas.0812723106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A.D., Powers A.F., Gestaut D.R., Gonen T., Davis T.N., Asbury C.L. 2007. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 9:832–837 10.1038/ncb1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Cheeseman I.M. 2011. Kinetochore assembly: if you build it, they will come. Curr. Opin. Cell Biol. 23:102–108 10.1016/j.ceb.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A., Carter A.P., Reck-Peterson S.L., Vale R.D. 2007. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 131:952–965 10.1016/j.cell.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., Cimini D. 2011. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 21:374–381 10.1016/j.tcb.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., McIntosh J.R. 2006. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25:4888–4896 10.1038/sj.emboj.7601353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Molodtsov M.I., Ataullakhanov F.I., McIntosh J.R. 2005. Force production by disassembling microtubules. Nature. 438:384–388 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., Straight A.F. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T.L. 1985. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA. 82:4404–4408 10.1073/pnas.82.13.4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A.J., McIntosh J.R. 1998. The dynamic behavior of individual microtubules associated with chromosomes in vitro. Mol. Biol. Cell. 9:2857–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoué S., Salmon E.D. 1995. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 6:1619–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D.C., Molk J.N., Bloom K.S., Salmon E.D. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 10.1083/jcb.200803027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K., Joglekar A., Hori T., Suzuki A., Fukagawa T., Salmon E.D. 2010. Vertebrate kinetochore protein architecture: protein copy number. J. Cell Biol. 189:937–943 10.1083/jcb.200912022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S., Wright A. 2010. Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol. 8:600–607 10.1038/nrmicro2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D.E., Mitchison T.J., Kirschner M.W. 1988. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 331:499–504 10.1038/331499a0 [DOI] [PubMed] [Google Scholar]
- Kumar S., Li M.S. 2010. Biomolecules under mechanical force. Phys. Rep. 486:1–74 10.1016/j.physrep.2009.11.001 [DOI] [Google Scholar]
- LaFountain J.R., Jr, Oldenbourg R., Cole R.W., Rieder C.L. 2001. Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell. 12:4054–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain J.R., Jr, Cohan C.S., Siegel A.J., LaFountain D.J. 2004. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol. Biol. Cell. 15:5724–5732 10.1091/mbc.E04-08-0750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S. 2010. The Dam1 complex confers microtubule plus end–tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K.S., Salmon E.D. 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195:573–582 10.1083/jcb.201106036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband D. 2000. Measuring the forces that control protein interactions. Annu. Rev. Biophys. Biomol. Struct. 29:1–26 10.1146/annurev.biophys.29.1.1 [DOI] [PubMed] [Google Scholar]
- Li X., Nicklas R.B. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632 10.1038/373630a0 [DOI] [PubMed] [Google Scholar]
- Lin S., Chen J.-L., Huang L.-S., Lin H.-W. 2005. Measurements of the forces in protein interactions with atomic force microscopy. Current Proteomics. 2:55–81 10.2174/1570164053507754 [DOI] [Google Scholar]
- Liu D., Vader G., Vromans M.J.M., Lampson M.A., Lens S.M.A. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P., Straight A., Coughlin P., Mitchison T.J., Salmon E.D. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382 10.1083/jcb.200301088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T., Westermann S. 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32:409–423 10.1038/emboj.2012.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D. 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184:373–381 10.1083/jcb.200808130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos I., Maiato H. 2011. Prevention and correction mechanisms behind anaphase synchrony: implications for the genesis of aneuploidy. Cytogenet. Genome Res. 133:243–253 10.1159/000323803 [DOI] [PubMed] [Google Scholar]
- Mazumdar M., Misteli T. 2005. Chromokinesins: multitalented players in mitosis. Trends Cell Biol. 15:349–355 10.1016/j.tcb.2005.05.006 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., Ataullakhanov F.I. 2008. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53 10.1038/16219 [DOI] [PubMed] [Google Scholar]
- Mitchison T.J., Salmon E.D. 1992. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119:569–582 10.1083/jcb.119.3.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov M.I., Ermakova E.A., Shnol E.E., Grishchuk E.L., McIntosh J.R., Ataullakhanov F.I. 2005. A molecular-mechanical model of the microtubule. Biophys. J. 88:3167–3179 10.1529/biophysj.104.051789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. 1965. Chromosome velocity during mitosis as a function of chromosome size and position. J. Cell Biol. 25:119–135 10.1083/jcb.25.1.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. 1983. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 97:542–548 10.1083/jcb.97.2.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. 1988. The forces that move chromosomes in mitosis. Annu. Rev. Biophys. Biophys. Chem. 17:431–449 10.1146/annurev.bb.17.060188.002243 [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Ward S.C., Gorbsky G.J. 1995. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 130:929–939 10.1083/jcb.130.4.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., Fukagawa T. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., Fukagawa T. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. 2000. Structural insights into microtubule function. Annu. Rev. Biochem. 69:277–302 10.1146/annurev.biochem.69.1.277 [DOI] [PubMed] [Google Scholar]
- Nogales E., Wang H.-W. 2006. Structural mechanisms underlying nucleotide-dependent self-assembly of tubulin and its relatives. Curr. Opin. Struct. Biol. 16:221–229 10.1016/j.sbi.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Pearson C.G., Maddox P.S., Salmon E.D., Bloom K. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266 10.1083/jcb.152.6.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., Asbury C.L. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Iskratsch T., Sheetz M.P. 2012. Finding the weakest link: exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 125:3025–3038 10.1242/jcs.095794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I.X., Yates J.R., III, Oegema K., Hyman A., Desai A. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 127:1179–1191 10.1016/j.cell.2006.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., Westermann S. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N., Bassett E.A., Rogers D.J., Black B.E. 2010. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 467:347–351 10.1038/nature09323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Rogers G.C., Scholey J.M. 2000. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2:922–930 10.1038/35046574 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Hori T., Nishino T., Usukura J., Miyagi A., Morikawa K., Fukagawa T. 2011. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J. Cell Biol. 193:125–140 10.1083/jcb.201012050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U. 2002. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 14:365–371 10.1016/S0955-0674(02)00328-9 [DOI] [PubMed] [Google Scholar]
- Taylor E.W. 1965. Brownian and saltatory movements of cytoplasmic granules and the movement of anaphase chromosomes. In Proceedings of the fourth international congress on Rheology. Part 4-Symposium on Biorheology. Copley A.L., editor Interscience, New York: 175–191 [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., Davis T.N. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K.S.K., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 184:383–390 10.1083/jcb.200811028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit N.T., Gestaut D.R., Tien J.F., Vollmar B.S., Gonen T., Asbury C.L., Davis T.N. 2012. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA. 109:16113–16118 10.1073/pnas.1209615109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk J.S., Gardner R., Stephens A.D., Yeh E., Bloom K. 2012. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol. Biol. Cell. 23:2560–2570 10.1091/mbc.E11-07-0651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher K., Schnitzer M.J., Block S.M. 1999. Single kinesin molecules studied with a molecular force clamp. Nature. 400:184–189 10.1038/22146 [DOI] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.-T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., Cimini D., Cameron L.A., Salmon E.D. 2012. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol. Biol. Cell. 23:1035–1046 10.1091/mbc.E11-09-0767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel J.W., Shuman H., Litvinov R.I. 2003. Protein-protein unbinding induced by force: single-molecule studies. Curr. Opin. Struct. Biol. 13:227–235 10.1016/S0959-440X(03)00039-3 [DOI] [PubMed] [Google Scholar]
- Welburn J.P.I., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R., III, Cheeseman I.M. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P.I., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Maresca T.J., Skoko D., Adams C.D., Xiao B., Christensen M.O., Heald R., Marko J.F. 2007. Micromanipulation studies of chromatin fibers in Xenopus egg extracts reveal ATP-dependent chromatin assembly dynamics. Mol. Biol. Cell. 18:464–474 10.1091/mbc.E06-09-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., Rieder C.L. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 17:973–980 10.1016/j.cub.2007.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]