Human ribosomal RNA processing is initiated by endonuclease cleavage and followed by 3′ to 5′ exonucleolytic processing by RRP6 and the exosome.
Abstract
Human ribosome production is up-regulated during tumorogenesis and is defective in many genetic diseases (ribosomopathies). We have undertaken a detailed analysis of human precursor ribosomal RNA (pre-rRNA) processing because surprisingly little is known about this important pathway. Processing in internal transcribed spacer 1 (ITS1) is a key step that separates the rRNA components of the large and small ribosomal subunits. We report that this was initiated by endonuclease cleavage, which required large subunit biogenesis factors. This was followed by 3′ to 5′ exonucleolytic processing by RRP6 and the exosome, an enzyme complex not previously linked to ITS1 removal. In contrast, RNA interference–mediated knockdown of the endoribonuclease MRP did not result in a clear defect in ITS1 processing. Despite the apparently high evolutionary conservation of the pre-rRNA processing pathway and ribosome synthesis factors, each of these features of human ITS1 processing is distinct from those in budding yeast. These results also provide significant insight into the links between ribosomopathies and ribosome production in human cells.
Introduction
The 18S, 5.8S, and 28S (25S in yeast) ribosomal RNAs (rRNAs) are transcribed as a single precursor by RNA polymerase I in the eukaryotic nucleolus. This precursor rRNA (pre-rRNA) undergoes processing and assembly steps to produce the large (LSU) and small (SSH) ribosomal subunits (Henras et al., 2008). Ribosome biogenesis requires more than 200 trans-acting factors including nucleases, helicases, small nucleolar RNPs, and 80 ribosomal proteins (Henras et al., 2008). This process consumes a large percentage of cellular energy and the rate of ribosome biogenesis determines the proliferative potential of the cell (Warner, 1999). Ribosome production is regulated in a cell cycle–dependent manner being down-regulated during differentiation and up-regulated in cancer. Ribosome biogenesis is down-regulated by tumor suppressor p19ARF and plays a direct role in activating the tumor suppressor p53 (Lessard et al., 2010; Chakraborty et al., 2011). Furthermore, several genetic diseases, including cartilage-hair hypoplasia (CHH), alopecia, neurological defects, and endocrinopathy syndrome, and Diamond Blackfan anemia, are linked to defects in ribosome production (Nousbeck et al., 2008; Narla and Ebert, 2010).
The vast majority of studies of ribosome biogenesis have been performed in the yeast Saccharomyces cerevisiae. The transcribed spacers are removed by a series of endonucleolytic and exonucleolytic processing steps. Key to pre-rRNA processing are the cleavages at sites A2 and A3 in internal transcribed spacer 1 (ITS1), which separate the RNAs for the LSU (28S and 5.8S) and SSU (18S). Cleavage at site A2, reportedly mediated by Rcl1 (Horn et al., 2011), is linked to the removal of the 5′ external transcribed spacer (ETS) and these processing steps are performed by the SSU processome complex (Phipps et al., 2011). One protein in this complex, Rrp5, is also required for A3 cleavage (Venema and Tollervey, 1996). After A2 cleavage, the remaining 5′ region of ITS1 is removed by Nob1 cleavage at site D at the 3′ end of 18S rRNA (Pertschy et al., 2009; Lamanna and Karbstein, 2011). Cleavage at A3 is mediated by the RNA–protein complex RNase MRP (Lygerou et al., 1996) and also requires Nop4 (RBM28 in humans; Bergès et al., 1994; Sun and Woolford, 1994). The 5′ end of 5.8S is generated by the 5′ to 3′ exonucleases Xrn2/Rat1 and Rrp17 (NOL12 in humans; Petfalski et al., 1998; Oeffinger et al., 2009) and also requires seven proteins, termed the A3 cluster, which includes Erb1 (BOP1 in humans; Granneman et al., 2011; Sahasranaman et al., 2011).
Much less is known about pre-rRNA processing in mammalian cells than in yeast. Indeed, many of the human pre-rRNA cleavage sites have not been clearly mapped and only one enzymatic activity has been documented (Mullineux and Lafontaine, 2012). Although many of the essential ribosome biogenesis factors characterized to date in yeast can be found in humans little has been done to determine whether these proteins perform the same functions in humans. Mammalian rRNA processing is proposed to involve alternative processing pathways, an additional cleavage site in the 5′ETS (A’; Mullineux and Lafontaine, 2012), and an additional processing step (21S to 18SE) in the removal of ITS1 (Rouquette et al., 2005; Fig. 1, A and B). Both endonucleolytic and exonucleolytic processing pathways have been postulated for 18SE production but no clear data exists for either pathway (Carron et al., 2011; Wang and Pestov, 2011). Although A2 is the primary cleavage that separates the rRNAs of the SSU and LSU in yeast, it would appear that site 2 cleavage is the initial cleavage in the mammalian ITS1 (Mullineux and Lafontaine, 2012). It has, however, been speculated that the endonucleolytic cleavage at site 2a in mammals, which generates 18SE, is equivalent to the A2 cleavage in yeast (Carron et al., 2011; Wang and Pestov, 2011). It is also unclear whether site 2 is equivalent to the A3 site, which in yeast is cleaved by RNase MRP (Lygerou et al., 1996). Processing of mammalian 5.8S rRNA, which involves the removal of ITS1 sequences from the 5′ end, is influenced by knockdown of Dicer or Drosha, the endonucleases linked to miRNA production, although neither protein appeared important for the primary ITS1 cleavages (Liang and Crooke, 2011).
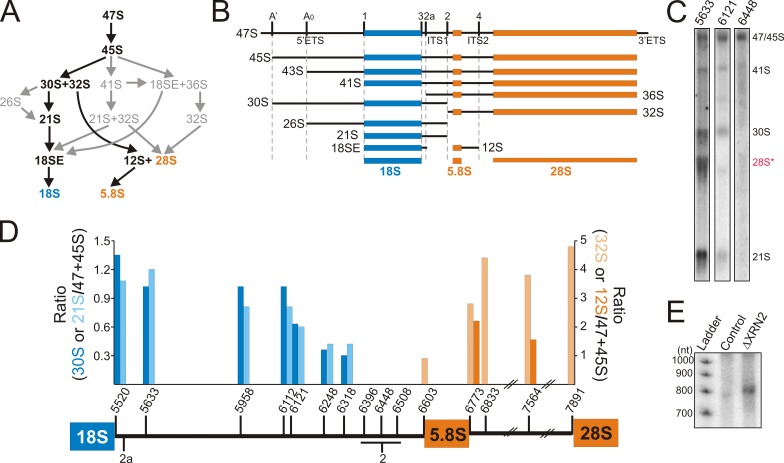
Figure 1.
Mapping the cleavages in human ITS1. (A) Alternative processing pathways of human rRNA, with the major pathway indicated in black. (B) Schematic representations of pre-rRNA intermediates, with the processing/cleavage sites, ETS and ITS, indicated. (C) HeLa cell RNA was separated by glyoxal/agarose gel electrophoresis, transferred to nylon membrane, and hybridized with a variety of ITS1/2 probes (indicated at the top of each lane). The RNAs detected are indicated on the right. Panels are reproduced in Fig. S1 A. (D) Ratios of the SSU (30S [dark blue] and 21S [light blue]) or LSU (32S [light orange] and 12S [dark orange]) precursors to the 45S/47S transcripts were calculated and plotted for each probe (x axis). A schematic representation of the region from the 3′ end of 18S rRNA to the 5′ end of 28S is shown. One set of data, which is representative of at least two experiments, is shown. (E) RNA was extracted from control HeLa cells and cells depleted of XRN2 by RNAi (indicated above each lane). Equal amounts of RNA were separated on an 8% acrylamide/7 M urea gel and analyzed by Northern blotting using the ITS1 6121 probe. “Ladder” indicates a 32P-labeled 100-bp ladder.
Human ribosome production is coupled to cellular growth rate and linked to both cancer and genetic disease (Narla and Ebert, 2010). Despite this, several fundamental questions remain about the early pre-rRNA processing steps in humans and, in particular, the processing pathway and the enzymes involved in ITS1 processing need to be determined. To address these points, we have systematically analyzed factors linked to ITS1 processing in human cells.
Results
The cleavage sites in human ITS1
The potential additional step in the 3′ processing of the 18S rRNA raises several important questions about human ITS1 processing (Rouquette et al., 2005). In particular, it was suggested that 18SE can be generated by either endonucleolytic or exonucleolytic processing of 21S (Carron et al., 2011). Northern blotting was therefore used to recharacterize the pre-rRNA intermediates containing ITS1 sequences and better define the cleavage sites. HeLa cell RNA was separated by glyoxal gel electrophoresis, transferred to nylon membrane, and then hybridized with a variety of probes recognizing ITS1, 5.8S, and ITS2. For each probe, the ratio of the smaller RNAs (30S, 32S, 21S, and 12S) to 45/47S was calculated and plotted.
All of the probes recognized the 47S/45S pre-rRNAs (Fig. 1, C and D; and Fig. S1 A). Interestingly, three ITS1 probes (6396, 6448, and 6508) failed to detect the smaller pre-rRNAs (32S, 30S, 21S, and 12S), suggesting this region is missing from both the LSU and SSU precursors. The 30S and 21S precursors of 18S rRNA were detected by probes hybridizing between 5520 and 6318, although weaker signals were reproducibly seen using probes between nucleotides 6121 and 6318. The 3′ ends of 30S and 21S pre-rRNAs are therefore very heterogeneous. The 32S and 12S pre-rRNAs were consistently detected using 5.8S and ITS2 probes. In contrast, probe 6603, at the 3′ end of ITS1, only weakly detected 32S and did not recognize 12S (Figs. 1 D and S1 A). We conclude that the majority of 32S and all 12S pre-rRNAs lack ITS1 sequence, with site 2 cleavage occurring in the region between nucleotides 6396 and 6508.
We next investigated whether fragments of ITS1 were generated by serial endonucleolytic cleavages. Yeast ITS and ETS fragments released by endonuclease cleavage are degraded by either the exosome or the 5′ to 3′ exonuclease Xrn2 (Rat1; Petfalski et al., 1998). XRN2 and the exosome component RRP6 were therefore depleted from HeLa cells using RNAi (Fig. S2 A) and the accumulation of ITS1 fragments analyzed by Northern blotting. Depletion of XRN2 resulted in the accumulation of an ∼800-nt fragment of ITS1 that extended from site 2a to position 6318 (Fig. 1 E). This fragment was barely detectable in control cells and no other fragments were detected after depletion of XRN2 and/or RRP6 (unpublished data). This indicated that there are two early endonucleolytic cleavages in ITS1 (at sites 2 and 2a), which together release the ∼800-nt fragment.
Only a small fraction of the 32S species extended to site 2, indicating rapid processing to the mature 5′ end of 5.8S. In yeast, cleavage upstream of 5.8S is very rapidly followed by 5′ to 3′ exonucleolytic digestion. Knockdown of XRN2 resulted in an increase of 32S containing the 3′ end of ITS1, indicating that this is also the case in humans (Fig. S1 B). The heterogeneity observed in the 3′ ends of 30S and 21S suggested that these precursors might be processed by 3′ to 5′ exonucleases acting from site 2, in addition to endonucleolytic cleavage at 2a.
RRP6 is required for 18S rRNA accumulation in human cells
The accumulation of the ∼800-nt ITS1 fragment clearly indicates that the 18SE precursor can be generated by endonucleolytic cleavage at 2a. However, previous analyses after depletion of the small subunit biogenesis factor ENP1 indicated that 18SE can be produced by both endonucleolytic and exonucleolytic processing pathways (Carron et al., 2011). The 3′ to 5′ exonucleolytic activity of the exosome is a likely candidate for carrying out this step. We therefore depleted ENP1, XRN2, or the exosome component RRP6 from HeLa cells using RNAi (Fig. S2 A), and then analyzed the levels of rRNA processing intermediates by Northern blotting (Fig. 2 A).
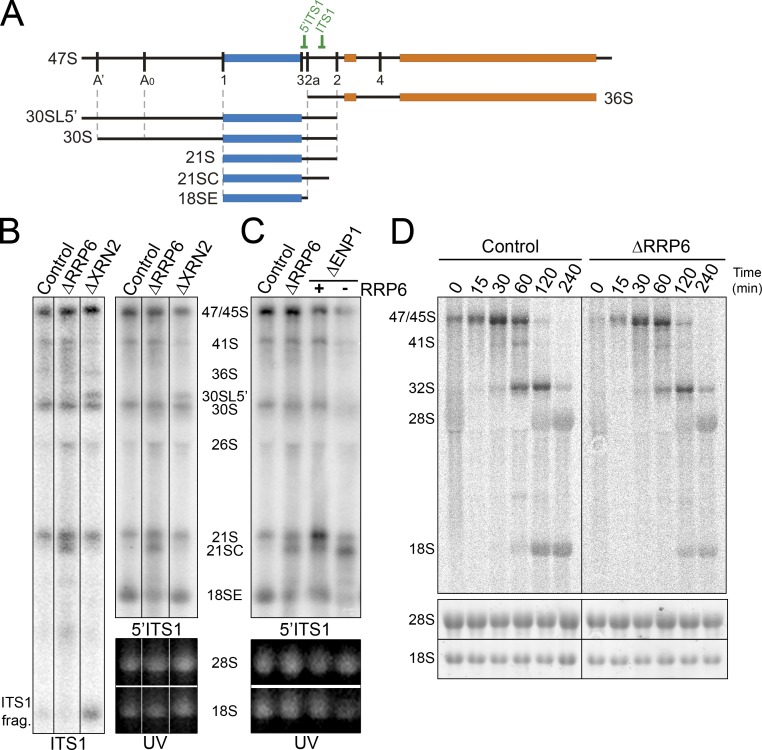
Figure 2.
RRP6 is required for ITS1 processing. (A) Schematic representation of the 47S pre-rRNA intermediate and relevant pre-rRNAs. The relative positions of Northern blot probes are indicated. (B and C) RNA was extracted from HeLa cells depleted of RRP6, XRN2, or ENP1 by RNAi or control cells and analyzed by ethidium bromide staining (UV) and Northern hybridization using probes specific to the 5′ end and middle of ITS1 as indicated. Positions of the various intermediates detected are indicated. (D) HeLa cells depleted of RRP6, or control cells, were pulse labeled with 3H methyl-methionine followed by incubation in normal media for varying lengths of time (indicated above each lane). RNA was recovered from the cells, separated using glyoxal/agarose gel electrophoresis, and visualized using a phosphorimager and ethidium bromide staining (UV). The control data are repeated in Fig. 6 E. Black lines in B indicate intervening lanes that have been removed.
As in mouse cells, depletion of XRN2 in HeLa cells caused the accumulation of 47S and 30SL5′, a 30S precursor with an additional 5′ fragment of the 5′ETS detected if the A’ cleavage is affected (Fig. 2 A), relative to the control cells (Fig. 2 B; Wang and Pestov, 2011). In addition, a significant increase in 26S was seen in some, but not all, experiments, implying that the order of cleavages in the 5′ETS is altered by depleting XRN2. Reproducible accumulation of 36S pre-rRNA was observed relative to control cells (Fig. 2 B, ITS1 probe) as previously reported in mouse cells (Wang and Pestov, 2011), indicating increased initial cleavage of ITS1 at site 2a before processing at site 2. Notably, depletion of XRN2 changed the levels of several pre-rRNA intermediates without significantly slowing down rRNA processing, suggesting that only the balance between competing pathways is altered. Depletion of ENP1 resulted in increased levels of 21S pre-rRNA, the appearance of 21SC, a 3′ shortened form of 21S (Carron et al., 2011), and significantly decreased 18SE levels (Fig. 2 C). Depletion of RRP6 did not alter the accumulation of the longer precursors but resulted in a more significant decrease in 18SE levels and the accumulation of 21SC (Fig. 2, B and C). In addition, a range of products, seen as a smear, was often observed between 21S and 18SE, indicating inefficient exonucleolytic processing of 21S pre-rRNA. Codepletion of RRP6 and ENP1 resulted in stronger accumulation of 21SC and more significant reduction in 21S and 18SE than either single knockdown (Fig. 2 C). This indicates that RRP6 functions together with certain rRNA processing factors in the processing of ITS1.
We next used metabolic labeling experiments to determine the effect of depleting RRP6 on the accumulation of mature rRNAs. HeLa cells were transfected with control siRNAs or siRNAs targeting the RRP6 mRNA. The cells were metabolically labeled using 3H methyl-methionine (Fig. 2 D) or 32P orthophosphate (Fig. S3, A and B) and harvested at various time points after the addition of unlabeled media. Total RNA was separated by glyoxal gel electrophoresis, transferred to nylon membrane, visualized using a phosphorimager, and quantified.
In control cells, 47S/45S pre-rRNAs were observed at the start of the chase (Fig. 2 D). With time, the 32S intermediate accumulated and this was followed by the mature 18S and 28S rRNAs. Because of their low relative abundances, the 41S, 30S, and 21S pre-rRNAs were barely visible and difficult to follow in these experiments. In both control and knockdown cells, 32S pre-rRNA and 28S rRNA accumulated at a similar rate and to similar levels. In contrast, significantly less 18S rRNA was produced during the time course in RRP6-depleted cells (approximately two- to threefold less at 3 h relative to 28S levels). We conclude that RRP6 is required for the efficient production of the 18S rRNA in human cells and that the majority of the SSU rRNA is produced via this pathway.
The exonuclease activity of RRP6 is required for 18SE production
To determine whether the exonucleolytic activity of RRP6 was required for 18SE production, stably transfected HEK293 cells were generated expressing either FLAG-tagged RRP6 or an exonuclease mutant, RRP6exo (Fig. S3, C and D). The catalytic site mutation (D313A) abolished exonuclease activity of recombinant RRP6exo in vitro (Fig. S3 E). The ORFs of wild-type and mutant RRP6 were altered to make the mRNAs resistant to RNAi-mediated depletion of RRP6 without changing the coding potential (Fig. S3 F). The cells were transfected with siRNAs targeting endogenous RRP6 and the expression of RRP6 was analyzed by Western blotting (Fig. 3 A). Control cells, expressing just the FLAG tag and transfected with control siRNAs, were also analyzed. After siRNA treatment the endogenous protein was no longer detectable, whereas the tagged RRP6 and RRP6exo proteins, which because of the presence of the tag run slower in the SDS gel, were not affected.
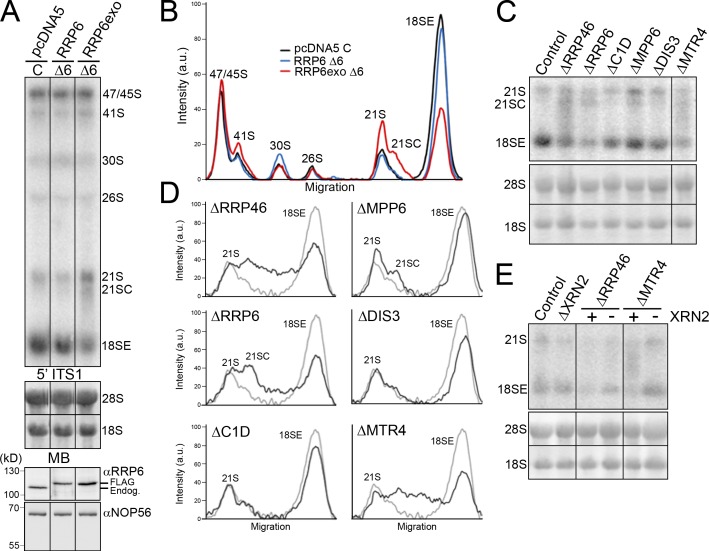
Figure 3.
The exonuclease activity of RRP6, the exosome core, and the RNA helicase MTR4 are required for 18SE production. (A) HEK293 cells stably transfected with plasmids expressing the FLAG tag alone (pcDNA5), FLAG-RRP6, or FLAG-RRP6exo were transfected with either control siRNAs (C) or siRNAs that target the endogenous RRP6 mRNA (Δ6). The FLAG-RRP6 cDNA sequences had been altered to render the mRNA resistant to the RRP6 siRNAs. Levels of the endogenous and plasmid-expressed RRP6 were monitored by Western blotting (bottom panels). Levels of pre-rRNAs were analyzed by Northern blotting using a probe hybridizing to the 5′ end of ITS1. (B) The levels of the pre-rRNA intermediates from A, which is representative of two experiments, are plotted. The identity of each peak is indicated. (C) RNA extracted from HeLa cells depleted of exosome or exosome-cofactor proteins (as indicated above each lane) by RNAi was analyzed by Northern blotting using a probe specific for the 5′ end of ITS1. (D) The levels of the pre-rRNA intermediates from C, which is representative of more than three experiments, are plotted for each knockdown (black) and the control (gray). The identity of each peak is indicated. (E) RNA extracted from control HeLa cells or HeLa cells depleted of RRP46 or MTR4, alone or together with XRN2 (indicated at top), was analyzed by Northern blotting using a probe against the 5′ end of ITS1. Positions of the mature and pre-rRNAs are indicated on the left. RNA loading was monitored by methylene blue staining of the 18S and 28S rRNAs (MB). In codepletion experiments, the presence (+) or depletion (−) of XRN2 is indicated. Black lines in A, C, and E indicate intervening lanes that have been removed.
HEK293 cells contain higher levels of 47S/45S and 18SE than the other precursors relative to HeLa cells, but pre-rRNA levels were unaltered by replacement of endogenous RRP6 by FLAG-RRP6 (Fig. 3, A and B). In contrast, 18SE levels were reduced and both 21S and 21SC levels were increased in cells expressing only RRP6exo. This demonstrates that the exonucleolytic activity of RRP6 is required for efficient conversion of 21S to 18SE.
The exosome core and exosome cofactors are required for 18SE production
The exosome is a multi-subunit complex and we assessed whether the nuclease DIS3, an exosome core component (RRP46), or exosome cofactors (MPP6, C1D, or MTR4; Lykke-Andersen et al., 2011) are required for 18SE accumulation. Individual proteins were depleted from HeLa cells using RNAi (Fig. S2 A) and the effects on rRNA processing were analyzed. No significant changes in the levels of the longer, 47S to 30S, pre-rRNAs were seen. Depletion of either RRP46 or the RNA helicase MTR4 resulted in a fourfold reduction in 18SE levels (Fig. 3, C and D). However, instead of the prominent band of 21SC seen when RRP6 was depleted, a broad range of differently sized products between 21S and 18SE, often visible as a smear, were seen upon depletion of RRP46 or MTR4. Depletion of DIS3 or C1D (the human homologue of yeast Rrp47/Lrp1) had no clear effect on 21S, 21SC, and 18SE levels, whereas depletion of MPP6 resulted in a slight accumulation of 21SC but no change in 18SE levels. Each of these proteins were, however, essential for 3′ processing of 5.8S, as previously reported (Fig. S2 B; Schilders et al., 2007; Tomecki et al., 2010). These data suggest that RRP6 acts catalytically in 18SE production, functioning together with the exosome core, the MTR4 helicase, and, to a lesser extent, the exosome cofactor MPP6. The lack of effect seen on DIS3 depletion is not entirely unexpected, as little of the protein is in the nucleolus where this processing step is predicted to occur (Tomecki et al., 2010).
The majority of 18SE appears to be generated by exonucleolytic processing of 21S by RRP6 and the exosome core, whereas endonucleolytic cleavage at site 2a was only observed after the depletion of XRN2. We therefore tested whether stimulation of the endonucleolytic pathway by depletion of XRN2 would make 18SE production independent of the exosome and MTR4. Using RNAi, RRP46 or MTR4 were depleted, alone or together with XRN2, and the effect on 18SE production was monitored by Northern blotting. As described above, depletion of XRN2 had no effect on 21S and 18SE levels, whereas depletion of RRP46 or MTR4 reduced the levels of 18SE more than fourfold (Fig. 3 E). Strikingly, codepletion of XRN2 with either RRP6 or MTR4 resulted in almost normal levels (80–90%) of 18SE.
We conclude that depletion of XRN2 favors endonucleolytic cleavage at site 2a before cleavage at site 2, reducing the requirement for the normally major pathway of exosome/MTR4 processing from site 2 to 2a.
Human endonuclease cleavage site 2a is analogous to yeast A2
Our data indicate that cleavage at site 2 normally separates the human LSU and SSU pre-rRNAs. In yeast, A2 cleavage performs the same function, but this site is putatively suggested to be homologous to the human 2a site (Carron et al., 2011; Wang and Pestov, 2011). Cleavage at A2 is dependent on factors linked to 18S processing, including SSU processome components (Henras et al., 2008). We therefore used RNAi to determine whether SSU processome proteins (fibrillarin, UTP24, RCL1, BMS1, and RRP5) and other 18S rRNA processing factors (NOB1, ENP1, and RPS19) are required for ITS1 processing in HeLa cells (Fig. S2 A). The requirement of factors for 2a endonucleolytic cleavage cannot readily be assessed because 36S pre-rRNA, the only intermediate specific to this pathway, is barely detectable in control cells. However, depletion of XRN2 stimulates processing through this pathway, so each knockdown was performed both individually and in combination with XRN2 to facilitate monitoring of 2a cleavage. Pre-rRNA processing in the knockdown cells was followed by Northern blotting (Fig. 4 A) and metabolic labeling (Fig. S4).
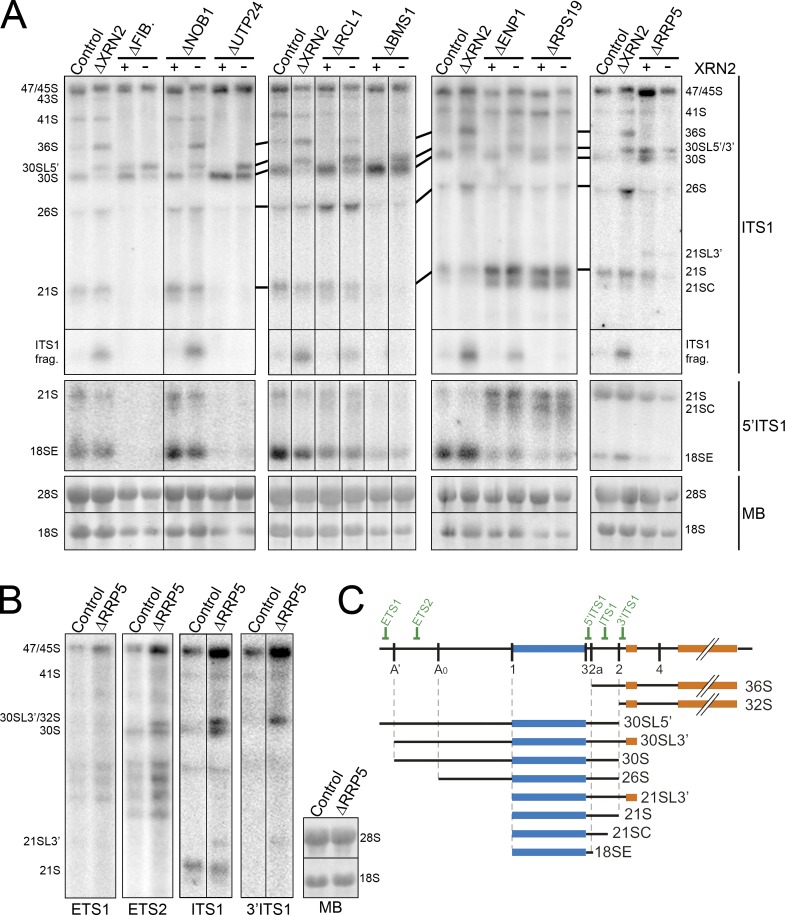
Figure 4.
SSU processome components are required for endonuclease cleavage at site 2a in ITS1. (A) RNA extracted from control HeLa cells or cells depleted of ribosome biogenesis factors either alone or together with XRN2 (indicated at top) was analyzed by Northern blotting using probes that recognized either the middle or 5′ end of ITS1 (indicated on the right). RNA loading was monitored by methylene blue staining of 18S and 28S rRNAs (MB). FIB., fibrillarin. The presence (+) or depletion (–) of XRN2 is indicated at the top of the relevant lanes. (B) RNA from control cells or cells depleted of RRP5 was analyzed by Northern blotting using probes specific for the 5′ETS and ITS1 as indicated. The positions of the mature and pre-rRNAs are indicated. (C) Schematic representation of the full-length pre-rRNA transcript and relevant pre-rRNAs. Positions of Northern probes are indicated. Black lines in A and B indicate intervening lanes that have been removed.
Depletion of fibrillarin, BMS1, or UTP24 resulted in a strong reduction of 41S, 18SE, and 21S and the accumulation of 30S (Fig. 4 A). Fibrillarin depletion also caused accumulation of 30SL5′, the 5′ extended form of 30S generated when A’ cleavage is blocked. Codepleting each of these proteins with XRN2 inhibited 36S production, indicating that they are all required for 2a cleavage. Decreasing the levels of ENP1, RPS19, or RCL1 resulted in reduced 18SE levels. Depletion of ENP1 or RPS19 also brought about 21S and 21SC accumulation, and RPS19 depletion caused slight accumulation of 30SL5′, indicating a mild A’ processing defect. Reducing RCL1 levels resulted in accumulation of 26S and 30S pre-rRNAs. When XRN2 was also depleted, 36S and ITS1 fragment accumulation was abolished in cells depleted for RPS19, but was merely reduced in cells where RCL1 or ENP1 had also been depleted. In yeast, Nob1 is the 18S rRNA 3′ endonuclease (Pertschy et al., 2009; Lamanna and Karbstein, 2011). Depletion of NOB1 resulted in 18SE accumulation (Fig. 4 A) but no other processing defects, consistent with a specific, conserved role in cleavage of site 3 at the 3′ end of 18S. In agreement with this, decreased levels of NOB1, fibrillarin, BMS1, UTP24, ENP1, RPS19, or RCL1 resulted in a reduction or complete loss of 18S rRNA production in metabolic labeling experiments (Fig. S4 A). Depletion of the box C/D small nucleolar RNP protein fibrillarin also reduced 28S rRNA production, likely reflecting U8 involvement in ITS2 processing (Peculis and Steitz, 1993).
In yeast, Rrp5 is important for both A2 and A3 cleavages (Venema and Tollervey, 1996). In HeLa cells, depletion of RRP5 led to a major increase in 45S and a reduction in 30S and 18SE levels (Fig. 4 A). Decreasing RRP5 levels had a greater impact on 18SE levels than on 21S, suggesting that it is also important for exonucleolytic processing to site 2a. Metabolic labeling experiments confirmed that 45S pre-rRNA accumulated in cells depleted of RRP5, whereas maturation to both 18S and 28S rRNAs was severely inhibited (Fig. S4). In Northern blots, novel bands above both 30S and 21S were also seen, which we termed 30SL3′ and 21SL3′, respectively. The 30SL3′ intermediate, which unfortunately runs at the same position as 30SL5′ and LSU 32S, lacked the 5′ end of the 5′ETS (present in 30SL5′; Fig. 4 C). It was not possible map the 3′ end of 30SL5′ because of the overlap with the LSU intermediate 32S, but 21SL3′ contained the 3′ end of ITS1 and 5.8S (Fig. 4 B) and from this we infer that 30SL3′ also includes the complete ITS1 and 5.8S regions. When RRP5 was codepleted with XRN2, 36S pre-rRNA and the ITS1 fragments were not detected. We conclude that decreasing the levels of RRP5 significantly affects both cleavages in ITS1, with some precursors produced in which both ITS1 cleavages are bypassed and cleavage of the primary transcript occurs in ITS2. Collectively, these data are consistent with the human 2a site being equivalent to the A2 cleavage site in yeast. Furthermore, the impairment of cleavage at both ITS1 sites on depletion of human RRP5 appears analogous to the inhibition of both A2 and A3 cleavages after depletion of yeast Rrp5 (Venema and Tollervey, 1996).
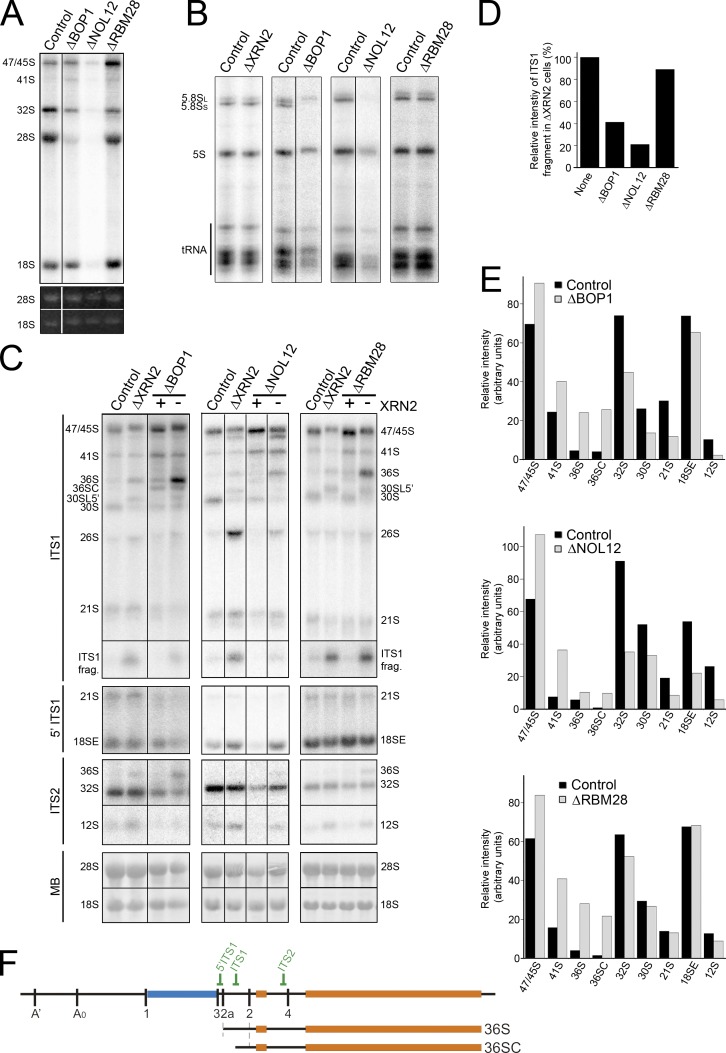
Depletion of BOP1, RBM28, and NOL12 impair cleavage at site 2 in ITS1
In yeast, the A3 endonuclease cleavage site is located 3′ of site A2. This suggested that human site 2 may be equivalent to yeast A3. In yeast, A3 cleavage requires Nop4 (Bergès et al., 1994; Sun and Woolford, 1994), which is homologous to human RBM28 (Nousbeck et al., 2008). Subsequent exonucleolytic digestion from A3 to the major 5′ end of 5.8S rRNA requires factors including Erb1 and Rrp17 (Oeffinger et al., 2009; Granneman et al., 2011; Sahasranaman et al., 2011), which are homologous to human BOP1 (Pestov et al., 2001) and NOL12, respectively. RBM28, BOP1, or NOL12 were depleted from HeLa cells using RNAi, either alone or in combination with XRN2, to test whether human site 2 is analogous to the yeast A3 cleavage. The effects on pre-rRNA processing were analyzed by Northern blotting and metabolic labeling experiments. Depletion of BOP1 resulted in clear defects in 28S and 5.8S accumulation, with loss of the normally major, short form of 5.8S rRNA (5.8SS) and accumulation of the longer form (5.8SL; Fig. 5, A and B). In addition, depletion of BOP1 also resulted in a reduced rate of 18S production. Interestingly, depletion of XRN2 had no effect on the ratio of 5.8S rRNA species (Fig. 5 B). BOP1 depletion resulted in reproducible increases in the levels of 47/45S, 41S, 36S, and 36SC pre-rRNA, a 5′ shortened form of 36S (Fig. 5, C, E, and F). Conversely, the levels of 32S, 30S, 21S, and 12S were reduced, whereas 18SE levels were only mildly affected probably because this intermediate can be generated directly by 2a cleavage. This indicates that processing at site 2a is only slowed, whereas site 2 cleavage is more impaired. Consistent with this, significantly less of the ITS1 fragment was observed after the codepletion of XRN2 (Fig. 5 D). After BOP1 depletion, 36S accumulation was greatly increased by codepletion of XRN2. The levels of 36SC pre-rRNA appeared reduced after XRN2 was codepleted with BOP1, suggesting that this represents an intermediate in 5′ to 3′ exonucleolytic processing of 36S to 32S pre-rRNA.
Figure 5.
BOP1, NOL12, and RBM28 are required for site 2 cleavage in ITS1. (A and B) HeLa cells depleted of BOP1, RBM28, or NOL12 using RNAi or control cells were pulse labeled with 32P orthophosphate and then incubated with normal media for 3 h. RNA was recovered, separated using glyoxal/agarose gel electrophoresis (A) or denaturing PAGE (B), and visualized using a phosphorimager and ethidium bromide staining (UV). Positions of the mature and precursor RNAs are indicated on the left. Note that reduced tRNA production observed after depletion of BOP1 and NOL12 was not consistently seen and likely reflects the exceptionally strong effect the siRNAs had on the cells. (C) RNA extracted from control HeLa cells or HeLa cells depleted of ribosome biogenesis factors either alone or together with XRN2 (indicated at top) was analyzed by Northern blotting using probes that recognize either ITS2, ITS1, or the 5′ end of ITS1 (as indicated on the left). RNA loading was monitored by methylene blue staining of the mature 18S and 28S rRNAs (MB). In codepletion experiments, the presence (+) or depletion (−) of XRN2 is indicated at the top of the relevant lanes. (D) The relative intensity of the ITS1 fragment is plotted for the XRN2 and XRN2 double knockdowns (C), which is representative of three experiments. (E) The relative intensity of the pre-rRNA intermediates detected in C, which is representative of three experiments, are plotted (black, control; gray, knockdown). (F) Schematic representations of 47S pre-rRNA and relevant pre-rRNAs. Relative positions of Northern blot probes are indicated. Black lines in A–C indicate intervening lanes that have been removed.
Depletion of NOL12 resulted in defects in 28S, 5.8S, and 18S rRNA production in metabolic labeling experiments, indicating a complete block in ITS1 processing (Fig. 5 A). NOL12 depletion also increased the levels of 47/45S, 41S, 36S, and 36SC pre-rRNAs, although the effect on 36S and 36SC was not as dramatic as seen after BOP1 depletion (Fig. 5, C and D). The levels of 32S, 30S, 21S, 12S, and 18SE were all significantly reduced, indicating that processing at both sites 2a and 2 was severely impaired. Consistent with this, ITS1 fragment levels were significantly reduced upon codepletion of XRN2 (Fig. 5 D). After codepletion of XRN2 with NOL12, 36SC levels were decreased and 36S accumulation was increased, but not to the extent seen in BOP1 and XRN2 codepletions. The presence of more 36SC than 36S in NOL12-depleted cells suggests that this probable 5′ to 3′ exonuclease is not required for the exonucleolytic processing at the 5′ end of 36S. Strikingly, codepletion of XRN2 with NOL12 restored 18SE levels to near wild-type levels, suggesting that stimulating site 2a cleavage could rescue 18SE production (Fig. 5 C).
In metabolic labeling experiments, depletion of RBM28 had no effect on 18S rRNA accumulation and resulted in only a mild decrease in 5.8S and 28S levels with no change in the ratio of 5.8S species (Fig. 5, A and B). Depletion of RBM28 resulted in increased levels of 45S, 41S, 36S, and 36SC, whereas 32S and 12S levels were slightly reduced. 30S, 21S, and 18SE levels were unaffected (Fig. 5, C and D). Codepletion of RBM28 with XRN2 caused significantly more 36S to accumulate than when only RBM28 levels were reduced. Compared with the yeast homologue of RBM28, which is essential for A3 cleavage (Bergès et al., 1994; Sun and Woolford, 1994), depletion of the human counterpart only slowed down ITS1 processing and, as seen with the depletion of XRN2, resulted in an increase in cleavage at site 2a before site 2.
We conclude that site 2 in the human ITS1 is analogous to site A3 in the yeast pre-rRNA. Interestingly, our data therefore suggest that, in humans, the A3-like cleavage is the primary endonucleolytic cleavage that separates the rRNAs of the large and small subunits.
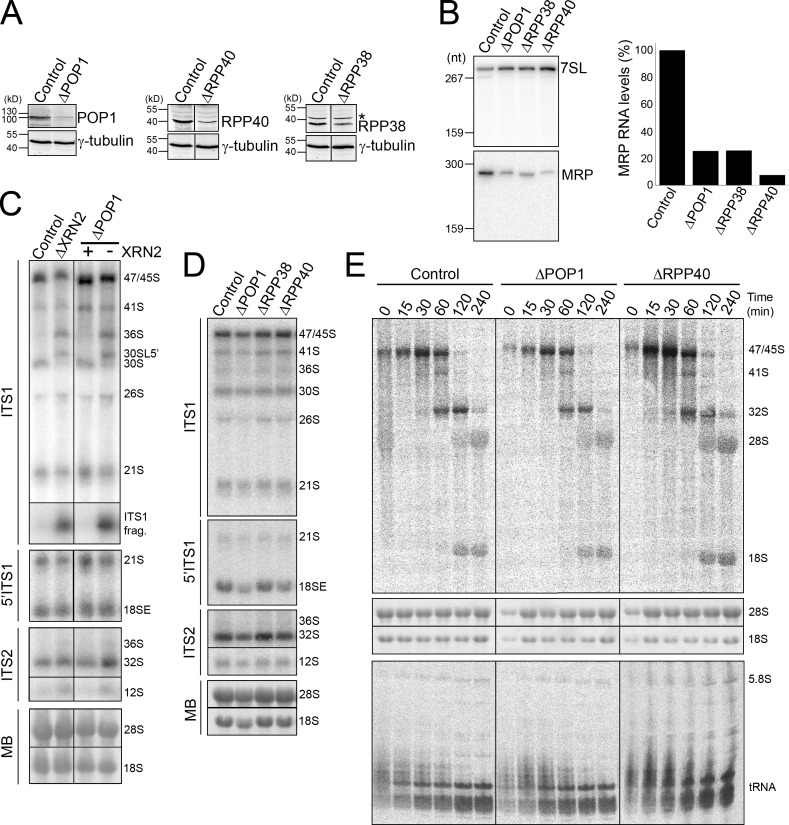
Depletion of RNase MRP proteins does not affect cleavage in ITS1
In yeast, A3 cleavage is performed by the endonuclease complex RNase MRP (Lygerou et al., 1996). Loss of this complex has only a minor effect on pre-rRNA processing and results in defective 5′ processing of 5.8S (Schmitt and Clayton, 1993). If human site 2, which is the major ITS1 processing site, is cleaved by RNase MRP, then we would expect that this endonuclease is far more important to pre-rRNA processing in humans than it is in yeast. The core RNase MRP/RNase P protein POP1, which is essential for the accumulation and function of both of these RNPs in yeast, was depleted from HeLa cells using RNAi, either alone or in combination with XRN2 (Fig. 6 A). Depletion of POP1 resulted in a substantial decrease in the levels of this protein and a four- to fivefold decrease in RNase MRP and P RNA levels (Figs. 6 B and S4 B). Knockdown of POP1 also significantly increases the levels of the viperin protein (Mattijssen et al., 2011). The viperin mRNA is a target for either RNase MRP or RNase P and lack of proteins common to these endonucleases results in increased expression of the viperin protein. Northern analyses showed that depletion of POP1 reproducibly resulted in accumulation of pre-tRNAiMet (Fig. S4 B), confirming the impaired function of RNase P, although this defect was not clearly visible using pulse chase labeling (Figs. 6 E and S4 C). A mild increase in mature tRNA levels was observed in the POP1-depleted cells, but this was substantially less than seen for the precursor. In contrast, depletion of POP1, either alone or together with XRN2, had no noticeable effect on the levels of any of the pre-rRNA intermediates or the ratio of 5.8SS and 5.8SL (Fig. 6, C and D; and Fig. S4 D). In metabolic labeling experiments the levels of the mature rRNAs were also not significantly affected by the depletion of POP1 and there was no change in the ratio of 5.8S species detected (Figs. 6 E and S4 C).
Figure 6.
Depleting levels of RNase MRP does not affect human ribosome biogenesis. Protein (A) and MRP RNA (B) levels in HeLa cells depleted of POP1, RPP38, or RRP40 using RNAi were analyzed by Western and Northern blotting, respectively. In A, the control in each panel is derived from reprobing the same sample. A representative experiment of three is shown. Antibodies or probes used are indicated on the right. Asterisk, nonspecific protein. 7SL RNA levels were used as a loading control. The levels of RNase MRP RNA shown in A were quantified and plotted. (C and D) RNA extracted from control HeLa cells or HeLa cells that had been depleted of RNase MRP subunits either alone or together with XRN2 (indicated at top) was analyzed by Northern blotting using probes that recognize either ITS2, ITS1, or the 5′ end of ITS1 (as indicated on the left). RNA loading was monitored by methylene blue staining of the mature 18S and 28S rRNAs (MB). In codepletion experiments, the presence (+) or depletion (−) of XRN2 is indicated at the top. (E) HeLa cells depleted of POP1 or RRP40 using RNAi or control cells were pulse labeled with 3H methyl-methionine and then incubated with normal media for varying time-points. RNA was recovered from the cells, separated using glyoxal/agarose gel electrophoresis (top) or denaturing PAGE (bottom) and visualized using a phosphorimager. Total RNA was visualized by ethidium bromide staining (middle panels). Positions of the mature and precursor RNAs are indicated. 5S rRNA is not detected using this method, indicating that it is not methylated. The left three panels of control data are repeated from Fig. 2 D. Black lines in A and C indicate intervening lanes that have been removed.
These results were unexpected and we therefore also analyzed the effect of depleting additional RNase MRP/P subunits, RPP38 and RPP40. Levels of these proteins were also significantly reduced by RNAi treatment and RNase MRP/P RNA levels were decreased (Fig. 6, A and B; and Fig. S4 B). Depletion of RPP40 and, to a lesser extent, RPP38 also caused accumulation of pretRNAiMet, as previously demonstrated for RPP38 (Cohen et al., 2003; Fig. S4 B). Interestingly, modest increases in mature tRNA levels were also seen upon depletion of these proteins, although we cannot currently explain this observation. Consistent with the depletion of POP1, reducing the levels of either RPP38 or RPP40 had no detectable effect on pre-rRNA levels or on accumulation of the mature rRNAs (Fig. 6, D and E; and Fig. S4, C and D). Simultaneous depletion of POP1 and RPP40/RPP38 also had no clear effect on pre-rRNA processing (Fig. S4, C and D; unpublished data) and no greater impact on tRNA processing than the single depletions. Site 2 cleavage plays an important role in human pre-rRNA processing so the observation that depletion of RNase MRP/P proteins by more than 10-fold had no effect on ITS1 processing therefore questions the role of human RNase MRP in ribosome biogenesis.
Discussion
Here we have characterized alternative ITS1 processing pathways in human cells. The major pathway involves a single endonucleolytic cleavage at site 2 followed by exonucleolytic processing to both the 5′ end of 5.8S and to site 2a to generate the 18SE precursor, which contains ∼25 nt of ITS1 at the 3′ end of 18S rRNA. The final ITS1 sequence is then removed by endonucleolytic cleavage at site 3 (Fig. 7). Alternatively, the 18SE precursor can be generated by endonucleolytic cleavage at site 2a. This minor pathway can compensate for a block in 18SE production by exonucleolytic processing after site 2 cleavage when stimulated by XRN2 depletion.
Figure 7.
Alternative processing pathways for human ITS1. Schematic representations of the yeast, human major, and human minor ITS1 processing pathways.
After cleavage at site 2, RRP6, a 3′ to 5′ exonuclease component of the exosome complex, processes to site 2a. Importantly, the exosome has not been demonstrated to be involved in ITS1 processing before in any organism. The core exosome and the TRAMP component MTR4 are also needed for the full activity/processivity of RRP6. The involvement of the RNA helicase MTR4 is not surprising given that this region of human ITS1 is ∼800 nt long (compared with ∼85 nt in yeast), 82% GC rich, and highly structured (Fig. S5). The requirement for RPS19, ENP1, RRP5, and RCL1 in this processing step indicates that the exosome also interacts with the SSU processome. Depletion of RRP6 severely reduced, but did not abolish, 18SE accumulation and 18S rRNA production. This may be a result of incomplete depletion of RRP6 but more likely suggests that other proteins, such as the Rex exonucleases, also contribute to 21S 3′ processing. Depletion of XRN2, which favors site 2a cleavage, can rescue 18SE production in the absence of the exosome or MTR4, indicating that the majority of rRNA is normally processed via this exonucleolytic pathway. In yeast, the A3 cleavage site is an entry point for Rrp6 (Allmang et al., 2000). Moreover, pre-rRNA deletions around site A2 do not block 18S synthesis (Allmang et al., 1996), which is consistent with the possibility of alternative pathways in yeast that might more closely resemble human pre-rRNA processing.
We found that SSU processome components are required for endonucleolytic cleavage at site 2a as is the case for site A2 cleavage in yeast. Surprisingly, site 2a cleavage is part of the minor rRNA processing pathway. 36S rRNA, the only precursor specific to this pathway, is rarely observed in a variety of human cell lines (HeLa, HEK293, TC7, and MCF7; unpublished data) and under different growth conditions, making it difficult to determine the fraction of pre-rRNA normally processed via endonucleolytic cleavage at 2a. As seen with the A2 cleavage in yeast, no precursors processed at 2a but still containing the 5′ETS sequence were observed, indicating that endonucleolytic cleavage at site 2a obligatorily occurs together with or after removal of the 5′ETS.
We found that site 2 cleavage in human cells was significantly inhibited by depletion of RRP5, NOL12, BOP1, or to a lesser extent RBM28, suggesting that it is analogous to the A3 cleavage in yeast. Depletion of NOL12, RRP5, BOP1, and RBM28 also affected site 2a cleavage. However, as it is not known what percentage of transcripts are naturally processed at 2a it is difficult to determine whether depletion of these proteins actually increases or decreases cleavage at this site. In the case of RRP5 depletion, both ITS1 cleavages could be bypassed, with primary cleavage occurring in ITS2. The observation that the putative exonuclease NOL12 is essential for both cleavages in ITS1 was particularly surprising. The 36SC pre-rRNA, seen after depletion of RBM28 or BOP1, is likely to be an intermediate in the 5′ exonucleolytic processing of the 36S precursor to 32S. Indeed, in either BOP1 or RBM28 knockdowns, 36S accumulation was increased relative to 36SC by codepletion of XRN2, an exonuclease likely to be involved in converting 36S to 32S. In yeast, blocking A3 cleavage or subsequent exonucleolytic processing from A3 to the 5′ end of 5.8S results in loss of 5.8SS and accumulation of 5.8SL. Depletion of human BOP1 resulted in the accumulation of only 5.8SL, suggesting that this processing mechanism is conserved. Similarities and differences between the functions of these proteins in humans and yeast are outlined in Fig. 7.
The lack of effect of depleting RNase MRP on site 2 cleavage and the 5′ processing of 5.8S was unexpected. The decrease in MRP/P protein levels significantly reduced RNase MRP/P RNA abundance and both inhibited cleavage of the viperin mRNA (Mattijssen et al., 2011) and affected processing of tRNAiMet (Fig. S5 A) and tRNATyr (unpublished data). It was surprising that the effect on tRNA processing, caused by depleting multiple components of the RNase P complex to this extent, was not stronger. These data are, however, consistent with previously published data analyzing RPP38 (Cohen et al., 2003). Indeed, the authors claimed that RPP38 is not essential for pre-rRNA processing after only observing very weak accumulation of 5.8S processing intermediates. We did not observe the accumulation of such precursors in RPP38, POP1, or RPP40 knockdowns (Fig. S5). As a result of the nature of RNAi knockdowns, low residual levels of proteins remained after depletion of RPP40, RPP38, or POP1 (<10% of each). In consequence, we cannot exclude the possibility that the residual RNase MRP activity is sufficient to maintain pre-rRNA processing. However, our data suggest that RNase MRP is not essential for ITS1 processing and that a distinct endonuclease is responsible for site 2 cleavage in human cells.
It is not clear why endonucleolytic cleavage at site 2a is not part of the major processing pathway in humans. In yeast, several pre-rRNA processing steps, including cleavage at A2, occur cotranscriptionally (Kos and Tollervey, 2010), whereas analyses of Miller spreads indicate that vertebrate pre-rRNA processing is mostly posttranscriptional. It is possible that 2a cleavage is inefficient in the absence of cotranscriptional processing. Alternatively, the close proximity of 2a to the 3′ end of 18S (∼25 nt) or extreme GC content of ITS1 may make this cleavage inefficient. Site 2a cleavage is affected by depletion of factors linked to cleavage at site 2, suggesting competition and/or cross talk between processing pathways. Alternative ITS1 processing pathways have also been described in Xenopus laevis oocytes (Savino and Gerbi, 1990), suggesting that this is a conserved feature of pre-rRNA processing that may aid overall cleavage efficiency. Our data indicate that the initial ITS1 cleavage is primarily linked to factors important for LSU biogenesis. Furthermore, the rRNA processing defects seen upon depletion of BOP1, RRP5, and NOL12 were strikingly similar to those seen upon ARF overexpression (Sugimoto et al., 2003) or depletion of the ARF-regulated transcription termination factor TTF-1 (Lessard et al., 2010). It is possible that higher eukaryotes have evolved the processing pathway we observe in humans so processing occurs posttranscriptionally and can be regulated by ARF.
CHH is caused by mutations in either the RNA or POP1 protein subunit of RNase MRP (Ridanpää et al., 2001; Glazov et al., 2011) and is commonly cited as an example of a disease resulting from defective ribosome biogenesis. However, reduction of RNase MRP RNA or POP1 levels to a greater extent than observed in many CHH patients had no noticeable effect on ribosome biogenesis, strongly suggesting that this disease actually arises from a defect in another function of this enzyme, such as mRNA cleavage. Mutations in the exosome component RRP40 (EXOSC3) have recently been shown to cause pontocerebellar hypoplasia and spinal motor neuron degeneration (PCH1; Wan et al., 2012). Our results demonstrate a novel, essential role for the exosome in ITS1 processing and from this we suggest that PCH1 is another ribosomopathy. This underlines the importance of studying ribosome biogenesis in human cells and not solely relying on data from the yeast model system to understand the causes of these genetic diseases.
Materials and methods
RNAi
siRNA duplex (listed in Table S1) transfection was performed using Lipofectamine RNAiMAX reagent (Invitrogen). Cells were harvested 60 h after transfection and then analyzed by Western (antibodies listed in Table S2) and Northern blotting.
The cDNA for human RRP6, altered to make the mRNA resistant to the siRNAs used to deplete endogenous RRP6, was cloned into the pcDNA5/FRT/TO vector (Invitrogen) containing 2× FLAG tags under the control of a tetracycline-regulated promoter. A single amino acid mutation in the catalytic site of RRP6 (RRP6exo; D313A) was also generated. The oligonucleotides used to make these constructs are listed in Table S3. The constructs were transfected into Flp-In T-Rex HEK293 cells and stably transfected cells selected as described by the manufacturer (Invitrogen). Cells expressing RRP6, RRP6exo, or the FLAG tag alone were incubated with 0.1 mg/ml tetracycline to induce protein expression, and then transfected with siRNAs to deplete endogenous RRP6. 48 h later, the cells were harvested and analyzed by Western and Northern blotting.
RNA analysis
RNA was extracted from cell pellets using TRI reagent (Sigma-Aldrich) and analyzed by glyoxal-agarose gel or urea-PAGE and transferred to nylon membrane. For Northern blot analysis, oligonucleotides (Table S4) were 5′ labeled with γ-[32P]ATP using T4 polynucleotide kinase and used as probes. Random prime-labeled probes hybridizing immediately upstream and downstream of the A’ cleavage in the 5′ETS (ETS1 and ETS2, respectively) were produced from PCR products (Turner et al., 2009). Random prime-labeled probes against the full-length RNase MRP RNA and the S domain of 7SL were also generated.
For metabolic labeling experiments using 3H methyl-methionine, the cells were incubated in methionine-free media (15 min) and then in methionine-free media containing 50 µCi/ml 3H-labeled methyl-methionine (30 min). The cells were then incubated in normal media containing 10× the normal concentration of methionine and harvested at the required time points (0, 15, 30, 60, 120, and 240 min). For metabolic labeling experiments using 32P orthophosphate, cells were grown in phosphate-free media (1 h) followed by phosphate-free media containing 15 µCi/ml 32P-labeled inorganic phosphate (1 h). Cells were then incubated in normal media and harvested at the required time points (0, 30, 60, 120, and 240 min). RNA was extracted using TRI reagent and analyzed by agarose-glyoxal or urea-PAGE as appropriate. Results were visualized using a phosphorimager (Typhoon FLA9000; GE Healthcare). All quantitation was normalized to the levels of mature 18S or 28S rRNA, as appropriate.
Immunofluorescence
HEK293 cells expressing either the FLAG tag only (pcDNA5 [Knox et al., 2011]), FLAG-RRP6, or FLAG-tagged, RNAi-resistant RRP6 or RRP6exo were grown on coverslips and induced with 0.1 mg/ml tetracycline for 36 h before being fixed with PBS containing 4% paraformaldehyde. The cells were washed with PBS and permeabilized in 0.2% Triton X-100 and then blocked in PBS/10% FCS. Cells were incubated with the anti-FLAG (rabbit; Sigma-Aldrich) and anti-fibrillarin (72B9; mouse) in PBS/10% FCS. Cells were washed with PBS and then incubated with the secondary antibody (Alexa Fluor 555–labeled donkey anti–mouse and Alexa Fluor 647–labeled donkey anti–rabbit) in PBS/10% FCS. The cells were washed with PBS containing DAPI and then mounted in Mowiol. All steps were performed at room temperature. Images were captured with an inverted microscope (Axiovert 200M; Carl Zeiss) with a Plan-Apochromat, 100×/1.40 oil DIC ×/0.17 objective (Carl Zeiss) and an AxioCam HRm camera, using Axiovision software.
Online supplemental material
Fig. S1 shows Northern blot ITS1 mapping data. Fig. S2 shows RNAi depletion of ribosome biogenesis factors from HeLa cells. Fig. S3 is a characterization of HEK293 stable cells lines expressing RRP6. Fig. S4 shows the requirement for SSU proteins and RNase MRP/P subunits in the formation of the mature rRNAs. Fig. S5 is a model of human ITS1 structure. Table S1 shows ribosome biogenesis factor siRNA sequences. Table S2 contains the antibodies used in this study. Table S3 shows the primers used to make RRP6 expression constructs. Table S4 shows Northern blot probe sequences. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207131/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Ulrike Kutay, Dimitri Pestov, Philip Mason, and Sydney Altman for providing antibodies, Kaspar Burger for advice on the metabolic labeling experiments, and Carla Onnekink for technical assistance. We would also like to thank Claudia Schneider for critically reading the manuscript.
This work was funded by the Biotechnology and Biological Sciences Research Council (N.J. Watkins), the Wellcome Trust (N.J. Watkins and D. Tollervey), and the Council for Chemical Sciences (NWO-CW) of the Netherlands Organization for Scientific Research (G.J.M. Pruijn).
Footnotes
Abbreviations used in this paper:
- CHH
- cartilage-hair hypoplasia
- ETS
- external transcribed spacer
- ITS
- internal transcribed spacer
- LSU
- large ribosomal subunit
- pre-rRNA
- precursor rRNA
- rRNA
- ribosomal RNA
- SSU
- small ribosomal subunit
References
- Allmang C., Henry Y., Morrissey J.P., Wood H., Petfalski E., Tollervey D. 1996. Processing of the yeast pre-rRNA at sites A(2) and A(3) is linked. RNA. 2:63–73 [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Mitchell P., Petfalski E., Tollervey D. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28:1684–1691 10.1093/nar/28.8.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergès T., Petfalski E., Tollervey D., Hurt E.C. 1994. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 13:3136–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carron C., O’Donohue M.F., Choesmel V., Faubladier M., Gleizes P.E. 2011. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 39:280–291 10.1093/nar/gkq734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A., Uechi T., Kenmochi N. 2011. Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip. Rev. RNA. 2:507–522 10.1002/wrna.73 [DOI] [PubMed] [Google Scholar]
- Cohen A., Reiner R., Jarrous N. 2003. Alterations in the intracellular level of a protein subunit of human RNase P affect processing of tRNA precursors. Nucleic Acids Res. 31:4836–4846 10.1093/nar/gkg691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov E.A., Zankl A., Donskoi M., Kenna T.J., Thomas G.P., Clark G.R., Duncan E.L., Brown M.A. 2011. Whole-exome re-sequencing in a family quartet identifies POP1 mutations as the cause of a novel skeletal dysplasia. PLoS Genet. 7:e1002027 10.1371/journal.pgen.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., Petfalski E., Tollervey D. 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 30:4006–4019 10.1038/emboj.2011.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359 10.1007/s00018-008-8027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D.M., Mason S.L., Karbstein K. 2011. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 286:34082–34087 10.1074/jbc.M111.268649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox A.A., McKeegan K.S., Debieux C.M., Traynor A., Richardson H., Watkins N.J. 2011. A weak C’ box renders U3 snoRNA levels dependent on hU3-55K binding. Mol. Cell. Biol. 31:2404–2412 10.1128/MCB.05067-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M., Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 37:809–820 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna A.C., Karbstein K. 2011. An RNA conformational switch regulates pre-18S rRNA cleavage. J. Mol. Biol. 405:3–17 10.1016/j.jmb.2010.09.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard F., Morin F., Ivanchuk S., Langlois F., Stefanovsky V., Rutka J., Moss T. 2010. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol. Cell. 38:539–550 10.1016/j.molcel.2010.03.015 [DOI] [PubMed] [Google Scholar]
- Liang X.H., Crooke S.T. 2011. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 39:4875–4889 10.1093/nar/gkr076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervey D., Séraphin B. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 272:268–270 10.1126/science.272.5259.268 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Tomecki R., Jensen T.H., Dziembowski A. 2011. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 8:61–66 10.4161/rna.8.1.14237 [DOI] [PubMed] [Google Scholar]
- Mattijssen S., Hinson E.R., Onnekink C., Hermanns P., Zabel B., Cresswell P., Pruijn G.J. 2011. Viperin mRNA is a novel target for the human RNase MRP/RNase P endoribonuclease. Cell. Mol. Life Sci. 68:2469–2480 10.1007/s00018-010-0568-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineux S.T., Lafontaine D.L. 2012. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie. 94:1521–1532 10.1016/j.biochi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Narla A., Ebert B.L. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 115:3196–3205 10.1182/blood-2009-10-178129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousbeck J., Spiegel R., Ishida-Yamamoto A., Indelman M., Shani-Adir A., Adir N., Lipkin E., Bercovici S., Geiger D., van Steensel M.A., et al. 2008. Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am. J. Hum. Genet. 82:1114–1121 10.1016/j.ajhg.2008.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Zenklusen D., Ferguson A., Wei K.E., El Hage A., Tollervey D., Chait B.T., Singer R.H., Rout M.P. 2009. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol. Cell. 36:768–781 10.1016/j.molcel.2009.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis B.A., Steitz J.A. 1993. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 73:1233–1245 10.1016/0092-8674(93)90651-6 [DOI] [PubMed] [Google Scholar]
- Pertschy B., Schneider C., Gnädig M., Schäfer T., Tollervey D., Hurt E. 2009. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 284:35079–35091 10.1074/jbc.M109.040774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov D.G., Stockelman M.G., Strezoska Z., Lau L.F. 2001. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 29:3621–3630 10.1093/nar/29.17.3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E., Dandekar T., Henry Y., Tollervey D. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps K.R., Charette J.M., Baserga S.J. 2011. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip. Rev. RNA. 2:1–21 10.1002/wrna.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpää M., van Eenennaam H., Pelin K., Chadwick R., Johnson C., Yuan B., vanVenrooij W., Pruijn G., Salmela R., Rockas S., et al. 2001. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 104:195–203 10.1016/S0092-8674(01)00205-7 [DOI] [PubMed] [Google Scholar]
- Rouquette J., Choesmel V., Gleizes P.E. 2005. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 24:2862–2872 10.1038/sj.emboj.7600752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasranaman A., Dembowski J., Strahler J., Andrews P., Maddock J., Woolford J.L., Jr 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 30:4020–4032 10.1038/emboj.2011.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R., Gerbi S.A. 1990. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 9:2299–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G., van Dijk E., Pruijn G.J. 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 35:2564–2572 10.1093/nar/gkm082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E., Clayton D.A. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M., Kuo M.L., Roussel M.F., Sherr C.J. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell. 11:415–424 10.1016/S1097-2765(03)00057-1 [DOI] [PubMed] [Google Scholar]
- Sun C., Woolford J.L., Jr 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R., Kristiansen M.S., Lykke-Andersen S., Chlebowski A., Larsen K.M., Szczesny R.J., Drazkowska K., Pastula A., Andersen J.S., Stepien P.P., et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 29:2342–2357 10.1038/emboj.2010.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.J., Knox A.A., Prieto J.L., McStay B., Watkins N.J. 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell. Biol. 29:3007–3017 10.1128/MCB.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., Tollervey D. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701–5714 [PMC free article] [PubMed] [Google Scholar]
- Wan J., Yourshaw M., Mamsa H., Rudnik-Schöneborn S., Menezes M.P., Hong J.E., Leong D.W., Senderek J., Salman M.S., Chitayat D., et al. 2012. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet. 44:704–708 10.1038/ng.2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Pestov D.G. 2011. 5′-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 39:1811–1822 10.1093/nar/gkq1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437–440 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.