Par6–aPKC recruitment to the premature apical membrane through Morg1 interaction with Par6 is required for definition of apical identity of epithelial cells.
Abstract
Formation of apico-basal polarity in epithelial cells is crucial for both morphogenesis (e.g., cyst formation) and function (e.g., tight junction development). Atypical protein kinase C (aPKC), complexed with Par6, is considered to translocate to the apical membrane and function in epithelial cell polarization. However, the mechanism for translocation of the Par6–aPKC complex has remained largely unknown. Here, we show that the WD40 protein Morg1 (mitogen-activated protein kinase organizer 1) directly binds to Par6 and thus facilitates apical targeting of Par6–aPKC in Madin-Darby canine kidney epithelial cells. Morg1 also interacts with the apical transmembrane protein Crumbs3 to promote Par6–aPKC binding to Crumbs3, which is reinforced with the apically localized small GTPase Cdc42. Depletion of Morg1 disrupted both tight junction development in monolayer culture and cyst formation in three-dimensional culture; apico-basal polarity was notably restored by forced targeting of aPKC to the apical surface. Thus, Par6–aPKC recruitment to the premature apical membrane appears to be required for definition of apical identity of epithelial cells.
Introduction
Cell polarization is crucial for diverse processes including cell fate determination, differentiation, and specialized cell functions that underlie morphogenesis. The plasma membrane of mammalian epithelial cells is asymmetrically organized into apical and basolateral domains; the two domains serve differently to integrate epithelial function. The apical membrane, facing a lumen, is separated from the basolateral one by tight junctions (TJs), which participate in epithelial barrier function (Goldstein and Macara, 2007; Bryant and Mostov, 2008; Prehoda, 2009; Knoblich, 2010; St Johnston and Ahringer, 2010).
Formation of apico-basal polarity in epithelial cells likely involves atypical protein kinase C (aPKC), which is considered to serve as a master enzyme in animal cell polarization (Goldstein and Macara, 2007; Bryant and Mostov, 2008; Prehoda, 2009; Knoblich, 2010; St Johnston and Ahringer, 2010). aPKC constitutively interacts with Par6, an evolutionarily conserved adaptor protein, which interaction is mediated via N-terminal PB1 (Phox and Bem1p 1) domains of both proteins (Noda et al., 2003; Sumimoto et al., 2007). In Par6, the PB1 domain is followed by a semi-CRIB (Cdc42/Rac interactive binding) motif and a PDZ (PSD95/Dlg/ZO-1) domain (Kemphues, 2000; Noda et al., 2003; Suzuki and Ohno, 2006; Sumimoto et al., 2007). During epithelial cell polarization in the fruit fly Drosophila melanogaster, the Par6–aPKC complex is shown to translocate from the cytoplasm to the apical surface, where the complex may be anchored via direct interaction of Par6 with the apical membrane-integrated protein Crumbs, the Pals1-related adaptor protein Stardust, or the small GTPase Cdc42 in the GTP-bound form (Goldstein and Macara, 2007; St Johnston and Ahringer, 2010). Par6 binding to Cdc42 requires both the semi-CRIB motif and the adjacent PDZ domain; this domain allows the semi-CRIB motif to efficiently interact with Cdc42 (Garrard et al., 2003; Peterson et al., 2004). On the other hand, the Par6 PDZ domain is capable of directly binding to the extreme C terminus of Crumbs in a canonical PDZ-binding manner (Lemmers et al., 2004) and also to an internal sequence of Pals1 (Penkert et al., 2004; Wang et al., 2004). In mammalian epithelial cells, it is established that Par3, a component of the well-known Par complex (containing Par3, Par6, and aPKC), is enriched solely at TJs; on the other hand, both Par6 and aPKC appear to be distributed not only at TJs but also throughout the apical surface (Yamanaka et al., 2003, 2006; Martin-Belmonte et al., 2007), although it has also been suggested that Par6 predominantly localizes to the TJs (Hurd et al., 2003; Deborde et al., 2008). Thus, it is likely that although the Par3-containing complex is accumulated at TJs, the Par6–aPKC binary complex (without Par3) localizes specifically to the apical membrane during polarization in mammalian epithelial cells. The significance of Par6–aPKC translocation to the apical surface, however, is not well understood, especially in mammalian epithelial cells. This may be partly because RNAi-mediated knockdown of aPKC leads to apoptosis of MDCK cells (Suzuki et al., 2004; Durgan et al., 2011) and RNAi-mediated depletion of Par6 is unsuccessful probably due to the protein stability and/or the presence of three distinct but closely related proteins (Noda et al., 2001). It seems thus important to find and characterize a molecule involved in apical translocation of Par6–aPKC.
Cdc42, a Par6-binding protein, is known as regulator of cell polarity in many systems including yeasts, fruit fly neuroblasts, and migrating animal cells (Prehoda, 2009; Slaughter et al., 2009). In Drosophila epithelial cells, wild-type Par6 localizes to the apical membrane, but a mutant protein defective in binding to Cdc42 delocalizes to the cytoplasm, resulting in impaired formation of apico-basal polarity (Hutterer et al., 2004). Although this finding suggests that Cdc42 localizes to the apical surface for anchoring of Par6, apical localization of Cdc42 in these cells has not been well evidenced. This may be because anti-Cdc42 antibodies suitable for immunostaining have been unavailable or because fixation conditions used have been unsuitable for immunostaining. Similarly, in monolayer culture of mammalian epithelial cells such as Madin-Darby canine kidney (MDCK) cells, localization of endogenous Cdc42 has not been well studied, although it has been reported that, in 3D culture of MDCK cells, GFP-fused Cdc42 is recruited to the apical surface and appears to participate in apical localization of aPKC (Martin-Belmonte et al., 2007). The role of Cdc42 in aPKC targeting to the apical surface, however, has been questioned using experiments of 3D culture of human colon carcinoma-derived Caco-2 cells (Jaffe et al., 2008).
The type I transmembrane protein Crumbs, another Par6 target, is known to serve as an evolutionarily conserved apical determinant (Bulgakova and Knust, 2009; Datta et al., 2011). The C-terminal cytoplasmic region of Crumbs contains a canonical PDZ-binding motif, which directly interacts with the Par6 PDZ domain (Lemmers et al., 2004) and also with the PDZ domain of Pals1, an adaptor protein that is enriched together with Patj at TJs but not in the apical surface (Makarova et al., 2003). In Drosophila epithelial cells, Par6 localization to the apical surface appears to require Crumbs (Kempkens et al., 2006). Its dominant homologue in mammalian epithelial cells is Crumbs3 (Crb3; Makarova et al., 2003; Lemmers et al., 2004). Crb3 has been shown to be capable of recruiting Par6 to the membrane in unpolarized mammalian cells (Hurd et al., 2003). It has recently been reported that depletion of Crb3 results in a failure of aPKC to localize to the forming apical membrane of MDCK cells at the two-cell stage in 3D culture (Schlüter et al., 2009). However, the relationship between Par6 binding to Crb3 and Cdc42 has remained unknown. Thus, the mechanisms for Par6–aPKC translocation to the apical membrane are still unclear.
In the present study, we have identified the WD40 protein Morg1 (mitogen-activated protein kinase organizer 1, also known as WDR83; Vomastek et al., 2004) as a novel Par6-binding protein. Morg1 participates in apico-basal polarization in MDCK cells: RNAi-mediated depletion of Morg1 impairs both TJ development in monolayer culture and cyst formation in 3D culture. Although depletion of Morg1 mislocalizes Par6–aPKC to the cytoplasm, forced targeting of aPKC to the apical surface restores apico-basal polarity in Morg1-depleted cells, providing evidence that Morg1-regulated translocation of Par6–aPKC plays a crucial role in polarization of mammalian epithelial cells. Morg1 interacts not only with Par6 but also with the apical protein Crb3, which facilitates Par6 binding to Crb3, leading to apical targeting of Par6–aPKC. We also show that endogenous Cdc42 is enriched at the apical membrane, and that this GTPase strongly promotes Par6 binding to Crb3, thereby stabilizing apical localization of Par6–aPKC for epithelial cell polarization.
Results
Morg1 directly interacts with Par6
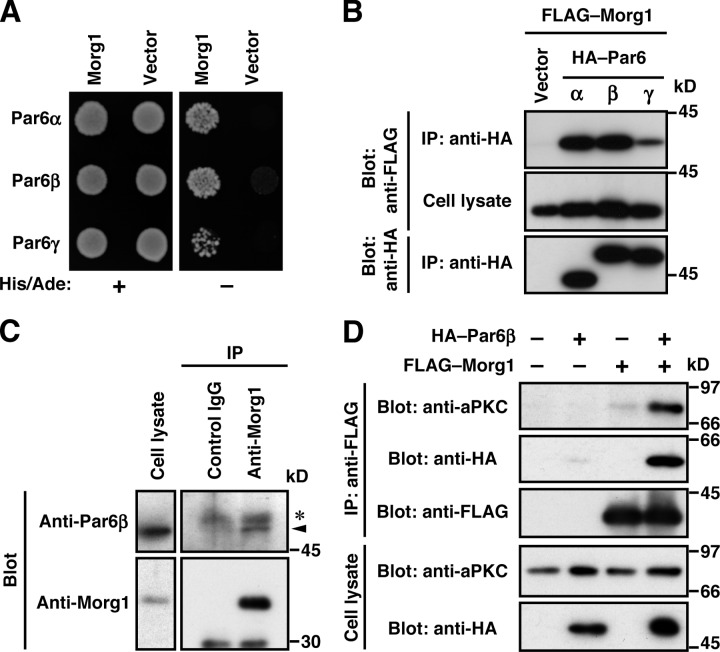
To identify a Par6-interacting protein that regulates Par6–aPKC translocation, we screened a human fetal cDNA library in the yeast two-hybrid system using human Par6α as a bait. A positive clone obtained encodes Morg1 (also known as WDR83) of 315 amino acid residues, which comprises seven repeats of WD40, a module known to be involved in a variety of protein–protein interactions (Xu and Min, 2011). Morg1 exhibited a two-hybrid interaction not only with Par6α but also with its homologous proteins Par6β and Par6γ (Fig. 1 A). These Par6 proteins interacted with Morg1 also when exogenously expressed in COS-7 cells (Fig. 1 B). To test the interaction between endogenous proteins, we prepared antibodies raised against Morg1 as well as those against Par6 (see Materials and methods; Fig. S1). As shown in Fig. 1 C, the anti-Morg1 antibodies coprecipitated Par6β with Morg1 from MDCK cells, indicating that endogenous Par6β interacts with endogenous Morg1 in these epithelial cells. It should be noted that this interaction does not prevent the constitutive association between Par6 and aPKC (Fig. 1 D).
Figure 1.
Morg1 interacts with Par6. (A) The yeast two-hybrid interaction between Morg1 and Par6. PJ69-4A cells transformed with a pair of pACT2–Morg1 and pGBK encoding Par6α, Par6β, or Par6γ were grown in the absence of histidine and adenine. (B and D) Proteins in the lysate of COS-7 cells expressing indicated proteins (Cell lysate) were immunoprecipitated (IP) and then analyzed by immunoblot (Blot) with the indicated antibodies. (C) Proteins in lysates of MDCK cells were immunoprecipitated with the anti-Morg1 or control IgG, and then analyzed by immunoblot with the indicated antibodies. The arrowhead indicates endogenous Par6β; asterisk denotes the heavy chain of IgG.
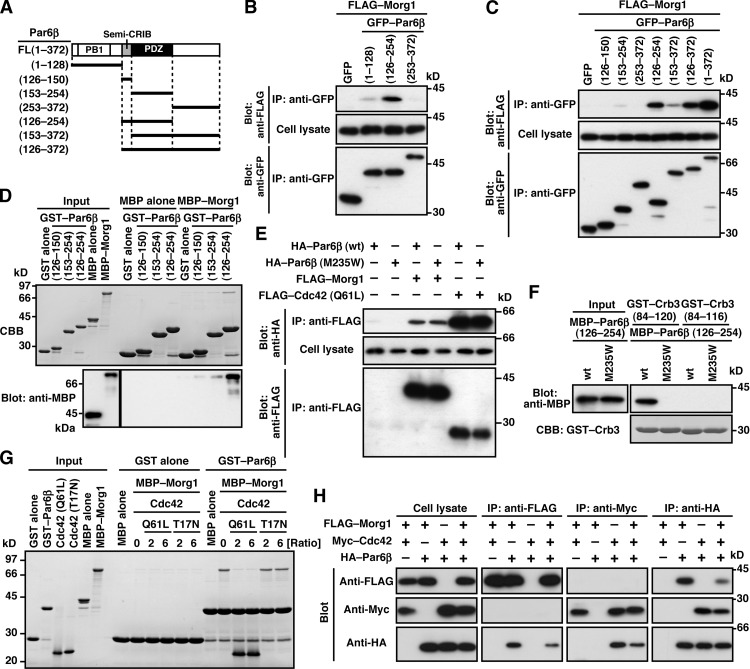
Morg1 expressed in COS-7 cells was efficiently coimmunoprecipitated with full-length Par6β, Par6β (126–254), and Par6β (126–372), each containing both semi-CRIB motif and PDZ domain (Fig. 2, A–C), whereas deletion of either module resulted in a markedly reduced interaction. The N-terminal PB1 domain also contributed to the interaction but to a lesser extent. Furthermore, purified MBP–Morg1 was effectively pulled down with GST–Par6β (126–254) but not with proteins lacking the semi-CRIB motif or the PDZ domain (Fig. 2 D). Thus, Morg1 directly interacts with Par6, which interaction requires both semi-CRIB motif and PDZ domain. This interaction does not appear to use the pocket for binding to canonical PDZ ligands (typically C-terminal ligands) in the Par6 PDZ domain because substitution of tryptophan for Met-235 in the pocket did not inhibit Par6 binding to Morg1 (Fig. 2 E; Yamanaka et al., 2003). On the other hand, the canonical PDZ interaction with the C terminus of Crb3, which required the C-terminal four amino acids (Fig. 2 F), was impaired by the M235W substitution (Fig. 2 F; Lemmers et al., 2004). Consistent with the previous structural analysis (Garrard et al., 2003; Peterson et al., 2004), the Met-235–containing pocket was not involved in Par6 interaction with GTP–Cdc42 (Fig. 2 E), which depends on both semi-CRIB motif and PDZ domain (Fig. S2 A; Joberty et al., 2000; Lin et al., 2000; Qiu et al., 2000; Noda et al., 2001) as the interaction with Morg1. It seems thus possible that Morg1 and Cdc42 bind to Par6 in a mutually exclusive manner. Indeed, in binding to GST–Par6β, MBP–Morg1 was replaced by a GTP-bound form of Cdc42 (Q61L), but not by a GDP-bound form of Cdc42 (T17N; Fig. 2 G). The replacement also occurred at the cellular level: Morg1 binding to Par6 was abrogated by coexpression of Cdc42 (Q61L) in COS-7 cells (Fig. 2 H). This raises the possibility that Morg1 and Cdc42 function at distinct stages of a cellular event that involves Par6.
Figure 2.
Morg1 and Cdc42 interact with Par6 in a mutually exclusive manner. (A) Schematic structure of Par6β and its truncated proteins used in the present study. (B, C, E, and H) Proteins in the lysate of COS-7 cells expressing indicated proteins (Cell lysate) were immunoprecipitated (IP) and then analyzed by immunoblot (Blot) with the indicated antibodies. (D) GST–Par6β (126–150, 153–254, or 126–254) or GST alone was incubated with MBP–Morg1 or MBP alone, and pulled down with glutathione-Sepharose 4B beads, followed by SDS-PAGE analysis with Coomassie brilliant blue (CBB) staining or immunoblot with anti-MBP antibodies. (F) GST–Crb3-(84–120) or GST–Crb3-(84–116) was incubated with MBP–Par6β-(126–254), and analyzed as in D. (G) GST–Par6β-(126–254) or GST alone was incubated with MBP–Morg1 in the presence of a two- or sixfold molar excess of Cdc42 (Q61L) or Cdc42 (T17N) relative to Morg1 (Ratio). Proteins pulled down with glutathione-Sepharose 4B beads were subjected to SDS-PAGE and stained with CBB.
Morg1 participates in TJ development by regulating apico-basal polarity
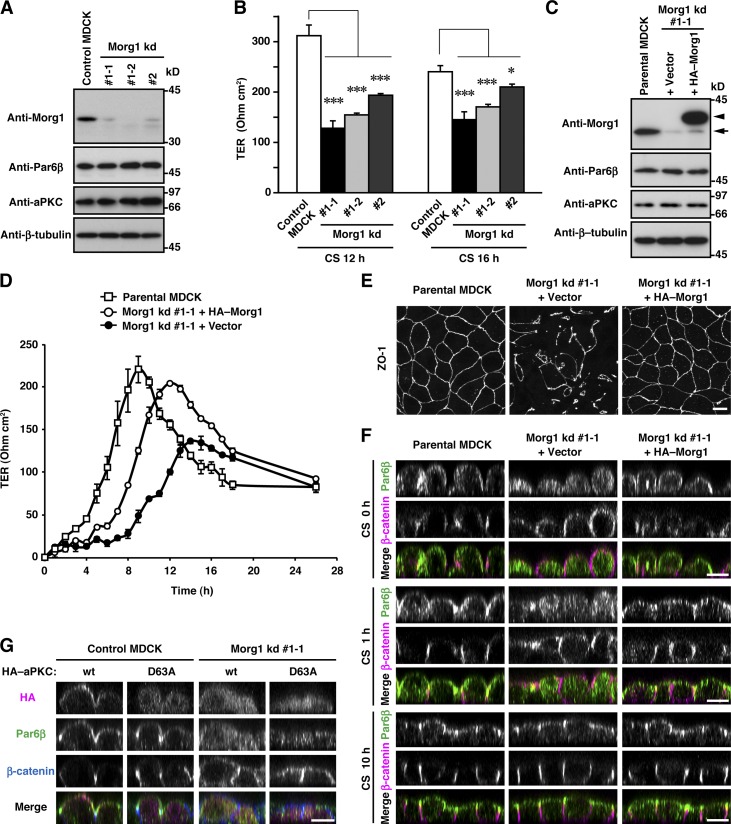
To investigate the function of Morg1 in mammalian epithelial cells, we generated two MDCK cell lines stably expressing a distinct short hairpin RNA (shRNA) against canine Morg1 (Morg1 kd #1 and Morg1 kd #2), in which the protein level of Morg1 was specifically reduced (Figs. 3 A and S1 A). Because Par6–aPKC is known to regulate epithelial apico-basal polarity for TJ development (Suzuki et al., 2001), we investigated the role for the Par6-binding protein Morg1 in TJ assembly: a calcium-switch assay was performed to monitor the transepithelial electrical resistance (TER), a functional measure of TJ integrity, in 2D monolayer culture (Chen and Macara, 2005). The increase in TER of Morg1-depleted cells was significantly delayed compared with control cells, indicating a role for Morg1 in functional TJ formation (Fig. 3 B). Transfection of Morg1 knockdown (kd #1-1) cells with an RNAi-resistant Morg1 cDNA (Fig. 3 C) restored development of functional TJs (Fig. 3 D), which was confirmed by immunostaining for ZO-1, a TJ marker protein (Fig. 3 E). Thus, Morg1 likely plays an important role in TJ development, a landmark of epithelial apico-basal polarization.
Figure 3.
Morg1 is required for Par6 translocation to the apical surface in mammalian epithelial cells. (A) Proteins in lysates of MDCK cells stably expressing Morg1-shRNA #1 (Morg1 kd #1-1 and kd #1-2), Morg1-shRNA #2 (Morg1 kd #2), or empty vector (Control) were analyzed by immunoblot with the indicated antibodies. (B) TER measurements of monolayers of Morg1-depleted cells 12 or 16 h after the calcium switch (CS). The values are means ± SD of three independent experiments. *, P < 0.02; ***, P < 0.001 (Dunnett’s test). (C) Lysates of Morg1 kd #1-1 cells stably transformed with the siRNA-resistant HA–Morg1 plasmid (Morg1 kd #1-1 + HA–Morg1) or empty vector (Morg1 kd #1-1 + Vector) were analyzed by immunoblot with the indicated antibodies. The arrow and arrowhead indicate endogenous Morg1 and HA–Morg1, respectively. (D) TER measurements of monolayers of parental MDCK or Morg1 kd #1-1 cells with or without HA–Morg1 after CS. (E) Representative images of Morg1 kd #1-1 cells with or without HA–Morg1. Cells were fixed 4 h after CS and stained with the anti-ZO-1 antibody. (F) Cross-sectional z-stack analysis of representative confocal images of Morg1 kd #1-1 cells with or without HA–Morg1. Cells were fixed 0, 1, or 10 h after CS, and stained with the indicated antibodies. (G) Cross-sectional z-stack analysis of representative confocal images of control MDCK or Morg1 kd #1-1 cells expressing HA–aPKC (wt or D63A). Cells were fixed 2 h after CS, and stained with the indicated antibodies. Bar, 10 µm.
Morg1 participates in translocation of Par6–aPKC to the apical surface
It seems possible that the Par6-binding protein Morg1 is involved in Par6 translocation because Morg1 contributes to TJ assembly (Fig. 3, D and E), an event that involves Par6 as a crucial regulator (Gao et al., 2002). To test this possibility, we investigated effects of Morg1 depletion on Par6 localization in MDCK cells. Even after calcium depletion from culture media of parental MDCK cells, Par6β staining was retained at the apical surface (Fig. 3 F). In Morg1 knockdown cells, Par6β was initially distributed to the cytoplasm and later to the apical surface; the delay was restored by expression of HA–Morg1, a protein translated from the RNAi-resistant mRNA (Fig. 3 F), indicating a crucial role for Morg1 in Par6 translocation. aPKCι/λ, as well as Par6, translocated to the apical membrane in a manner dependent on Morg1 (Fig. 3 G). aPKC translocation appears to require its interaction with Par6: aPKCι/λ (D63A), defective in binding to Par6 (Ito et al., 2001; Noda et al., 2003), failed to localize to the apical membrane even in cells normally expressing Morg1 (Fig. 3 G). Intriguingly, Morg1 depletion led to mislocalization of the basolateral marker β-catenin to the apical membrane; the delay of β-catenin relocalization to the basolateral membrane was restored upon expression of HA–Morg1 (Fig. 3 F). Thus, Morg1 likely regulates Par6–aPKC translocation to define the apical identity, thereby preventing basolateral proteins from associating with the apical cortex.
Cdc42 also participates in apical localization of Par6–aPKC
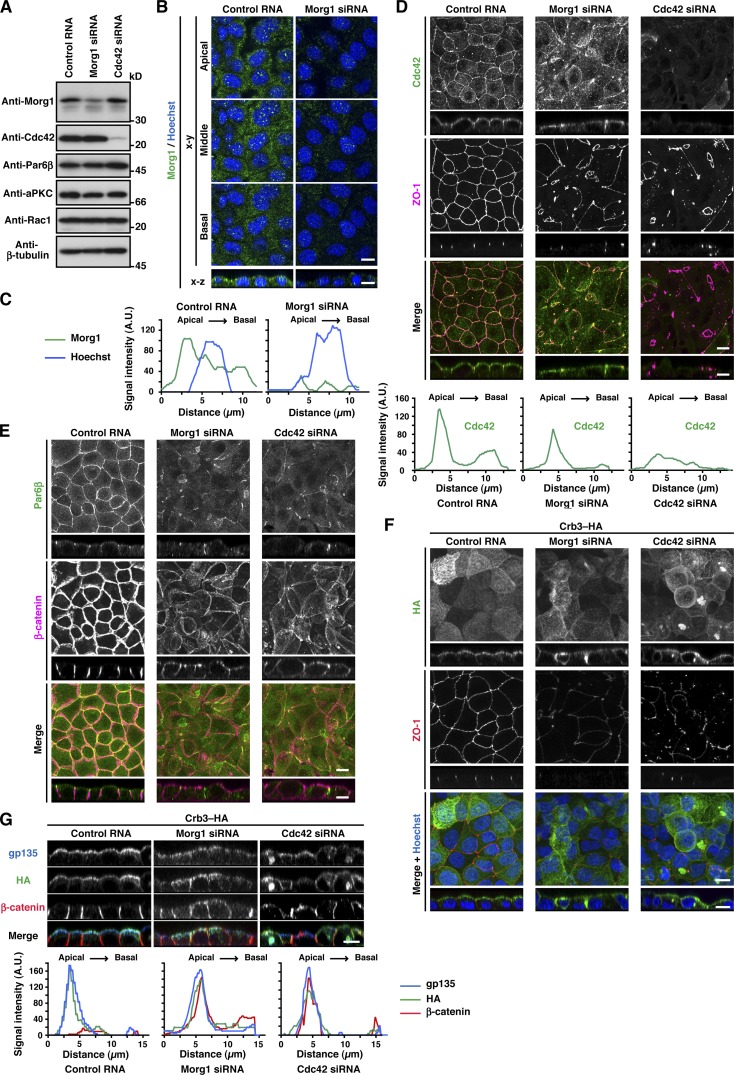
The two Par6 partners Morg1 and Cdc42, intrinsically expressed in MDCK cells (Fig. 4 A), showed different intracellular localization. Morg1 was predominantly distributed to the cytoplasm (Fig. 4 B), although careful microscopic examination revealed that Morg1 is slightly enriched on the apical side (Figs. 4 C and S3). In contrast to Morg1, Cdc42 mainly localizes to the apical surface (Fig. 4 D and S3): the localization was demonstrated by immunostaining of cells fixed with trichloroacetic acid, as described in the Materials and methods. Because the role of endogenous Cdc42 in Par6 targeting in mammalian epithelial cells has not been well studied especially under 2D culture conditions, we studied the role by treating MDCK cells with small interfering RNAs (siRNAs) specific to Cdc42. The treatment resulted in specific depletion of Cdc42 (Fig. 4 A) and a failure in TJ development (Figs. 4 [D and F] and S2 [C and D]). In Cdc42-depleted cells, Par6 failed to localize to the apical surface (Fig. 4 E), whereas β-catenin was redistributed over the entire plasma membrane (Fig. 4 E). Thus, similar to Morg1, Cdc42 participates in apical localization of Par6, thereby controlling cell polarization. Because apical localization of Cdc42 was independent of Morg1 (Fig. 4 D), Cdc42 appears to function at a distinct stage in Par6 translocation; Cdc42 may anchor Par6 to the apical membrane via direct interaction.
Figure 4.
Cdc42 as well as Morg1 regulates Par6 recruitment to the apical surface to define apical identity. (A) Proteins in lysates of MDCK cells transfected with negative control RNA, Morg1-specific siRNA, or Cdc42-specific siRNA were analyzed by immunoblot with the indicated antibodies. (B) Representative confocal images of MDCK cells transfected with negative control RNA or Morg1-specific siRNA. Cells were fixed 4 h after the calcium switch (CS) and stained as indicated. (C) Representative fluorescence intensity of Morg1 and Hoechst staining along the apical–basal axis. (D and E) Representative confocal images of MDCK cells transfected with control RNA, Morg1 siRNA, or Cdc42 siRNA. Cells were fixed 4 h after CS and stained with the indicated antibodies. Representative fluorescence intensity of Cdc42 staining along the apical–basal axis (D). (F and G) Representative confocal images of MDCK cells stably expressing Crb3–HA. Cells were transfected with the indicated RNA, fixed 4 h after CS, and stained with the indicated antibodies. Bar, 10 µm. Statistical analyses of data in C, D, and G are shown in Fig. S3.
Morg1 interacts with Crb3 to facilitate Par6–aPKC binding to Crb3 for membrane localization
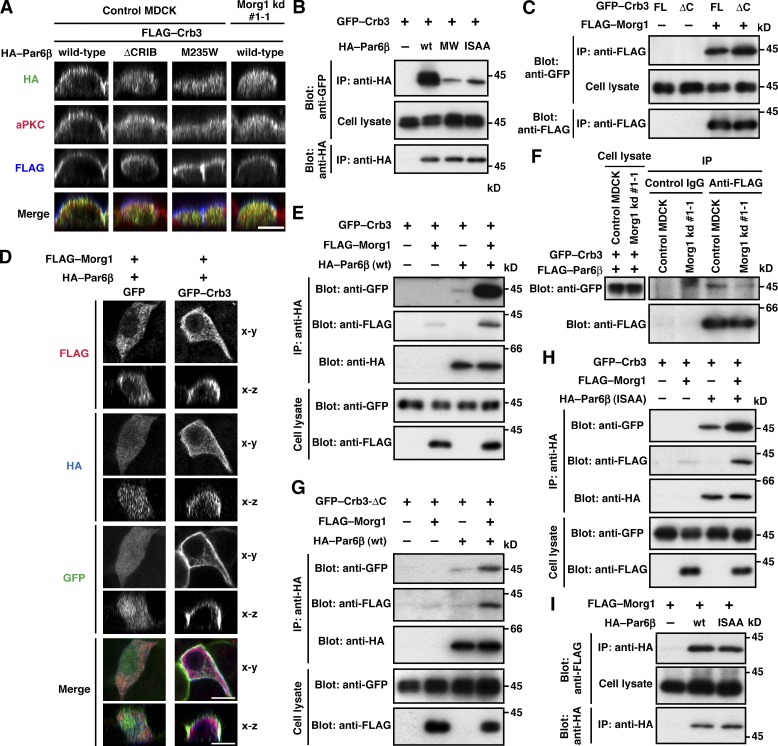
Crb3, another candidate for Par6-anchoring proteins (Schlüter et al., 2009), was present at the apical membrane of MDCK cells under monolayer culture conditions (Figs. 4 [F and G] and S3). Crb3 was at least partly retained at the apical surface even in Morg1-depleted cells (Fig. 4, F and G), consistent with the idea that Crb3 as well as Cdc42 serves as an anchor for Par6–aPKC. To investigate the role of Crb3 in Par6 localization, we coexpressed GFP–Crb3 with wild-type Par6β or a mutant protein carrying the M235W substitution in the PDZ domain, which is capable of binding to Morg1 and Cdc42 but defective in interacting with Crb3 (Fig. 2) in MDCK cells. As shown in Fig. 5 A, wild-type Par6β colocalized with Crb3 to membranes. In contrast, Par6β (M235W) failed to interact with full-length Crb3 (Fig. 5 B) and mislocalized to the cytoplasm (Fig. 5 A). These findings indicate that Crb3 serves as a Par6-anchoring protein. The anchoring function may be expected because Crb3 is a membrane-integrated protein that is initially transported to the pre-apical surface during cell polarization (Datta et al., 2011).
Figure 5.
Morg1 facilitates Par6 binding to Crb3. (A) Cross-sectional z-stack analysis of representative confocal images of MDCK cells expressing GFP–Crb3 and various mutants of HA–Par6β. Cells were stained with the indicated antibodies. (B, C, E, and G–I) Proteins in the lysate of COS-7 cells expressing indicated proteins (Cell lysate) were immunoprecipitated (IP) and then analyzed by immunoblot (Blot) with the indicated antibodies. MW and ISAA denote the M235W and I133A/S134A substitutions, respectively. (D) Representative confocal images of MDCK cells expressing GFP–Crb3, HA–Par6β, and FLAG–Morg1, visualized by GFP (green) and with the anti-FLAG (red) and HA (blue) antibodies. (F) Proteins in cell lysates of control MDCK or Morg1 kd #1-1 cells expressing FLAG–Par6β and GFP–Crb3 were analyzed as in B. Bar, 10 µm.
Par6 binding to Crb3 is known to require the PDZ domain but not the semi-CRIB region (Lemmers et al., 2004). However, a mutant Par6β lacking this region (Par6β-ΔCRIB), as well as Par6β (M235W), was distributed to the cytoplasm in GFP–Crb3-expressing cells (Fig. 5 A), indicating that semi-CRIB also has a role in apical translocation of Par6. Because semi-CRIB is also required for Par6 interaction with Morg1 and Cdc42 (Fig. 2), it seems possible that these two proteins control Par6 binding to Crb3. To test this possibility, we first studied the role for Morg1 in Par6–Crb3 interaction. Indeed, Morg1 did interact with Crb3 (Fig. 5 C), which is consistent with the observation that exogenous expression of Crb3 promoted apical enrichment of Morg1 (Fig. 5 D). Intriguingly, Morg1 enhanced Par6 binding to Crb3 to a large extent (Fig. 5 E). On the other hand, the binding was reduced in Morg1-depleted cells (Fig. 5 F), which is consistent with the finding that Morg1 depletion disrupts apical localization of Par6 in cells expressing GFP–Crb3 (Fig. 5 A). Morg1 can function even in the absence of direct interaction between Par6 and Crb3; indeed, Morg1 facilitated Par6 binding to Crb3-ΔC (Fig. 5 G), a mutant protein incapable of directly binding to Par6 (Fig. 2 F). It is thus likely that Morg1 tethers Par6 to Crb3, i.e., Morg1 allows Crb3 to anchor Par6–aPKC at the apical surface. In addition, Morg1 exerted the tethering effect independently of Cdc42 (Fig. 5 H): Morg1 facilitated Crb3 interaction with Par6β (I133A/S134A), which is able to bind to Morg1 (Fig. 5 I) but not to Cdc42 (Atwood et al., 2007). Thus, Morg1 appears to regulate apical translocation of Par6–aPKC by facilitating Par6 interaction with the apical transmembrane protein Crb3.
Cdc42 replaces Morg1 and further stabilizes Par6–aPKC binding to Crb3 for membrane localization
We next investigated the role of the apically localized small GTPase Cdc42 (Fig. 4) in interaction of Par6 with Crb3. As shown in Fig. 6 A, expression of the constitutively active form of Cdc42 (Q61L), but not the dominant-negative form of Cdc42 (T17N), led to a marked increase in interaction between Par6 and Crb3. On the other hand, Par6β (I133A/S134A) interacted only weakly with Crb3 in cells, whereas wild-type Par6β strongly bound to Crb3, probably because of the function of endogenous Cdc42 (Fig. 5 B). The effect of Cdc42 appears to be due to its direct binding to Par6, because purified Cdc42 (Q61L) also enhanced interaction between the recombinant proteins Par6 and Crb3 (Fig. 6 B). These findings indicate that Cdc42 markedly increases Par6 binding to Crb3, which likely allows the Par6–aPKC complex to fully localize to the apical membrane.
Figure 6.
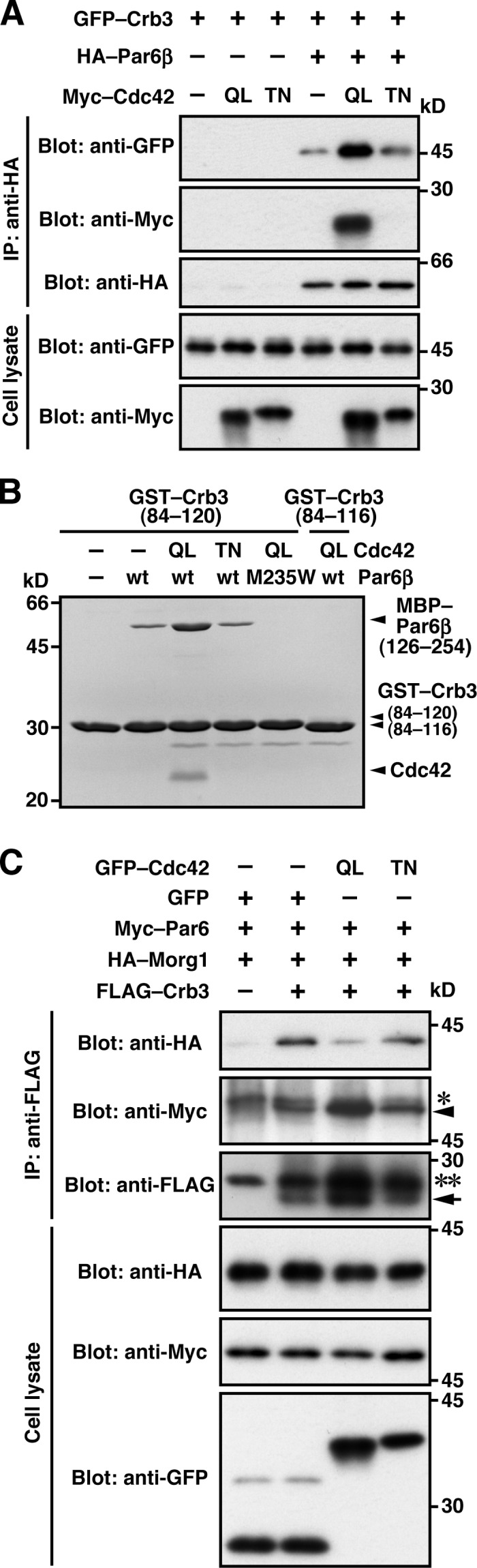
Cdc42 facilitates Par6 binding to Crb3. (A and C) Proteins in the lysate of COS-7 cells expressing indicated proteins (Cell lysate) were immunoprecipitated (IP) and then analyzed by immunoblot (Blot) with the indicated antibodies. In C, the arrow and arrowhead indicate the positions of FLAG–Crb3 and Myc–Par6, respectively; Single and double asterisks denote the heavy and light chains of IgG, respectively. (B) GST–Crb3-(84–120 or 84–116) was incubated with MBP–Par6β-(126–254) in the presence of Cdc42 (Q61L or T17N), and pulled down with glutathione-Sepharose 4B beads, followed by SDS-PAGE analysis with CBB staining.
Because Morg1 and Cdc42 each facilitate Par6–Crb3 interaction as described in the previous section, we next tested whether they function together or separately. As shown in Fig. 6 C, Par6 binding to Crb3 in the presence of Morg1 was further enhanced by GTP–Cdc42 but not by GDP–Cdc42. Importantly, the GTP–Cdc42 form simultaneously induced dissociation of Morg1 from the Crb3 complex (Fig. 6 C). This may be in agreement with the present observation that Cdc42 effectively replaced Morg1 in binding to Par6 (Fig. 2, G and H), whereas the Cdc42–Par6 interaction was not affected by depletion of Morg1 (Fig. S2 B). Thus, it seems likely that Morg1 recruits the Par6–aPKC dimer from the cytoplasm to the apical membrane via interaction with Crb3, and Cdc42 replaces Morg1 at the apical membrane and further stabilizes the complex containing Crb3 and Par6–aPKC.
Morg1 participates in cyst formation by regulating apico-basal polarity
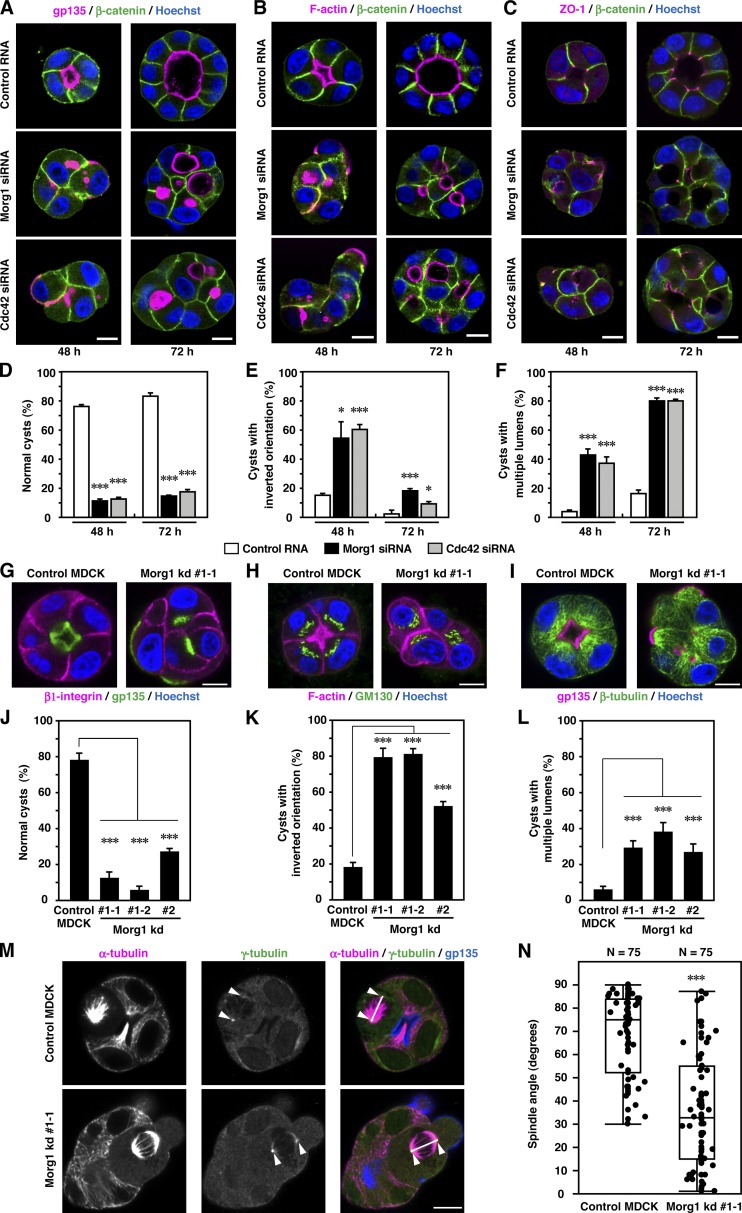
The 3D culture system of MDCK cells provides an excellent model of epithelial morphogenesis in vitro (Datta et al., 2011): culture of a cell embedded in a gel of ECM leads to formation of a spherical cyst with a central lumen, which is enclosed by a polarized cell monolayer. To assess the role for Morg1 in epithelial morphogenesis, we depleted Morg1 with specific siRNA from MDCK cells (Fig. S4, A and B) and cultured them in an ECM-containing Matrigel (Figs. 7 and S4). After culture for 48 h, most control cells formed normal cysts with a solitary lumen, the surface of which was positive for the apical marker gp135 and F-actin (Fig. 7, A, B, D, and J). The basolateral marker β-catenin was exclusively present at sites of cell–cell contact and sites facing the ECM, whereas the TJ marker ZO-1 existed at the site between apical and basolateral membranes (Fig. 7 C). Under Morg1-depleted conditions, “inverted” cysts containing one or more cells that exhibit an inverted polarity with the apical markers facing the ECM (Fig. 7, A, E, G, and K) as well as cysts with multiple lumens and gp135-rich vesicles/vacuoles (Fig. 7 A) were increased (Fig. 7, F and L). Formation of these abnormal cysts appears to be attributable to Morg1 depletion because exogenous expression of RNAi-resistant Morg1 mRNA significantly restored normal cyst development (Fig. S4, C–F). Thus, Morg1 likely controls apico-basal polarity during cyst development. Consistent with this, depletion of Morg1 also affected other polarity-dependent processes: mislocalization of the basolateral marker β1-intergrin (Fig. 7 G), the Golgi apparatus protein GM130 (Fig. 7 H), and microtubule cytoskeleton (Fig. 7 I); and spindle orientation defect in early mitotic cells (Fig. 7, M and N). Similarly, Cdc42 knockdown induced both inversion of cell polarity in cysts (Figs. 7 [A and E]; and S2 E) and formation of cysts with multiple lumens (Fig. 7, A and F).
Figure 7.
Morg1 controls apico-basal polarity during cyst development. (A–C) Representative confocal images of MDCK cells transfected with control RNA, Morg1 siRNA, or Cdc42 siRNA. Cells were grown for 48 or 72 h in 3D culture and stained as indicated. (D–F) Quantification of normally oriented cysts with a solitary lumen (D, normal cysts), cysts with inverted orientation (E), or cysts with multiple lumens (F) in 3D culture of MDCK cells transfected with the indicated RNA. Values are means ± SD from three independent experiments (n ≥ 100 cysts/experiment). *, P < 0.05 (Welch’s t test); ***, P < 0.001 (Student’s t test). (G–I) Representative confocal images of control MDCK or Morg1 kd #1-1 cells. Cells were grown for 48 h in 3D culture and stained as indicated. (J–L) Quantification of normally oriented cysts with a solitary lumen (J, normal cysts), cysts with inverted orientation (K), or cysts with multiple lumens (L) in Morg1-depleted cells. Values are means ± SD from three independent experiments (n ≥ 100 cysts/experiment). ***, P < 0.001 (Dunnett’s test). (M) Representative confocal microscopy images of control MDCK or Morg1 kd #1-1 cells in mitosis. Cells were grown for 48 h in 3D culture and stained with the indicated antibodies. Arrowheads and white lines indicate spindle poles and spindle axes, respectively. (N) Scatter diagrams and box-and-whisker plots of metaphase spindle angles in control MDCK and Morg1 kd #1-1 cells (n = 25 cysts/experiment). ***, P < 0.001 (Wilcoxon rank sum test). Bar, 10 µm.
Morg1 contributes to apical development in early cyst morphogenesis
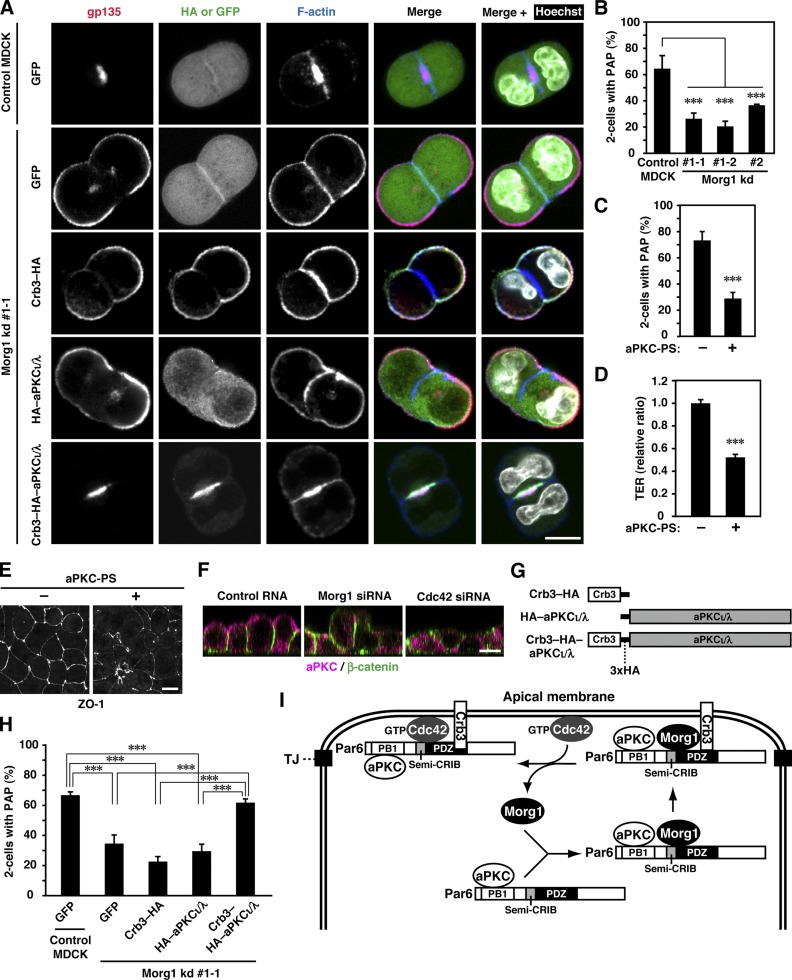
Using the 3D culture system we also studied the role of Morg1 at the two-cell stage of epithelial morphogenesis. In 3D culture, individual MDCK cells divided to predominantly form two-cell aggregates (Schlüter et al., 2009), where the apical marker gp135 as well as F-actin was concentrated at a plasma membrane domain, designated as pre-apical patch (PAP; Fig. 8, A and B). PAP, a structure formed between two divided cells, serves as a precursor of the apical membrane facing a lumen of the cyst (Ferrari et al., 2008). In two-cell aggregates of Morg1-depleted cells, gp135 mislocalized to the plasma membrane facing ECM (Fig. 8 B), which leads to defective formation of PAP at sites facing another cell (Fig. 8 A). Thus, Morg1 likely contributes to apical development at the early stage of epithelial cyst morphogenesis.
Figure 8.
Forced targeting of aPKC to the apical surface rescues apico-basal polarization in Morg1-depleted cells during cystogenesis. (A) Representative confocal images of Morg1 kd #1-1 cells expressing GFP, Crb3–HA, HA–aPKCι/λ, or Crb3–HA–aPKCι/λ in 3D culture. Cells were fixed and stained with the indicated antibodies. (B) Quantification of two-cell aggregates with PAP, a gp135-positive structure, in 3D culture of Morg1-depleted cells. Values are means ± SD from three independent experiments (n ≥ 75 aggregates/experiment). ***, P < 0.001 (Dunnett’s test). (C) Quantification of two-cell aggregates with PAP in 3D culture of MDCK cells treated with (+) or without (−) 40 µM of aPKC-PS. Values are means ± SD from three independent experiments (n ≥ 50 aggregates/experiment). ***, P < 0.001 (Student’s t test). (D) TER measurements of 2D monolayers of MDCK cells 8 h after the calcium switch (CS) in the presence of (+) or absence (−) of aPKC-PS. TER values of each sample were expressed as percentage of those of control cells. Values are means ± SD from three independent experiments. ***, P < 0.001 (Student’s t test). (E) Representative microscopy images of MDCK cells treated with (+) or without (−) aPKC-PS in 2D culture. Cells were fixed 4 h after CS and stained with the anti-ZO-1 antibody. (F) Cross-sectional z-stack analysis of representative confocal images of MDCK cells transfected with the indicated RNA in 2D culture. Cells were fixed 4 h after CS and stained with the indicated antibodies. (G) Schematic structure of aPKCι/λ-fusion proteins used in the present study. (H) Quantification of two-cell aggregates with PAP in 3D culture of Morg1 kd #1-1 cells expressing GFP, Crb3–HA, HA–aPKCι/λ, or Crb3–HA–aPKCι/λ. Values are means ± SD from three independent experiments (n ≥ 100 aggregates/experiment). ***, P < 0.001 (Tukey-Kramer test). (I) A proposed model for Morg1-mediated targeting of Par6–aPKC to the apical surface. For detail, see the text. Bar, 10 µm.
Forced targeting of aPKC to the apical surface restores apico-basal polarity in Morg1-depleted cells
We finally investigated the role for aPKC, tightly dimerized with Par6 (a Morg1 target), in cyst morphogenesis. PAP formation of MDCK two-cell aggregates was significantly blocked by aPKC-PS (Fig. 8 C), a myristoylated pseudosubstrate that specifically inhibits aPKC. This inhibitor also disturbed assembly of functional TJs in MDCK monolayers (Fig. 8, D and E). These findings indicate a crucial role of aPKC activity in epithelial cell polarization. Because Morg1 depletion abrogated not only formation of a solitary lumen (Fig. S4 G) but also apical recruitment of Par6–aPKC (Figs. 3, F and G; 4 E; and 8 F), it seems possible that disturbed cell polarization in Morg1-depleted cells is due to the failure in apical recruitment of aPKC. This possibility may be supported by the observation that knockdown of Cdc42, a protein involved in anchoring of Par6–aPKC to the apical membrane, prevents both apical targeting of Par6–aPKC (Figs. 4 E and 8 F) and formation of a solitary lumen (Fig. S4 G). To clarify the role for Morg1-mediated aPKC recruitment in epithelial cell polarization, we performed a “forced” targeting of aPKC to the apical surface by expressing aPKCι/λ as a protein fused to the apical transmembrane protein Crb3 (Crb3–HA–PKCι/λ) in Morg1-depleted cells (Fig. 8 G). Expression of Crb3–HA–PKCι/λ in Morg1-depleted cells culminated in correct formation of PAP at sites of cell–cell contact (Fig. 8, A and H) and a solitary lumen at the 3–4-cell stage (Fig. S5 A). On the other hand, PAP or lumen formation was not restored by untargeted PKCι/λ (HA–PKCι/λ). The restoration of cell polarity in Morg1-depleted cells was also observed by expression of the aPKC partner Par6 fused to Crb3 (Fig. S5, B and C). These findings indicate that apical targeting of aPKC by itself is crucial for apico-basal polarization of epithelial cells, and that the targeting is normally regulated by Morg1.
Discussion
In the present study we show that Morg1, a WD40 protein that is well conserved from insects to mammals (Vomastek et al., 2004; Hopfer et al., 2006), plays a crucial role in mammalian epithelial morphogenesis by regulating translocation of Par6–aPKC from the cytoplasm to the apical surface. Depletion of Morg1 results in mislocalization of Par6–aPKC to the cytoplasm and defective formation of apico-basal polarity (Figs. 3, 4, and 8), as indicated by disruption of tight junction development in monolayer culture (Fig. 3) and cyst formation in 3D culture (Figs. 7 and 8). The idea that the primary role for Morg1 is to promote apical translocation of Par6–aPKC is also supported by the finding that apico-basal polarity is restored by forced targeting of aPKC to the apical membrane in Morg1-depleted cells (Fig. 8). Morg1 is considered to be essential for mammalian development because Morg1-deficient mice exhibit embryonic lethality at embryonic day 10.5 (Hammerschmidt et al., 2009). It should be noted that several WD40 proteins, such as RACK1 and a subunit of PP2A, also participate in targeting of their binding partners to an intracellular-specific site (Ron et al., 1999; Nunbhakdi-Craig et al., 2002).
The present study also highlights the fact that epithelial cell polarity requires the apically targeted activity of aPKC, an evolutionarily conserved enzyme for polarization in a variety of cells (Knoblich, 2010; St Johnston and Ahringer, 2010). Apico-basal polarity is disrupted when apical localization of Par6–aPKC is prevented by depletion of Morg1 or Cdc42 (Figs. 3 and 4) and when cells are treated with an inhibitor of aPKC activity (Fig. 8). Anchoring of Par6–aPKC to the apical surface appears to be mediated by the apical transmembrane protein Crb3. Although Par6 directly interacts via the PDZ domain with the C-terminal region of Crb3 (Fig. 6; Lemmers et al., 2004), Par6 (M235W), a mutant protein capable of binding to Morg1 and Cdc42 (Fig. 2) but not to Crb3 (Fig. 5), fails to localize to the plasma membrane in cells expressing GFP–Crb3 (Fig. 5). Furthermore, aPKC fused to Crb3 is targeted to the apical membrane, and thus rescues apico-basal polarity in Morg1-depleted cells (Fig. 8). Crb3 is considered to be transported to the region for future apical membranes at an earliest stage of epithelial cell polarization and to function as an apical membrane–initiating factor (Datta et al., 2011). This may be also supported by the present finding that Crb3 is retained at the apical surface even in Morg1- or Cdc42-depleted cells, where Par6–aPKC is largely excluded from the apical membrane (Figs. 3, 4, and 8). However, Crb3 by itself appears to be insufficient to define apical identity because the basolateral protein β-catenin associates not only with the basolateral membrane but also with the apical surface in Morg1- or Cdc42-depleted cells (Figs. 3 and 4). Definition of apical identity in epithelial cells likely requires Crb3-mediated anchoring of Par6–aPKC to the apical membrane.
In addition to the role as a Par6-anchoring protein, Crb3 is also directly involved in the mechanism by which Morg1 targets Par6–aPKC toward the apical surface. Morg1 interacts with Crb3 independently of Par6 and Cdc42 (Fig. 5) and localizes to the apical surface in cells overexpressing Crb3 (Fig. 5), which may explain the reason why Morg1 transports Par6–aPKC solely to the apical membrane, not to the basolateral membrane. Importantly, Morg1 facilitates Par6 binding to Crb3 (Fig. 5), by which Morg1 likely functions in apical translocation of Par6–aPKC.
The small GTPase Cdc42 appears to be another protein involved in anchoring Par6–aPKC to the apical membrane. Here we demonstrate, by fixation of cells with trichloroacetic acid, that endogenous Cdc42 mainly localizes to the apical membrane as well as TJs in monolayer culture of MDCK cells (Fig. 4). Although this fixation is not suitable for analysis of cells in 3D culture (unpublished data), GFP-fused Cdc42 has been shown to localize to the apical membrane in 3D culture (Martin-Belmonte et al., 2007). Apical transport of Cdc42 appears to be independent of Morg1 (Fig. 4), and is instead considered to be mediated by annexin 2, a protein that interacts with phosphatidylinositol 4,5-bisphosphate, an apically enriched phospholipid (Martin-Belmonte et al., 2007). It has been shown that Cdc42 is involved in translocation of Par6–aPKC from the cytoplasm to the apical membrane in Drosophila epithelial cells (Hutterer et al., 2004; Georgiou et al., 2008; Harris and Tepass, 2008; Leibfried et al., 2008) and in 3D culture of mammalian cells (Martin-Belmonte et al., 2007). The involvement of Cdc42 may be supported by the present observations that Par6-ΔCRIB, lacking the semi-CRIB motif required for binding to Cdc42 but not to Crb3 (Lemmers et al., 2004), is mainly distributed to the cytoplasm (Fig. 5), and that depletion of Cdc42 results in mislocalization of aPKC in monolayer culture of MDCK cells (Fig. 8). On the other hand, Cdc42 does not seem to be sufficient for apical localization of Par6–aPKC because Par6 (M235W), which is able to interact with Cdc42 (Fig. 2) but not with Crb3 (Fig. 5), localizes to the cytoplasm in MDCK cells under the conditions where wild-type Cdc42 is fully targeted to the membrane (Fig. 5).
Cdc42 has been shown to directly bind to the semi-CRIB motif adjacent to the PDZ domain of Par6, thereby increasing the affinity of Par6-PDZ for its target hexapeptide VKESLV as a canonical C-terminal ligand (Peterson et al., 2004). However, an intrinsic Par6-PDZ–binding protein that contributes to formation of apico-basal polarity in a Cdc42-dependent manner has not been identified. The present study shows that binding of Par6-PDZ to the Crb3 C-terminal region with the sequence EERLI-COOH is directly facilitated by Cdc42 (Fig. 6), which likely allows Par6–aPKC to fully localize to the apical membrane. A similar Cdc42-mediated recruitment of Par6–aPKC by Crumbs has been postulated at the nascent subapical membrane of Drosophila photoreceptor cells (Walther and Pichaud, 2010).
The adaptor protein Pals1, a homologue of Drosophila Stardust, is known to play an important role in apico-basal polarization of mammalian epithelial cells (Roh et al., 2002; Straight et al., 2004). Whereas Pals1 utilizes an L27 domain to interact with the Pals1-associated TJ protein Patj, an internal region near the N terminus of Pals1 is capable of directly binding to the Par6-PDZ domain (Penkert et al., 2004; Wang et al., 2004). In contrast to Crb3 and Cdc42, however, Pals1 does not seem to participate in apical localization of Par6–aPKC because Pals1 predominantly localizes together with Patj to TJs but not to the apical membrane in mammalian epithelial cells (Roh et al., 2002; Straight et al., 2004). In addition, although Par6 binding to the apical transmembrane protein Crb3 is directly enhanced by Cdc42 (Fig. 6), this GTPase does not affect Par6 binding to Pals1 (Penkert et al., 2004; Peterson et al., 2004). Because both Crb3 and Cdc42 are involved in apical localization of Par6–aPKC (Figs. 4, 6, and 8), it seems possible that Pals1 functions mainly at TJs to regulate apico-basal polarity in a manner independent of Par6–aPKC.
The main apical polarity proteins that are evolutionarily well conserved have traditionally been grouped into two complexes: the Par complex containing Par3/its Drosophila homologue Bazooka, Par6, and aPKC; and the Crb complex composed of Crb, Pals1/Stardust, and Patj. The interplay between these proteins has recently been recognized as more dynamic than previously expected. Indeed, the present study shows that Crb3 likely interacts with Par6 rather than Pals1 at the apical membrane (Figs. 5 and 8). On the other hand, Par3 as well as Pals1 is known to be accumulated solely at TJs in mammalian epithelial cells (Roh et al., 2002; Yamanaka et al., 2003, 2006; Straight et al., 2004; Martin-Belmonte et al., 2007). Because Par6–aPKC is distributed not only at TJs but also throughout the apical surface (Figs. 3, 4, and 8; Yamanaka et al., 2003, 2006; Martin-Belmonte et al., 2007), the Par6–aPKC binary complex (lacking Par3) appears to specifically localize to the apical membrane in mammalian epithelial cells. Distinct localization of the Par6–aPKC binary complex and the Par3-containing complex is also indicated in fruit fly epithelial cells: in cdc42 mutant cells the tight junctional localization of Par6 and aPKC is lost, whereas Bazooka remains localized at junctional sites (Georgiou et al., 2008).
Based on the present findings we propose a model for Morg1-mediated translocation of the Par6–aPKC complex to the apical surface (Fig. 8 I). In the cytoplasm of epithelial cells, the soluble protein Morg1 directly interacts with Par6 via binding to the semi-CRIB and PDZ region. During cell polarization, Morg1 mediates translocation of the Par6–aPKC complex to the apical surface. The apical targeting is probably mediated via Morg1 interaction with the apical transmembrane protein Crb3. Indeed, Morg1 facilitates Par6 binding to Crb3. The translocated Par6–aPKC complex is anchored there by Par6-PDZ–mediated direct binding to Crb3, which is fortified by Cdc42, an apically localized small GTPase. Cdc42 interacts with the semi-CRIB motif of Par6 to induce a conformational change (Peterson et al., 2004), thereby increasing the affinity of the PDZ domain for the Crb3 C terminus. At the apical membrane Cdc42 simultaneously promotes dissociation of Morg1 from the complex containing Crb3 and Par6–aPKC (Fig. 6), probably via replacing Morg1 in binding to Par6 (Fig. 2). Morg1 released from the apical complex may redistribute immediately to the cytoplasm to interact with another Par6–aPKC dimer; indeed, Morg1 is predominantly detected in the cytoplasm. At the apical surface, aPKC activity plays a crucial role in epithelial cell polarization by defining apical identity. aPKC may phosphorylate its target proteins to restrict their localization, thereby defining apico-basal polarity in epithelial cells. Phosphorylation by aPKC is known to prevent basolateral proteins such as Lgl (Betschinger et al., 2003; Plant et al., 2003; Yamanaka et al., 2003) and Par1 (Hurov et al., 2004; Suzuki et al., 2004) from mislocalizing to the apical membrane. aPKC phosphorylates not only Par3 (Nagai-Tamai et al., 2002) but also Bazooka for apical exclusion of this Par3 homologue in Drosophila epithelial cells (Morais de Sá et al., 2010). aPKC is also known to phosphorylate two threonine residues of Crumbs to regulate epithelial cell polarity in Drosophila (Sotillos et al., 2004). Although these residues are conserved in mammalian Crb3, it is presently unknown whether aPKC phosphorylates Crb3 or not. The more detailed mechanism for aPKC function in mammalian epithelial cells should be addressed in future studies.
Materials and methods
Plasmids
The cDNAs for human Par6α, Par6β, and Par6γ were obtained by RT-PCR using RNAs prepared from the human neuroblastoma SH-SY5Y cells, the human renal cell carcinoma KPK1 cells, and the human renal cell carcinoma SN12C cells, respectively; and the cDNA for human aPKCι/λ was obtained by RT-PCR using RNAs prepared from SH-SY5Y cells (Noda et al., 2001, 2003). The cDNA for wild-type Cdc42 was obtained by RT-PCR using RNAs prepared from the human leukemia K562 cells, and mutations leading to the T17N and Q61L substitution were introduced by PCR-mediated site-directed mutagenesis (Noda et al., 2001; Miyano et al., 2009). The cDNA encoding full-length human Morg1 (amino acid residues 1–315) was obtained by PCR using a human fetal brain cDNA library (Takara Bio Inc.). The cDNA encoding full-length human Crb3a (amino acid residues 1–120) was obtained by PCR using kidney cDNAs (Human Multiple Tissue cDNA Panel; Takara Bio Inc.). The DNA fragments encoding various lengths of Par6β and Crb3 were prepared by PCR using their respective full-length cDNAs. Mutations leading to the indicated amino acid substitutions in Par6β and Crb3 were introduced by PCR-mediated site-directed mutagenesis. The cDNAs were ligated to the following expression vectors: pACT2 and pGBK, a modified pGBT vector, for yeast two-hybrid experiments (Ito et al., 2001; Yamaguchi et al., 2007); pGEX-6P (GE Healthcare) for expression as GST fusion protein in Escherichia coli; pMAL-c2 (New England Biolabs, Inc.) for expression as protein fused to maltose-binding protein (MBP) in E. coli; pEF-BOS or pcDNA3.1(+) (Invitrogen) for expression in mammalian cells; and pEGFP-C1 (Takara Bio Inc.) for expression as GFP in mammalian cells. For expression of GFP–CRB3, pcDNA3.1(+) for a fusion protein containing the signal sequence of human Crb3, GFP, and a fragment of Crb3 lacking the signal sequence in this order was prepared. For expression of FLAG–Crb3, the FLAG peptide was inserted into the fusion protein instead of GFP. All the constructs were sequenced for confirmation of their identities.
Antibodies
Rabbit anti-Morg1 sera were raised against the C-terminal 14-amino acid peptide of canine Morg1, and monospecific anti-Morg1 antibodies were affinity purified using a HiTrap NHS-activated HP column (GE Healthcare) conjugated with the immunogen (Fig. S1 A). Rabbit antisera for Par6β were raised against the C-terminal 19-amino acid peptide of human Par6β (Fig. S1, B and C). The anti-gp135 monoclonal antibody 3F2 (Ojakian and Schwimmer, 1988) was generously gifted from G.K. Ojakian (SUNY Downstate Medical Center, Brooklyn, NY). Anti-Cdc42, anti-GM130, and anti–β-catenin monoclonal antibodies were purchased from BD; an anti–β1-integrin monoclonal antibody (AIIB2) from Developmental Studies Hybridoma Bank (Iowa City, IA); an α-tubulin monoclonal antibody (YL1/2) from EMD Millipore; anti–γ-tubulin polyclonal antibodies from Abcam; anti–β-catenin polyclonal antibodies, an anti-ZO-1 monoclonal antibody, anti-aPKC polyclonal antibodies (for immunoblot), and an anti-aPKC monoclonal antibody (for cell staining) from Santa Cruz Biotechnology, Inc.; anti–β-tubulin and anti-FLAG monoclonal antibodies from Sigma-Aldrich; anti-Myc and anti-HA monoclonal antibodies from Roche; an anti-HA monoclonal antibody from Covance; anti-MBP polyclonal antibodies from New England Biolabs, Inc.; a rabbit normal immunoglobulin fraction from Dako; and rat and mouse monoclonal antibodies against GFP from Nacalai Tesque.
Two-hybrid experiments
To obtain novel Par6-binding proteins, the yeast two-hybrid screening with a human fetal brain cDNA library (Takara Bio Inc.) was performed in the reporter strain PJ69-2A using full-length human Par6α as a bait (Izaki et al., 2005; Yamaguchi et al., 2007). Among 3.4 × 105 clones screened, a positive clone obtained encodes full-length Morg1 as revealed by sequencing analysis. For analysis of interaction between Morg1 and Par6, PJ69-4A cells were transformed with combinations of pACT2–Morg1 and pGBK encoding Par6α, Par6β, or Par6γ. After selection for the Trp+ and Leu+ phenotypes, transformed cells were tested for their ability to grow on plates lacking both histidine and adenine in the presence of 10 µM 3-aminotriazole.
An in vitro pull-down binding assay using purified proteins
GST- or MBP-tagged proteins were prepared as described previously (Miyano et al., 2009). In brief, recombinant proteins were expressed in the E. coli strain BL21, and cells were homogenized by sonication; after centrifugation, GST- or MBP-tagged proteins were purified using glutathione-Sepharose 4B (GE Healthcare) or amylose resin (New England Biolabs, Inc.), respectively. Recombinant Cdc42 (Q61L) and Cdc42 (T17N), both of which lack the C-terminal residues 189–191 and carry the C188S substitution for protein stabilization, were expressed as GST fusion protein in the E. coli strain BL21, and proteins were applied to glutathione-Sepharose 4B. GST-free, purified Cdc42 (Q61L) and Cdc42 (T17N) were obtained from glutathione-Sepharose 4B by cleavage of the GST moiety with PreScission Protease (GE Healthcare). GST-Par6β and MBP-Morg1 were incubated for 20 min at 4°C in 500 µl of a solution containing 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.05% Triton X-100, and 20 mM Tris-HCl, pH 7.6; unless otherwise indicated, 0.5 nmol of GST-Par6β and 1.3 nmol of MBP-Morg1 were used. GST–Crb3-(84–120) or GST–Crb3-(84–116) was incubated with MBP–Par6β (126–254) for 20 min at 4°C in 300 µl of the solution described above. Proteins were pulled down with glutathione-Sepharose 4B beads and subjected to SDS-PAGE, followed by protein staining with Coomassie brilliant blue (CBB).
Immunoprecipitation assay
COS-7 cells were transfected using LipofectAMINE (Invitrogen) with indicated cDNAs and cultured for 24 or 48 h in DMEM with 10% fetal calf serum (FCS). MDCKII cells were transfected using Amaxa Nucleofector (Lonza) with indicated cDNAs and cultured in Eagle’s minimal essential medium (MEM) with 10% FCS. Cells were lysed at 4°C with a lysis buffer (150 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.5% Triton X-100, 10% glycerol, and 50 mM Tris-Cl, pH 7.6) containing Protease Inhibitor Cocktail (Sigma-Aldrich). The lysates were precipitated with the indicated antibodies in the presence of protein G–Sepharose (GE Healthcare). The precipitants were analyzed by immunoblot with the indicated antibodies. The blots were developed using ECL-plus (GE Healthcare) for visualization of the antibodies.
Generation of Morg1-deficient MDCK cell lines
For expression of shRNA for Morg1 in canine kidney epithelial MDCKII cells, the following double-stranded synthetic oligomers were ligated to pSUPER.neo+gfp (OligoEngine): Morg1-shRNA#1, 5′-GATCCCCGAGTACACAGGCCACAAGATTCAAGAGATCTTGTGGCCTGTGTACTCTTTTTA-3′ (sense) and 5′-AGCTTAAAAAGAGTACACAGGCCACAAGATCTCTTGAATCTTGTGGCCTGTGTACTCGGG-3′ (anti-sense); Morg1-shRNA#2, 5′-GATCCCCGGCTCGTTTGACAACAGTATTCAAGAGATACTGTTGTCAAACGAGCCTTTTTA-3′ (sense) and 5′-AGCTTAAAAA GGCTCGTTTGACAACAGTATCTCTTGAATACTGTTGTCAAACGAGCCGGG-3′ (anti-sense). With these plasmids, MDCKII cells were transfected using Amaxa Nucleofector and selected in the presence of 600 µg/ml G418. The selection established two MDCK clones stably expressing Morg1-shRNA#1 (Morg1 kd #1-1 and #1-2), one clone expressing Morg1-shRNA#2 (Morg1 kd #2), and one clone containing pSUPER.neo+gfp vector (Vector). A clone expressing both Morg1-shRNA and RNAi-resistant Morg1 mRNA (Morg1 kd #1-1 + Morg1) was obtained by transfection of Morg1 kd #1-1 cells with the pcDNA3.1/Hygro(+) plasmid encoding a mutant Morg1, followed by selection with both 500 µg/ml G418 and 250 µg/ml hygromycin B. In the RNAi-resistant Morg1 cDNA, six nucleotides of the Morg1-shRNA#1 target region (5′-GAGTACACAGGCCACAAGA-3′) were mutated without affecting the amino acid sequence as follows: 5′-GAATATACCGGGCATAAAA-3′ (mutated nucleotides are shown in bold).
Knockdown with siRNA
As double-strand siRNA targeting canine Cdc42 and canine Morg1, 25-nucleotide modified synthetic RNAs (Stealth RNAi; Invitrogen) were used: Cdc42-siRNA-1, 5′-CCACUGUCCAAAGACUCCUUUCUUG-3′ (sense) and 5′-CAAGAAAGGAGUCUUUGGACAGUGG-3′ (antisense); Cdc42-siRNA-2, 5′-GGACCCAAAUUGAUCUCCGAGAUGA-3′ (sense) and 5′-UCAUCUCGGAGAUCAAUUUGGGUCC-3′ (antisesnse); and Morg1-siRNA 5′-CCAGGGATGGCATATCCAGTGTGAA-3′ (sense) and 5′-UUCACACUGGAUAUGCCAUCCCUGG-3′ (anti-sense). Medium GC Duplex of Stealth RNAi Negative Control Duplexes #2 (Invitrogen) was used as a negative control. MDCKII cells plated at 3 × 104/cm2 were transfected with 20 nM siRNA using LipofectAMINE 2000 (Invitrogen) and cultured for 24 h in MEM containing 10% FCS.
Calcium switch assay
MDCKII cells (5 × 106) were seeded on a 35-mm glass-bottom dish (MatTek Corporation) and grown in MEM with 10% FCS, which contains 1.8 mM calcium (normal calcium medium), for formation of confluent monolayers. The monolayer cells were cultured for 16 h in the low calcium medium containing 2.1 µM calcium (S-MEM; Invitrogen) supplemented with dialyzed 10% FCS, and then incubated for 2 or 4 h in the normal calcium medium. For measurement of TER (Matter and Balda, 2003), MDCKII cells (5 × 106) were seeded on a 12-mm Transwell filter (0.4 µm pore size; Corning). The change in TER after addition of calcium was monitored with a Millicel-ERS (EMD Millipore). TER values were calculated by subtracting the blank value from an empty filter and were expressed in ohm·cm2.
Immunofluorescence microscopy
For staining of Par6β, aPKC, and β-catenin, MDCKII cells were fixed for 20 min in 3.7% formaldehyde, and permeabilized for 30 min with 0.5% Triton X-100 in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.4) containing 3% bovine serum albumin (BSA). For staining of Cdc42, ZO-1, and Morg1, cells were fixed for 10 min at 4°C with 10% trichloroacetic acid. For 3D culture, MDCKII cells were trypsinized to a single cell suspension of 1.6 × 104 cells/ml in 5% Matrigel, containing laminin, type IV collagen, and entactin (BD), and 250 µl of the suspension was plated in an 8-well cover glass chamber (Iwaki) precoated with 60 µl of Matrigel, according to the method of Martin-Belmonte et al. (2007). For staining of gp135, β-catenin, and HA-tagged proteins, MDCKII cells plated in Matrigel were fixed for 30 min in 4% paraformaldehyde, and subsequently permeabilized for 1 h in PBS containing 0.5% Triton X-100 and 3% BSA. For ZO-1 staining, MDCKII cells in Matrigel were fixed for 30 min in 4% paraformaldehyde and then for 5 min in 100% methanol on ice, followed by blocking for 2 h with PBS containing 3% BSA. Indirect immunofluorescence analysis was performed using the following as secondary antibody: Alexa Fluor 405–labeled anti–mouse IgG antibodies, Alexa Fluor 488–labeled goat anti–rabbit or anti–mouse IgG antibodies (Invitrogen), Alexa Fluor 546–labeled goat anti–mouse IgG antibodies (Invitrogen); and Alexa Fluor 594–labeled goat anti–rat IgG antibodies or Alexa Fluor 633–labeled goat anti–rat IgG antibodies (Invitrogen). Actin filaments were stained with Alexa Fluor 594 or 647 phalloidin (Invitrogen). Nuclei were stained with Hoechst 33342 (Invitrogen). Confocal images were captured at room temperature on a confocal microscope (model LSM510 or LSM780; Carl Zeiss) and analyzed using the LSM Image Examiner or ZEN (Carl Zeiss), respectively. The microscopes were equipped with a Plan Apochromat 63×/1.4 NA oil immersion objective lens or a C-Apochromat 40×/1.2 NA W Corr water immersion objective lens. For analysis of cyst morphogenesis of MDCKII cells, more than 100 cysts were tested: a normal cyst had a single lumen surrounded by cells that exhibit both intense actin staining at the apical surface and β-catenin staining at the surface facing the ECM; on the other hand, a cyst with actin at the surface facing the ECM was designated as with inverted orientation, and a cyst containing more than two lumens with intense actin staining was designated as with multiple lumens. To discriminate lumens from cytoplasmic gp135-rich vesicles/vacuoles, we defined a lumen as a gp135-rich, F-actin–rich structure that makes a contact with β-catenin–containing membranes via ZO-1–positive junctions.
Forced protein targeting to the apical surface was performed by expressing as a fusion to the C-terminally deleted Crb3 (amino acids 1–116), as described by Zheng et al. (2010). Crb3–HA lacks the C-terminal PDZ-binding motif, but instead harbors the HA tag at the C terminus; and Crb3–HA–aPKCι/λ and Crb3–HA–Par6β additionally contain aPKCι/λ and Par6β, respectively, at the C terminus of Crb3–HA. For analysis of two-cell aggregates expressing GFP, Crb3–HA, HA–aPKCι/λ, Crb3–HA–aPKCι/λ, or Crb3–HA–Par6β, more than 75 aggregates were tested: an aggregate possessing a gp135-positive surface between the cells was designated as two cells with apical surface. Fluorescent intensities were measured by counting gradient values using LSM Image Examiner (Durgan et al., 2011; Ishiuchi and Takeichi, 2012).
Measurement of spindle angle
The measurement of spindle angle was performed according to the method of Jaffe et al. (2008) and Zheng et al. (2010). In brief, MDCK cysts grown for 48 h were fixed and stained with anti–γ-tubulin, anti–α-tubulin, and anti-gp135 antibodies. The acute angle between the spindle axis and the line connecting the centroid of the lumen and the center of the spindle was analyzed.
Statistical analysis
Statistical differences were determined as follows: data for Morg1 knockdown cell lines were analyzed by one-way ANOVA with Dunnett’s multiple comparison of means test; data for cells transfected with siRNA or treated with aPKC-PS were analyzed by two-tailed Student’s or Welch’s t test; data for analysis of spindle orientation were analyzed by two-sided Wilcoxon rank sum test; and data for Morg1 kd #1-1 cells expressing various constructs of Crb3 were analyzed by one-way ANOVA with Tukey-Kramer’s multiple comparison of means test.
Online supplemental material
Fig. S1 shows specificity of anti-Morg1 and anti-Par6β antibodies. Fig. S2 shows that Cdc42 is required for formation of apico-basal polarity in MDCK cells. Fig. S3 shows statistical analysis of apical localization of Morg1, Cdc42, gp135, Crb3, and β-catenin in Morg1- or Cdc42-depleted cells. Fig. S4 shows that Morg1 regulates apico-basal polarity of MDCK cells in 3D culture. Fig. S5 shows that Crb3–Par6 rescues apico-basal polarity in Morg1-depleted cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201208150/DC1.
Supplementary Material
Acknowledgments
We thank G.K. Ojakian for the gp135 (3F2) antibody and N. Tanimizu for helpful discussion; N. Kubo, Y. Kage, and N. Morinaga for technical assistance; M. Nishinou for secretarial assistance; and technical supports from Research Support Center, Kyushu University Graduate School of Medical Sciences, and from the Laboratory for Technical Support, Medical Institute of Bioregulation, Kyushu University.
This work was supported in part by in part by MEXT (the Ministry of Education, Culture, Sports, Science and Technology) KAKENHI grant no. 20117002; by Targeted Proteins Research Program (TPRP) from MEXT; and by JSPS (Japan Society for the Promotion of Science) KAKENHI grant no. 24590385.
Footnotes
Abbreviations used in this paper:
- aPKC
- atypical protein kinase C
- Crb3
- Crumbs3
- Morg1
- mitogen-activated protein kinase organizer 1
- PAP
- pre-apical patch
- TER
- transepithelial electrical resistance
- TJ
- tight junction
References
- Atwood S.X., Chabu C., Penkert R.R., Doe C.Q., Prehoda K.E. 2007. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 120:3200–3206 10.1242/jcs.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- Bryant D.M., Mostov K.E. 2008. From cells to organs: building polarized tissue. Nat. Rev. Mol. Cell Biol. 9:887–901 10.1038/nrm2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova N.A., Knust E. 2009. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 122:2587–2596 10.1242/jcs.023648 [DOI] [PubMed] [Google Scholar]
- Chen X., Macara I.G. 2005. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 7:262–269 10.1038/ncb1226 [DOI] [PubMed] [Google Scholar]
- Datta A., Bryant D.M., Mostov K.E. 2011. Molecular regulation of lumen morphogenesis. Curr. Biol. 21:R126–R136 10.1016/j.cub.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborde S., Perret E., Gravotta D., Deora A., Salvarezza S., Schreiner R., Rodriguez-Boulan E. 2008. Clathrin is a key regulator of basolateral polarity. Nature. 452:719–723 10.1038/nature06828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan J., Kaji N., Jin D., Hall A. 2011. Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J. Biol. Chem. 286:12461–12474 10.1074/jbc.M110.174235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A., Veligodskiy A., Berge U., Lucas M.S., Kroschewski R. 2008. ROCK-mediated contractility, tight junctions and channels contribute to the conversion of a preapical patch into apical surface during isochoric lumen initiation. J. Cell Sci. 121:3649–3663 10.1242/jcs.018648 [DOI] [PubMed] [Google Scholar]
- Gao L., Joberty G., Macara I.G. 2002. Assembly of epithelial tight junctions is negatively regulated by Par6. Curr. Biol. 12:221–225 10.1016/S0960-9822(01)00663-7 [DOI] [PubMed] [Google Scholar]
- Garrard S.M., Capaldo C.T., Gao L., Rosen M.K., Macara I.G., Tomchick D.R. 2003. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J. 22:1125–1133 10.1093/emboj/cdg110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou M., Marinari E., Burden J., Baum B. 2008. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18:1631–1638 10.1016/j.cub.2008.09.029 [DOI] [PubMed] [Google Scholar]
- Goldstein B., Macara I.G. 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 13:609–622 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt E., Loeffler I., Wolf G. 2009. Morg1 heterozygous mice are protected from acute renal ischemia-reperfusion injury. Am. J. Physiol. Renal Physiol. 297:F1273–F1287 10.1152/ajprenal.00204.2009 [DOI] [PubMed] [Google Scholar]
- Harris K.P., Tepass U. 2008. Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183:1129–1143 10.1083/jcb.200807020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer U., Hopfer H., Jablonski K., Stahl R.A., Wolf G. 2006. The novel WD-repeat protein Morg1 acts as a molecular scaffold for hypoxia-inducible factor prolyl hydroxylase 3 (PHD3). J. Biol. Chem. 281:8645–8655 10.1074/jbc.M513751200 [DOI] [PubMed] [Google Scholar]
- Hurd T.W., Gao L., Roh M.H., Macara I.G., Margolis B. 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 5:137–142 10.1038/ncb923 [DOI] [PubMed] [Google Scholar]
- Hurov J.B., Watkins J.L., Piwnica-Worms H. 2004. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14:736–741 10.1016/j.cub.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., Knoblich J.A. 2004. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell. 6:845–854 10.1016/j.devcel.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T., Takeichi M. 2012. Nectins localize Willin to cell-cell junctions. Genes Cells. 17:387–397 10.1111/j.1365-2443.2012.01593.x [DOI] [PubMed] [Google Scholar]
- Ito T., Matsui Y., Ago T., Ota K., Sumimoto H. 2001. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. EMBO J. 20:3938–3946 10.1093/emboj/20.15.3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaki T., Kamakura S., Kohjima M., Sumimoto H. 2005. Phosphorylation-dependent binding of 14-3-3 to Par3β, a human Par3-related cell polarity protein. Biochem. Biophys. Res. Commun. 329:211–218 10.1016/j.bbrc.2005.01.115 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Kaji N., Durgan J., Hall A. 2008. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 183:625–633 10.1083/jcb.200807121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G., Petersen C., Gao L., Macara I.G. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- Kemphues K. 2000. PARsing embryonic polarity. Cell. 101:345–348 10.1016/S0092-8674(00)80844-2 [DOI] [PubMed] [Google Scholar]
- Kempkens O., Médina E., Fernandez-Ballester G., Ozüyaman S., Le Bivic A., Serrano L., Knust E. 2006. Computer modelling in combination with in vitro studies reveals similar binding affinities of Drosophila Crumbs for the PDZ domains of Stardust and DmPar-6. Eur. J. Cell Biol. 85:753–767 10.1016/j.ejcb.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. 2010. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11:849–860 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., Fricke R., Morgan M.J., Bogdan S., Bellaiche Y. 2008. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr. Biol. 18:1639–1648 10.1016/j.cub.2008.09.063 [DOI] [PubMed] [Google Scholar]
- Lemmers C., Michel D., Lane-Guermonprez L., Delgrossi M.H., Médina E., Arsanto J.P., Le Bivic A. 2004. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol. Biol. Cell. 15:1324–1333 10.1091/mbc.E03-04-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., Pawson T. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547 10.1038/35019592 [DOI] [PubMed] [Google Scholar]
- Makarova O., Roh M.H., Liu C.J., Laurinec S., Margolis B. 2003. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene. 302:21–29 10.1016/S0378111902010843 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Gerke V., Mostov K. 2007. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 128:383–397 10.1016/j.cell.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K., Balda M.S. 2003. Functional analysis of tight junctions. Methods. 30:228–234 10.1016/S1046-2023(03)00029-X [DOI] [PubMed] [Google Scholar]
- Miyano K., Koga H., Minakami R., Sumimoto H. 2009. The insert region of the Rac GTPases is dispensable for activation of superoxide-producing NADPH oxidases. Biochem. J. 422:373–382 10.1042/BJ20082182 [DOI] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mirouse V., St Johnston D. 2010. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 141:509–523 10.1016/j.cell.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A., Ohno S. 2002. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 7:1161–1171 10.1046/j.1365-2443.2002.00590.x [DOI] [PubMed] [Google Scholar]
- Noda Y., Takeya R., Ohno S., Naito S., Ito T., Sumimoto H. 2001. Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells. 6:107–119 10.1046/j.1365-2443.2001.00404.x [DOI] [PubMed] [Google Scholar]
- Noda Y., Kohjima M., Izaki T., Ota K., Yoshinaga S., Inagaki F., Ito T., Sumimoto H. 2003. Molecular recognition in dimerization between PB1 domains. J. Biol. Chem. 278:43516–43524 10.1074/jbc.M306330200 [DOI] [PubMed] [Google Scholar]
- Nunbhakdi-Craig V., Machleidt T., Ogris E., Bellotto D., White C.L., III, Sontag E. 2002. Protein phosphatase 2A associates with and regulates atypical PKC and the epithelial tight junction complex. J. Cell Biol. 158:967–978 10.1083/jcb.200206114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojakian G.K., Schwimmer R. 1988. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 107:2377–2387 10.1083/jcb.107.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert R.R., DiVittorio H.M., Prehoda K.E. 2004. Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat. Struct. Mol. Biol. 11:1122–1127 10.1038/nsmb839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson F.C., Penkert R.R., Volkman B.F., Prehoda K.E. 2004. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol. Cell. 13:665–676 10.1016/S1097-2765(04)00086-3 [DOI] [PubMed] [Google Scholar]
- Plant P.J., Fawcett J.P., Lin D.C.C., Holdorf A.D., Binns K., Kulkarni S., Pawson T. 2003. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5:301–308 10.1038/ncb948 [DOI] [PubMed] [Google Scholar]
- Prehoda K.E. 2009. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb. Perspect. Biol. 1:a001388 10.1101/cshperspect.a001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R.G., Abo A., Steven Martin G. 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10:697–707 10.1016/S0960-9822(00)00535-2 [DOI] [PubMed] [Google Scholar]
- Roh M.H., Makarova O., Liu C.J., Shin K., Lee S., Laurinec S., Goyal M., Wiggins R., Margolis B. 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 157:161–172 10.1083/jcb.200109010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Jiang Z., Yao L., Vagts A., Diamond I., Gordon A. 1999. Coordinated movement of RACK1 with activated betaIIPKC. J. Biol. Chem. 274:27039–27046 10.1074/jbc.274.38.27039 [DOI] [PubMed] [Google Scholar]
- Schlüter M.A., Pfarr C.S., Pieczynski J., Whiteman E.L., Hurd T.W., Fan S., Liu C.J., Margolis B. 2009. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol. Biol. Cell. 20:4652–4663 10.1091/mbc.E09-02-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter B.D., Smith S.E., Li R. 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1:a003384 10.1101/cshperspect.a003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillos S., Díaz-Meco M.T., Caminero E., Moscat J., Campuzano S. 2004. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. J. Cell Biol. 166:549–557 10.1083/jcb.200311031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Ahringer J. 2010. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 141:757–774 10.1016/j.cell.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Straight S.W., Shin K., Fogg V.C., Fan S., Liu C.J., Roh M., Margolis B. 2004. Loss of PALS1 expression leads to tight junction and polarity defects. Mol. Biol. Cell. 15:1981–1990 10.1091/mbc.E03-08-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H., Kamakura S., Ito T. 2007. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci. STKE. 2007:re6 10.1126/stke.4012007re6 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Ohno S. 2006. The PAR-aPKC system: lessons in polarity. J. Cell Sci. 119:979–987 10.1242/jcs.02898 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Yamanaka T., Hirose T., Manabe N., Mizuno K., Shimizu M., Akimoto K., Izumi Y., Ohnishi T., Ohno S. 2001. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J. Cell Biol. 152:1183–1196 10.1083/jcb.152.6.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Hirata M., Kamimura K., Maniwa R., Yamanaka T., Mizuno K., Kishikawa M., Hirose H., Amano Y., Izumi N., et al. 2004. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14:1425–1435 10.1016/j.cub.2004.08.021 [DOI] [PubMed] [Google Scholar]
- Vomastek T., Schaeffer H.J., Tarcsafalvi A., Smolkin M.E., Bissonette E.A., Weber M.J. 2004. Modular construction of a signaling scaffold: MORG1 interacts with components of the ERK cascade and links ERK signaling to specific agonists. Proc. Natl. Acad. Sci. USA. 101:6981–6986 10.1073/pnas.0305894101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther R.F., Pichaud F. 2010. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 20:1065–1074 10.1016/j.cub.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Wang Q., Hurd T.W., Margolis B. 2004. Tight junction protein Par6 interacts with an evolutionarily conserved region in the amino terminus of PALS1/stardust. J. Biol. Chem. 279:30715–30721 10.1074/jbc.M401930200 [DOI] [PubMed] [Google Scholar]
- Xu C., Min J. 2011. Structure and function of WD40 domain proteins. Protein Cell. 2:202–214 10.1007/s13238-011-1018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Ota K., Ito T. 2007. A novel Cdc42-interacting domain of the yeast polarity establishment protein Bem1. Implications for modulation of mating pheromone signaling. J. Biol. Chem. 282:29–38 10.1074/jbc.M609308200 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Sugiyama Y., Ishiyama C., Suzuki A., Hirose T., Iwamatsu A., Shinohara A., Ohno S. 2003. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr. Biol. 13:734–743 10.1016/S0960-9822(03)00244-6 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Izumi N., Suzuki A., Mizuno K., Ohno S. 2006. Lgl mediates apical domain disassembly by suppressing the PAR-3-aPKC-PAR-6 complex to orient apical membrane polarity. J. Cell Sci. 119:2107–2118 10.1242/jcs.02938 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Zhu H., Wan Q., Liu J., Xiao Z., Siderovski D.P., Du Q. 2010. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J. Cell Biol. 189:275–288 10.1083/jcb.200910021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.