Meiosis I chromosome migration is biphasic, with an early, slow phase requiring Fmn2 and a later, fast and highly directed phase requiring the Arp2/3 complex.
Abstract
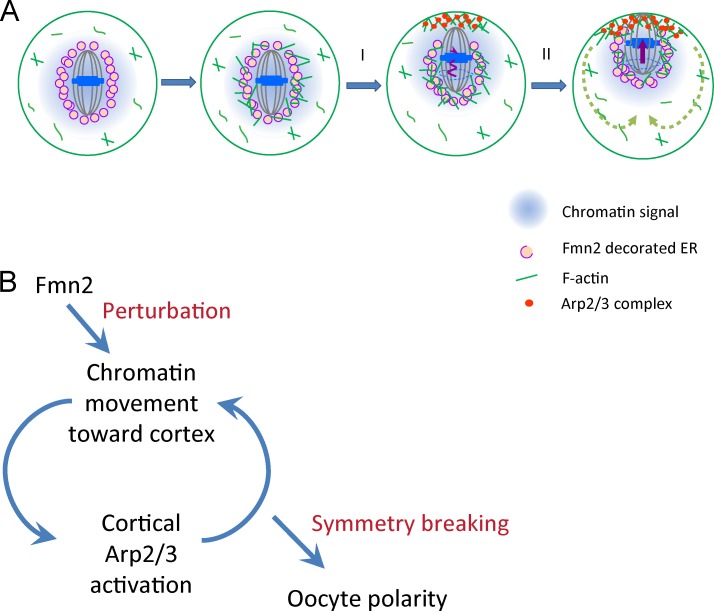
Polar body extrusion during oocyte maturation is critically dependent on asymmetric positioning of the meiotic spindle, which is established through migration of the meiosis I (MI) spindle/chromosomes from the oocyte interior to a subcortical location. In this study, we show that MI chromosome migration is biphasic and driven by consecutive actin-based pushing forces regulated by two actin nucleators, Fmn2, a formin family protein, and the Arp2/3 complex. Fmn2 was recruited to endoplasmic reticulum structures surrounding the MI spindle, where it nucleated actin filaments to initiate an initially slow and poorly directed motion of the spindle away from the cell center. A fast and highly directed second migration phase was driven by actin-mediated cytoplasmic streaming and occurred as the chromosomes reach a sufficient proximity to the cortex to activate the Arp2/3 complex. We propose that decisive symmetry breaking in mouse oocytes results from Fmn2-mediated perturbation of spindle position and the positive feedback loop between chromosome signal-induced Arp2/3 activation and Arp2/3-orchestrated cytoplasmic streaming that transports the chromosomes.
Introduction
Polar body extrusion during maturation of mammalian oocytes encompasses two extremely asymmetric meiotic cell divisions that depend on subcortical spindle positioning and establishment of a cortical actomyosin domain overlying the spindle (Maro and Verlhac, 2002; Brunet and Maro, 2005). Upon induction of meiotic resumption in vitro, germinal vesicle (GV) breakdown (GVBD) and the assembly of the MI spindle occur frequently in the oocyte center, and there is no sign of any cortical actomyosin asymmetry (Longo and Chen, 1985; Deng et al., 2007; Fig. 1 A). Shortly after the alignment of chromosomes in the MI spindle, the chromosomes–spindle complex migrates to a subcortical location, whereby the chromatin signals the assembly of a cortical actomyosin cap (Deng et al., 2007). The ability of the chromatin to signal the cortex is inversely related to the distance between the chromosomes and the cortex and requires the chromosomes to be within 15–25 µm from the cortex (compared with the oocyte radius of ∼35 µm).
Figure 1.
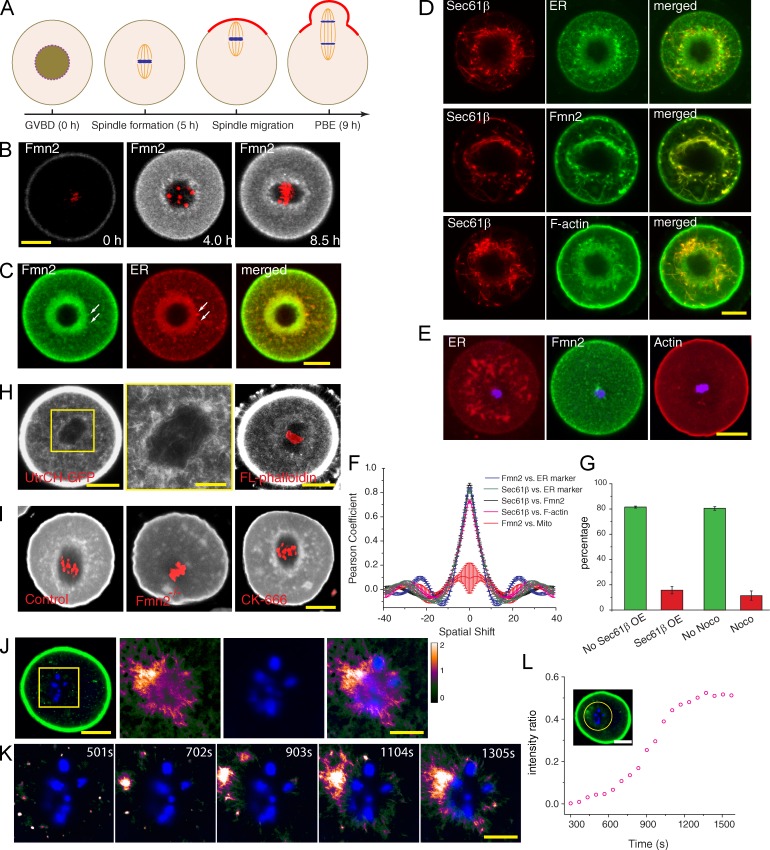
Fmn2 association with ER at spindle periphery and local actin polymerization. (A) Timeline of events in an unperturbed MI mouse oocyte. PBE, polar body extrusion. (B) Localization of Fmn2-AcGFP before and after GVBD (time stamps according to Video 1). Red, chromosomes. (C) Colocalization of Fmn2 with ER stained with blue-white DPX. Arrows point to some distinct structures with colocalization. (D) Overexpression of Sec61-β–mKate2 induces ER tubularization and accumulation of Fmn2 and F-actin along tubular ER. (E) Nocodazole treatment at GVBD + 0.5 h abolished normal Fmn2 distribution and actin enrichment around the chromosomes. (F) Plots of SEM and mean Pearson coefficient (n = 3) as a function of spatial shift in units of micrometers from spatial correlation analysis. (G) Percentage (mean and SEM from three experiments) of oocytes succeeded in chromosome migration when overexpressing Sec61-β–mKate2 (as in D; no Sec61-β overexpression [OE], n = 57; Sec61-β overexpression, n = 71) or treated with nocodazole at GVBD + 0.5 h to block ER congregation (as in E; no nocodazole [Noco], n = 59; nocodazole, n = 65). P < 0.005. (H) F-actin in the MI oocyte (GVBD + 6 h) visualized by strong expression of UtrCH-GFP (left) or microinjection of trace fluorescent phalloidin (right). A magnified view of the boxed region on the left is shown in the middle. (I) Staining of fixed oocytes (GVBD + 6 h) with Alexa Fluor 633–labeled phalloidin. (J) Right three images are time projections of a difference video with chromatin staining (blue) from the box of the UtrCH-GFP image on the left. (K) Montage of new F-actin growth near chromosomes after cytochalasin D washout. Full view is shown in Fig. S1 J. (L) Increase of intensity ratio of chromosome region (circled area; the data shown are from a representative video shown in Fig. S1 J, n = 5). Bars: (B–E, H [left and right], I, J [left], and L) 20 µm; (H [middle], J [right], and K) 10 µm.
Movement of the MI chromosomes to the cortex does not require an intact spindle but is fully dependent on actin (Longo and Chen, 1985; Verlhac et al., 2000) and Fmn2 (Leader et al., 2002; Higgs, 2005; Dumont et al., 2007; Azoury et al., 2008; Chesarone et al., 2010). Two distinct models were proposed to explain the mechanism underlying spindle migration. One model posits that myosin II associated with a spindle pole pulls on an actin filament network (Schuh and Ellenberg, 2008), whereas another hypothesized that actin polymerization stimulated by spindle peripheral Fmn2 pushes the spindle toward the cortex (Li et al., 2008). Further complicating the matter is a recent study that inhibition of the Arp2/3 complex also prevented MI spindle positioning (Sun et al., 2011). The Arp2/3 complex, which nucleates branched actin network, was recently shown to be a target of the chromatin signal and orchestrate a cytoplasmic steaming in meiosis II (MII) to maintain subcortical spindle positioning (Yi et al., 2011). In this study, we investigated the distinct roles for these actin nucleators during MI chromosome migration.
Results and discussion
ER-associated Fmn2 at the spindle periphery is required for MI chromosome migration
Previous studies noted two distinct sites of Fmn2 localization—the cortex and spindle periphery (Li et al., 2008; Schuh and Ellenberg, 2008; Azoury et al., 2011). To better understand the role of Fmn2 localization in MI chromosome migration, we constructed a C-terminal Aequorea coerulescens GFP (AcGFP)–tagged Fmn2, which provides brighter signal compared with the tags used previously. Injected Fmn2-AcGFP mRNA successfully rescued the spindle migration defect in Fmn2−/− oocytes (Fig. S1 A). Fmn2-AcGFP showed cortical localization in mouse oocyte before GVBD (Fig. 1 B). After GVBD, Fmn2 protein level rose, as shown previously (Azoury et al., 2011), and in addition to the cortical localization, Fmn2 gradually accumulated around the forming bipolar spindle (Fig. 1 B, Fig. S1 B, and Video 1). Fmn2-AcGFP accumulation at the spindle periphery was not caused by autofluorescence in the GFP channel (Fig. S1, C and D). Consistent with a previous study (Azoury et al., 2011), during spindle migration, cortical Fmn2-AcGFP was gradually cleared from the cortex in front of the approaching spindle (Fig. S1 E and Video 2), concurrent with the exclusion of microvilli from the cortical cap region (Longo and Chen, 1985). Indeed, cortical Fmn2 appeared to be associated with microvilli (Fig. S2 D).
ER and mitochondria are known to accumulate to spindle periphery at this stage of oocyte maturation (Van Blerkom and Runner, 1984; Calarco, 1995; Mehlmann et al., 1995; FitzHarris et al., 2007). Indeed, Fmn2-AcGFP showed colocalization with the ER dye blue-white DPX (Fig. 1 C). Spatial correlation analysis indicated a high degree of colocalization between Fmn2 and the ER tracker (Fig. 1 F). Thin-sectioning EM confirmed that in MI oocytes, ER surrounding the spindle organizes into vesicular structures (Fig. S2 A, red arrows), whereas mitochondria were further away from the spindle (Fig. S2 A, yellow arrows; and Fig. S1 G). Immunogold labeling showed Fmn2-AcGFP to be associated with the ER vesicles (Fig. S2 B) that contain Sec61-β, an ER resident protein (Fig. S2 C). Immunogold labeling using a published Fmn2 antibody (Azoury et al., 2011) confirmed the association of endogenous Fmn2 with the ER membrane at the spindle periphery (Fig. S2 E).
Expression of the ER protein Sec61-β tagged with mKate2 in oocytes further confirmed the colocalization of Fmn2 with ER (Fig. 1, D and F), but unexpectedly, Sec61-β–mKate2 overexpression induced extensive tubular ER structures from the spindle periphery that were decorated with Fmn2 (Fig. 1 D). F-actin accumulated along the Sec61-β–mKate tubules, whereas cortical actin was not apparently affected (Fig. 1, D and F). The induction of abnormal ER and actin structures at the spindle periphery by Sec61-β–mKate2 severely disrupted spindle migration (Fig. 1 G). Because previous work showed that the accumulation of ER to the spindle periphery is microtubule dependent (Mehlmann et al., 1995; FitzHarris et al., 2007), we treated oocytes with nocodazole immediately after GVBD (GVBD + 0.5 h; Fig. 1 A). This treatment disrupted ER accumulation and Fmn2-AcGFP localization to the central region surrounding the MI chromosomes, but cortical Fmn2 was not affected (Fig. 1 E). The chromosomes failed to migrate to the cortex in 88.7% of these (compared with 18.7% of control) oocytes (Fig. 1 G). Interestingly, Fmn2 does not colocalize with the dispersed ER, suggesting that other signals available in the vicinity of the chromosomes, or a normal ER organization unperturbed by the nocodazole, is required for the association of Fmn2 with ER vesicles.
Our previous work using the F-actin probe FITC-Lifeact showed an accumulation of F-actin around the MI spindle (Li et al., 2008), but other studies using utrophin calponin homology domain (UtrCH)-GFP, another F-actin probe (Burkel et al., 2007), reported a prominent cytoplasmic actin network that appeared to connect the spindle with the cortex. We confirmed that oocytes with high-level expression of UtrCH-GFP indeed showed a dense cytoplasmic actin network; however, spindle migration and polar body extrusion failed in 84.3% (n = 53) of the oocytes in which this was observed (Fig. 1 H). Even in these oocytes, an increased UtrCH-GFP intensity around the spindle was evident (Fig. S1, H and I). F-actin accumulation around the spindle was also observable by microinjecting trace amount of fluorescent phalloidin (Fig. 1 H, right) or by phalloidin staining of fixed oocytes (Fig. 1 I, left). Absence of this actin population in Fmn2-null oocytes indicates that it is dependent on Fmn2 (Fig. 1 I, middle), but it is not affected by CK-666, an inhibitor of the Arp2/3 complex (Fig. 1 I, right; Nolen et al., 2009).
To observe the site of actin polymerization in MI oocytes, cytochalasin D was used to reduce actin density in the cytoplasm observed with UtrCH-GFP. After drug washout, recovery of F-actin was monitored by time-lapse imaging. New actin filaments could be observed to grow at the spindle vicinity, with filaments radiating out from the chromosomal region (Fig. 1 K and Fig. S1 J). A difference video (Brugués et al., 2012) was generated by subtracting the fluorescence image of one frame with that of the previous frame. The time-projected image of the difference video highlights the preferential increase in fluorescence surrounding the chromosomes (Fig. 1, J and L). We conclude that Fmn2 is recruited to the Sec61-β–containing ER structures at the spindle periphery and promotes local actin polymerization and that a normal ER organization is required for spindle migration.
MI spindle and chromosomes migrate in a biphasic manner
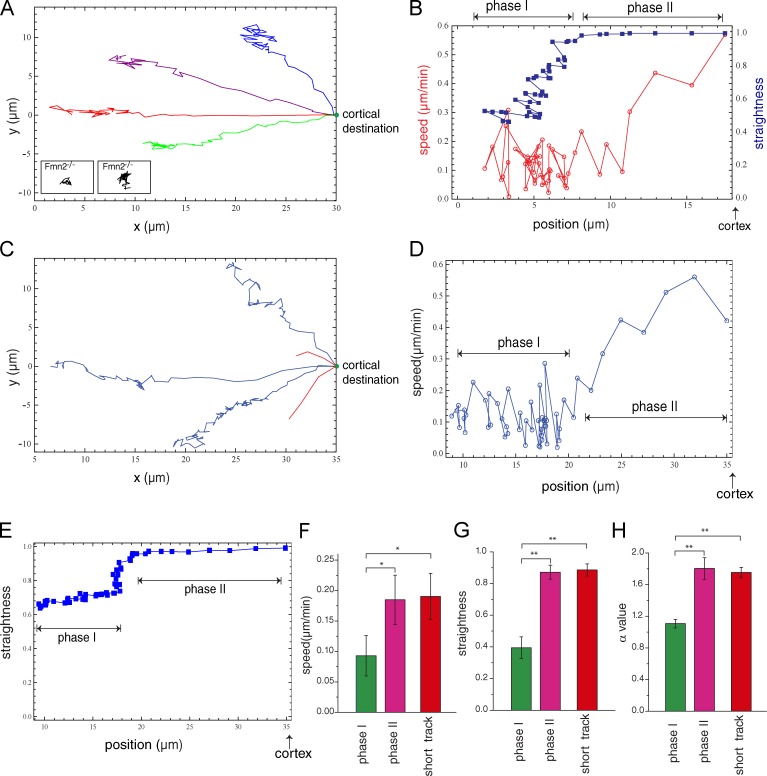
Because before the start of spindle migration the localization of Fmn2 or F-actin does not exhibit any distinct asymmetry, it was unclear how directional movement of the spindle could be initiated as a result of Fmn2-nucleated actin filaments. To shed light on this, we tracked the movement of the chromosomes toward the cortex by time-lapse confocal imaging. The resulting trajectories indicate two distinct phases of the movement (Fig. 2 A and Video 3). In the first phase, spanning the initial 5–10 µm of migration, the trajectories showed low straightness and instantaneous speed (Fig. 2 B and Fig. S3, C–E). Mean square displacement (MSD) analysis suggests that this phase can be described as a confined random walk (α = 0.7; Fig. S3 E). This first phase was followed by an abrupt transition into a second phase, in which the chromosomes moved in a much straighter trajectory toward the cortex at higher speed (Fig. 2 B and Fig. S3, C–E). Trajectories of migration of the spindleless chromosomes exhibited the same biphasic characteristics (Fig. 2, C–H; and Video 4), suggesting that similar mechanisms underlie the migration of the intact spindle or the spindleless chromosome cluster. It is useful to note that in these latter experiments, the spindle was disrupted by using the microtubule inhibitor colcemid, which was added around the time of spindle migration (GVBD + 5 h) but well after ER accumulation to the spindle periphery.
Figure 2.
MI spindle/chromosomes migrate in a biphasic manner. (A) Trajectories of chromosome migration with intact spindle in wild-type oocytes (examples from 11 oocytes analyzed). Insets show two example trajectories of chromosome migration in Fmn2−/− oocytes at the same spatial scale. (B) Speed and straightness as a function of the chromosome position along the axis connecting the initial and end positions of migration corresponding to a trajectory shown in A. “Cortex” refers to the chromosome position when the spindle contacts the cortex. (C) Trajectories of chromosome migration after spindle disassembly from an initial position near the cell center (blue traces) or near the cortex (red traces). Examples are from 12 oocytes analyzed. (D and E) Speed (D) and straightness (E) as a function of the chromosome position of a blue trajectory shown in C. (F–H) Quantification of speed (F), straightness (G), and α value of the MSD analysis (H) of trajectories from a near-center or off-center (short track) initial position (n = 5). Histograms show means and SEM. *, P < 0.05; **, P < 0.005.
Evidence that Fmn2-mediated actin assembly exerts an initial pushing force on the spindle
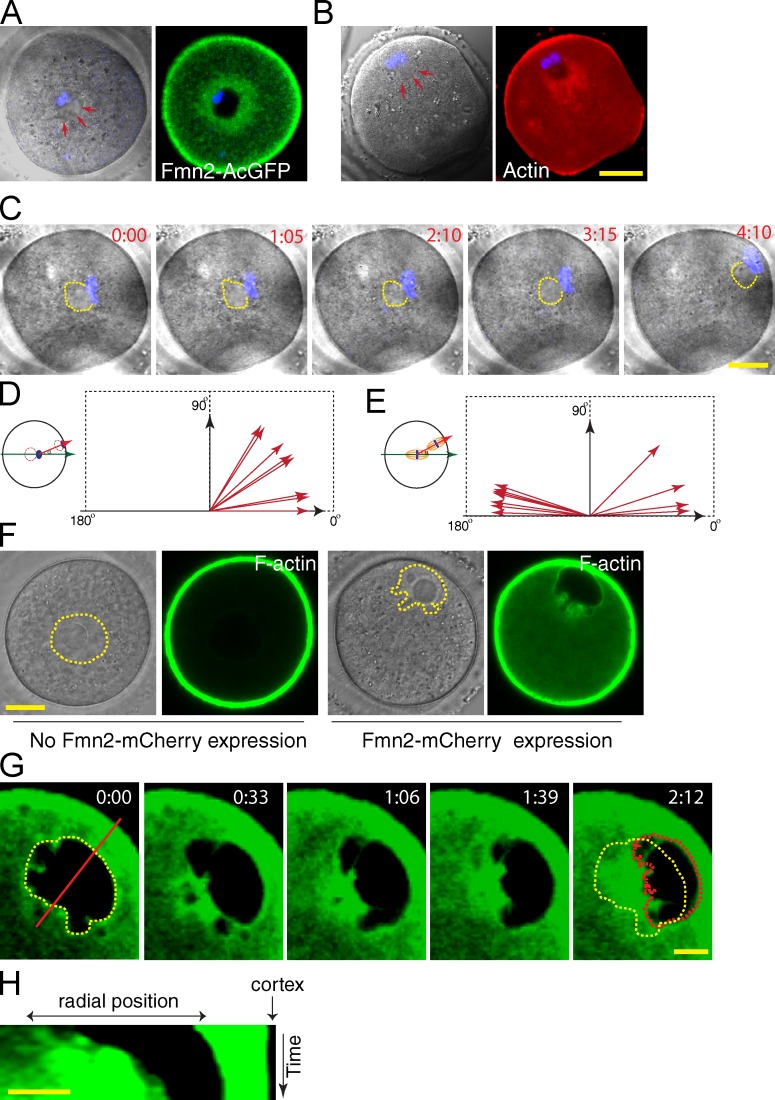
Consistent with previous observations (Azoury et al., 2008; Li et al., 2008; Schuh and Ellenberg, 2008), in the Fmn2−/− oocyte, no chromosome motion could be observed (Fig. 2 A, insets), suggesting that Fmn2 is required for the first phase of chromosome movement. We hypothesize that Fmn2-nucleated actin polymerization at the spindle periphery produces a randomly distributed pushing force on the spindle to drive the first phase of migration. To test this, we asked whether an initially asymmetric Fmn2 distribution would bias the direction of chromosome movement. We took advantage of the observation that spindle disruption immediately before migration initiation caused Fmn2 to distribute asymmetrically relative to the chromosomes. In these oocytes, Fmn2 accumulated around a structure with distinct appearance under transmitted light imaging (referred to hereafter as spindle remnant), and the chromosomes reside on one side of this structure (observed in 68/76 of the colcemid-treated oocytes; Fig. 3 A and Fig. S2, H and I).
Figure 3.
Fmn2-mediated actin assembly exerts a pushing force. (A) Fmn2 around the spindle remnant (outline pointed by arrows). (B) Distribution of actin filaments around the spindle remnant during spindle migration. (C) Time-lapse images of a representative oocyte undergoing chromosome migration in the presence of colcemid. Numbers indicate hours and minutes. (D) Angular distribution (n = 8) of chromosome migration (after spindle disassembly) relative to the spindle remnant–chromosome axis. (E) Angular distribution (n = 11) of spindle migration relative to the long spindle axis. (F) Expression of Fmn2-mCherry in oocytes that failed GVBD causes GV (outlined area in the transmitted light images or the dark area in the GFP channel) deformation and migration to the cortex. An enrichment of F-actin (labeled with UtrCH-GFP) can be observed in the area of deformation. (G and H) Time-lapse images of a representative oocyte undergoing GV (the dark area excluding UtrCH-GFP signal) migration upon Fmn2-mCherry expression. Yellow outlines the initial position of the GV, and red outlines the end position. (H) Kymograph generated along the red line in G. Bars: (A–C and F) 20 µm; (G and H) 10 µm.
Time-lapse imaging revealed that chromosomes always migrated in front of the spindle remnant (Fig. 3 C, a representative example). To quantify this, a polar axis was drawn from the spindle remnant toward the chromosomes located initially at the oocyte center (Fig. 3 D). The angular separation between this axis and that of chromosome movement was <59.15° in all cases, with a mean value of 30.50 ± 7.94° (n = 8; Fig. 3 D), distinct from a random distribution (Pearson correlation r < 0.005). This shows that the Fmn2 asymmetry relative to chromosomes roughly predicts the direction of chromosome movement that is consistent with Fmn2 orchestrating a pushing force behind the chromosomes. Consistently, actin filaments could be observed to concentrate around the spindle remnant behind the migrating chromosomes (Fig. 3 B). As a negative control, the same angular analysis was performed for the intact spindle moving from the center of an oocyte. Because of a lack of initial Fmn2 and F-actin asymmetry, the spindle axis was randomly chosen toward one of the poles in the first frame of the video. As shown in Fig. 3 E, the spindle migrates along its long axis with a deviation of <13.56 ± 3.60° (n = 11), but the orientation was unbiased.
In a small fraction (∼25%) of the oocytes that expressed Fmn2-mCherry, GVBD failed to occur after the oocytes were released from the prophase I arrest and invaginations appeared on the GV membrane (Fig. 3 F and Fig. S2 J), similar to that reported previously (Azoury et al., 2011). Both Fmn2-mCherry and F-actin accumulated at the GV invagination sites (Fig. S2 J) and the GV in 53 of 65 such oocytes, but not in control oocytes (Fig. 3 F and Fig. S2 K), eventually moved to the cortex. Significantly, in all oocytes in which GV moved to the cortex, the side of the membrane facing the cortex was smooth and slightly convex, whereas the opposite side, where actin filaments accumulated, was concave (Fig. 3, F and G). Time-lapse imaging confirmed that GV moved toward the cortex, with the leading side smoothly convex and trailing side invaginating inwardly (Fig. 3, G and H; and Video 5), suggesting that Fmn2-mediated actin polymerization exerted a pushing force at the backside of the GV during its migration. A similar nuclear morphology was reported recently in a study of nuclear migration in Drosophila melanogaster oocytes as a result of a microtubule-based pushing force (Zhao et al., 2012).
Fast phase of spindle migration depends on the Arp2/3 complex and cytoplasmic streaming
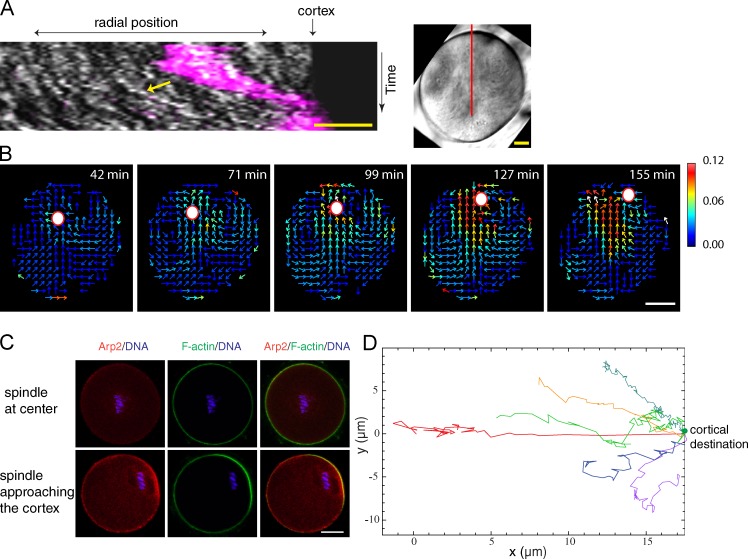
While tracking chromosome movement, we noticed that cytoplasmic streaming became visible during the second phase of chromosome movement, with cytoplasmic particles moving at a similar speed as the chromosomes, and streaming continued even after chromosomes touched the cortex (Fig. 4 A and Video 4). In the case of an intact spindle, cytoplasmic streaming also initiated during the fast migration phase, continued after first polar body extrusion, and maintained MII spindle at the cortex (Video 6 and Video 7). We did not observe any cortical indentation toward which the spindle migrates (Schuh and Ellenberg, 2008), but instead, the cortical region above the spindle often protrudes slightly outward (Fig. S2 L and Video 7). The presence of a pushing force on the MI spindle was indicated by shortening of the spindle immediately upon the leading pole reaching the cortex (Fig. S2, F and G; and Video 2).
Figure 4.
Fast migration phase depends on the Arp2/3 complex and cytoplasmic streaming. (A) Kymograph (left) along the red line shown in the right image (a time projected image from a transmitted light video). Chromosomes are shown in magenta; cytoplasmic particles are shown in white (yellow arrow). (B) Montages of vector maps of cytoplasmic streaming. The heat bar unit is given in micrometers per minute. (C) Localization of Arp2/3 complex by anti-Arp2 immunostaining. (D) Chromosome trajectories in a control (red) or CK-666–treated oocytes in which the chromosomes moved to the cortex (all other colors). Examples are from 15 oocytes analyzed. Bars: (A) 10 µm; (B and C) 20 µm.
The pattern of the cytoplasmic streaming (Fig. 4 B) was reminiscent of the Arp2/3-dependent cytoplasmic streaming observed previously in MII oocytes (Yi et al., 2011), and the streaming speed increased as the chromosome to cortex distance decreased (Fig. 4 B). Immunofluorescence staining using an anti-Arp2 antibody revealed no cortical Arp2/3 localization when the chromosomes were centrally located, but as the chromosomes approached the cortex, an Arp2 cap became apparent, colocalizing with a cortical actin cap (Fig. 4 C). To test Arp2/3 complex’s role in the two phases of chromosome movement, we treated the oocytes with CK-666. Time-lapse imaging showed that 48% of CK-666–treated oocytes (n = 35) failed to move their chromosomes/spindle to the cortex, whereas the rest eventually succeeded after a prolonged period (>8.5 h compared with ∼5 h in control oocytes). In the first group, trajectory analysis showed a random, nondirectional movement with parameters resembling the first phase of spindle motion in untreated oocytes (Fig. S3, F and G). However, because a small percentage of the spindles (21.4%, n = 27) from the control group also failed to reach the cortex, we focused on the group that eventually reached the cortex. In these oocytes, movement parameters for the entire trajectory resembled those of the first migration phase in untreated oocytes. Cytoplasmic streaming was not observed in 18/18 of these oocytes, and the second phase of straight and accelerated movement was absent (Fig. 4 D; Fig. S3, A–E; and Video 8), suggesting that the Arp2/3 complex and cytoplasmic streaming are required for the second but not the first phase of chromosome migration.
In a small fraction of the oocytes whose spindle was disrupted with colcemid, the position of the chromosomes was strongly skewed toward the cortex. In these oocytes, cytoplasmic streaming and the straight/fast phase of chromosome movement started at the outset of chromosome migration (Fig. 2, C and F–H; and Video 4), suggesting that the first phase could be bypassed as long as the chromosomes are close enough to the cortex to activate the Arp2/3 complex. To ask whether the role of Fmn2 is simply to bring the chromosomes closer to the cortex, we examined Fmn2−/− oocytes in which the chromosomes were naturally located close to the cortex. Surprisingly, the spindle did not move in such oocytes (n = 23). In these oocytes, despite the proximity of chromosomes to the cortex, the Arp2/3 complex did not localize (Fig. S3 I), and cytoplasmic streaming was not observed (Fig. S3 H). This observation indicates that Fmn2 may have a role in the communication between the chromatin and the cortex to enable Arp2/3 complex activation or localization, although Fmn2 is not required for all features of oocyte cortical polarity (Dumont et al., 2007).
Collectively, the aforementioned results lead us to propose a biphasic model for MI chromosome migration (Fig. 5 A). In the first phase, Fmn2 protein is recruited to the spindle periphery via ER vesicles, and local actin polymerization produces a random and heterogeneous pushing force on the spindle. We assume that the net magnitude of this force is relatively small and direction stochastic, but it could nonetheless displace the chromosomes from a central location toward the cortex. In the second phase, when the distance between the chromosomes and the cortex falls within ∼25 µm, a threshold for the chromatin signal to reach the cortex, Arp2/3 is activated at the proximal cortex to initiate cytoplasmic streaming, which applies a directional pushing force on the spindle. The magnitude of this force increases as the chromosomes migrate toward the cortex as a result of proximity-enhanced cortical activation by the chromatin signal (Deng et al., 2007), resulting in an irreversible commitment of the migration toward the cortical destiny and thus decisive symmetry breaking. We note that although this model does not require an initial Fmn2 asymmetry, an Fmn2 asymmetry was often observed as the spindle moved closer to the cortex (Fig. S1 E, an example), which may help enhance the directionality of the movement.
Figure 5.
Model of biphasic spindle movement and symmetry breaking. (A) Fmn2- and Arp2/3-regulated actin-based forces drive the first and second phase of chromosome migration, respectively. (B) A conceptual model of how symmetry breaking occurs in MI oocytes. See Discussion for detailed explanations.
Our model could explain two well-known characteristics of MI chromosome migration: (1) the migration tends to occur along the long axis of the spindle and (2) an initially off-centered spindle tends to move toward the proximal cortex. The first tendency could be an outcome of the difference in the drag force on the spindle that favors pole-led, as opposed to sideways, movement, whereas the second could be a result of the expected higher probability for the chromosomes to first reach the threshold, through a random walk, in the direction of the initial proximal cortex and undergo fast migration toward it. Our model also does not exclude a proposed role for myosin-II in chromosome migration (Schuh and Ellenberg, 2008). Recent studies have shown that myosin-II promotes actin depolymerization in motile (Wilson et al., 2010; Mseka and Cramer, 2011) and dividing cells (Mendes Pinto et al., 2012) and in vitro (Murrell and Gardel, 2012; Reymann et al., 2012). As such, myosin-II could play a role at the spindle poles, as suggested, to facilitate actin turnover in a treadmilling network that drives the cytoplasmic streaming (Yi et al., 2011). In summary (Fig. 5 B), our model implies that the Fmn2-mediated initial movement generates a “perturbation” on chromosome position, whereas decisive symmetry breaking and the continued maintenance of oocyte polarity are accomplished through the positive feedback loop between chromatin-induced cortical activation and cortical Arp2/3-orchestrated chromosome movement via cytoplasmic streaming.
Materials and methods
Oocyte dissection and culture
All animals used in this work were handled in accordance with guidelines defined by Institutional Animal Care and Use Committee of the Stowers Institute. MI oocytes were dissected from the ovaries of 5–9-wk-old wild-type or Fmn2-null female mice (provided by P. Leder, Harvard Medical School, Boston, MA) and cultured in M16 medium (EMD Millipore) containing 0.2 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich) at 37°C in a 5% CO2 atmosphere. To track the chromosome migration, we first sorted for oocytes with GV roughly at the cell center (except for observation of “short track” trajectories). To allow meiosis resumption, the oocytes were washed and then cultured in 3-isobutyl-1-methylxanthine–free M16 medium. In the case of drug treatment experiments, the medium was supplemented with 50 µM CK-666 (Chemdiv), 5 µg/ml cytochalasin D (Sigma-Aldrich), or 100 ng/ml colcemid (Invitrogen).
Plasmid construction and mRNA in vitro transcription
Full-length Fmn2 ORF was inserted into the pGEMHE vector in frame with AcGFP. Sec61-β ORF (provided by T. Rapoport, Harvard Medical School, Boston, MA) was inserted into pGEMHE-mKate2. Fmn2-mCherry and F-actin probe pCS2+-UtrCH-EGFP were gifts from J. Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany) and W. Bement (University of Wisconsin-Madison, Madison, WI), respectively. Capped mRNA was synthesized from linearized plasmid templates using T7 or SP6 mMessage mMachine (Ambion), poly-A tailed with Poly(A) Tailing kit, and purified with MEGAclear kit (Ambion).
Microinjection and time-lapse confocal microscopy
Microinjection was performed in M2 medium (EMD Millipore) using micromanipulators (Narishige). Typically, 10–12 pl (∼4% of the oocyte volume) of 0.5–1.5-µg/µl mRNA was injected into the oocytes. Time-lapse imaging was performed with a microscope (LSM 510 META; Carl Zeiss) equipped with a Plan Apochromat 40×, 1.2 NA water immersion objective. AcGFP/EGFP was excited with 488-nm Argon laser and detected with a 505–550-nm band pass. mCherry/mKate2 was excited with a 561-nm laser line and detected with a 575–615-nm band pass.
Immunofluorescence and immuno-EM
Mouse oocytes were immunostained as described previously (Yi et al., 2011). In brief, the oocytes were fixed in 3% paraformaldehyde in PBS and blocked with 0.1 M glycine solution. After permeabilizing with 0.1% Triton X-100 and blocking with blocking buffer (0.3% BSA and 0.01% Tween 20 in PBS), the oocytes were subjected to consecutive primary and secondary antibody incubation in the blocking buffer. The primary antibodies used are rabbit anti-Arp2 (Santa Cruz Biotechnology, Inc.; 1:200) and mouse anti–α-tubulin (Sigma-Aldrich; 1:2,000). Image acquisition of fixed oocytes was performed using a 40×, 1.3 NA oil objective on the aforementioned confocal microscope. For phalloidin staining presented in Fig. 1 I, the oocytes were quickly fixed with 3% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% Triton X-100 for 20 min, and then stained with Alexa Fluor 633–labeled phalloidin for 40 min. The oocytes were washed three times in M16 medium drops and subjected to confocal microscope imaging.
EM and immuno-EM were performed as described previously (Li et al., 2008). In brief, for ultrastructural observation, oocytes were fixed with 2% glutaraldehyde in 0.1 M Hepes, pH 7.3, 0.05% saponin, and 0.2% tannic acid (freshly filtered) for 40 min. After removal of the fixative by several washes in 0.1 M Hepes, the samples were postfixed with 1% OsO4 (aqueous) for 10 min, washed, and then dehydrated through a graded series of ethanol to 100%, infiltrated, and embedded in Epon resin. The resin was polymerized at 37 and 60°C for 48 h. Ultrathin sections (∼50–70 nm) were cut on an ultramicrotome (EM UC6; Leica) using diamond knives and then stained with 2% uranyl acetate and 1% lead citrate for 10 and 5 min, respectively.
For on-section labeling immuno-EM, the oocytes were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde in PBS, pH 7.4, for 4–6 h and washed three times in 0.1 M PBS for 15 min each. The samples were then dehydrated, infiltrated, and embedded in London resin white resin. The sections were stained with rabbit anti–Sec61-β (EMD Millipore) and/or mouse anti-GFP (A6455; Invitrogen) or rabbit anti-Fmn2 (provided by M.-H. Verlhac, Centre National de la Recherche Scientifique/Université Pierre et Marie Curie, Paris, France) and mouse anti-KDEL (Santa Cruz Biotechnology, Inc.) followed by 5- or 10-nm gold-conjugated goat anti–rabbit or anti–mouse secondary antibodies (Sigma-Aldrich; 1:20 dilution).
Colocalization analysis
Colocalization analysis was performed on dual-color confocal images using a spatial correlation approach (van Steensel et al., 1996). All processing and analysis was performed with custom plugins written for ImageJ (National Institutes of Health; see Supplemental material). Images were first scaled down with bilinear interpolation to match the largest pixel size and cropped to a 128 × 128–pixel region encompassing the oocyte. To calculate the colocalization profile of molecules around spindle periphery, we only focus on the signals within this region. Therefore, we replaced all pixels outside of a boundary drawn just inside the cortex with the median value of the intensity inside the boundary. Although this procedure introduces a small amount of spatial correlation, it is small relative to true positive correlation and displays a much broader spatial profile, which doesn’t interfere with the signal from spindle region correlation. The spatial Pearson product–moment correlation function was calculated as follows:
Here, covxy[m,n] is the covariance of m and n variables over both spatial coordinates and varxy[m] is the similarly defined variance. Subscripts g and r refer to independent detection channels (e.g., green and red). Sharply peaked correlation functions approaching 1 are indicative of strong spatial localization on subcellular length scales, whereas broader peaks indicate larger scale or background colocalization. Zero values indicate randomly localized signals, whereas negative values are indicative of spatial exclusion. The spatial correlation images were shifted so that the origin was in the center, and a circular average was calculated to represent correlation versus radial shift. Correlation profiles from at least three images were averaged to obtain the final correlation plots, with error bars representing SEM for each spatial shift.
Chromosomes/spindle tracking, trajectory analysis, and MSD analysis
To track the trajectory of the chromosomes/spindle, the images were aligned to eliminate the sample drift. Single multicolor z slices were registered based on the transmitted light channel using a modified version of the StackReg plugin (Thévenaz et al., 1998) for ImageJ. Centroid tracking of the chromosomes was performed after Gaussian smoothing to eliminate noise and simple thresholding segmentation in ImageJ. The trajectories were then exported and analyzed with Mathematica (Wolfram Research). Straightness of the trajectory (or any partial trajectory up to a certain time point) was defined as ratio D/L of the displacement (D) from the initial point to the end point over the path length (L) between the same points. The instantaneous speed of the cell was computed as a ratio of the displacement between consecutive time steps at a given point of the trajectory over the time between the steps. For mean square displacement (MSD) analysis, the MSD was defined using the equation in which τ defines the time step between the trajectory points r(t) and r(t + τ). The value of MSD was calculated with Mathematica using a sliding window of five time points along the trajectory. The resulting data were fitted to the equation MSD(τ) = 4Dτα using the nonlinear least squares method in Mathematica. Here, D is the diffusion coefficient, and the exponent, α, indicates nonrandom diffusion. Values of α < 1 correspond to a restricted subdiffusion, whereas values of α > 1 indicate superdiffusive motion. The trajectories of spindle/chromosomes that started near the cell center were characterized by a biphasic structure in which the first phase corresponds to the random motion with low values of α and directionality, whereas the second phase was highly directed (α = ∼1.8). The boundary between these two phases was found using the dependence of the straightness on the distance from the cortex, which exhibits a sharp increase between the phases.
For the straightness analysis of chromosome migration in the presence of colcemid (Fig. 3 D), the trajectory orientation was defined as an angle between two vectors—the first one points from the centroid of the spindle remnant to the chromosomes at the initial moment of time, whereas the second one points from initial chromosome position to its final position at the cell cortex. The angular distribution was quite narrow and was compared with 1,000 randomly generated uniform distributions in the range of angles between 0 and 180°. The Pearson correlation coefficient (r) between the observed distribution and the random ones was shown to be <0.005.
Spatiotemporal image correlation spectroscopy (STICS) analysis and velocity field measurement
STICS analysis was performed as previously described (Yi et al., 2011) using a custom ImageJ plugin for ImageJ (see Supplemental material). In brief, registration was performed as described for chromosome tracking analysis to eliminate sample drift. The sections were then scaled by a factor of 2 with bilinear interpolation and rotated so that the final chromosome position was at the top of the image. The cytoplasmic region excluding the cortex was selected, and all other regions were replaced by the average intensity of the cytoplasmic region. Then, the average intensity image for the entire video was subtracted from each frame, adding back the spatial average intensity of each frame. 32 × 32–pixel (9 × 9 µm) regions overlapping by 16 pixels were cross correlated with the same regions in images collected 17 min later, and maxima of the cross correlation images were determined as described previously (Yi et al., 2011). The vectors of these maxima were used to determine the velocity vectors for each region. As a modification of the previous method, we performed running STICS analysis for overlapping 10 frame segments separated by five frames from one another. In that way, STICS vector maps were generated at 30-min intervals throughout the spindle migration process (Fig. 4 B). The average streaming velocities were calculated by averaging velocity magnitudes from the STICS analysis over a specified region of interest. For tracking of the chromosomes and spindle, chromosome coordinates at each time point were determined by segmentation and center of mass calculation.
Kymograph, montage, and difference video
Before kymograph generation, images were aligned as discussed for the chromosomes/spindle tracking. Kymographs were typically averaged over a width of five pixels to portray representative behavior. To generate the montage and the difference video, first the data series was aligned to eliminate oocyte motion as described for the chromosomes/spindle tracking. Fluctuations in background and cytosolic fluorescence were eliminated by dividing from each image a cytosolic average for that image. This value was calculated as an average from a cytosolic (away from the spindle) region. For difference videos, each frame was subtracted from the next frame, as previously described (Brugués et al., 2012). For example, time point 1 was subtracted from time point 2, and then, time point 2 was subtracted from time point 3, etc. To generate the image shown in Fig. 1 J, the difference video was summed over time. This was overlaid with a simple temporal sum projection of the Hoechst signal.
Statistical analysis
Statistical analysis of the data was performed in Excel (Microsoft) or OriginPro 8.0 (OriginLab). P-values were determined using the Student’s t test.
Online supplemental material
Fig. S1 shows additional data on Fmn2 and F-actin organization. Fig. S2 shows Fmn2 immuno-EM and additional evidence that Fmn2 and actin exert a pushing force. Fig. S3 shows additional data on biphasic movement and Arp2/3′s role. Video 1 shows Fmn2 dynamics before spindle migration. Video 2 shows Fmn2 cortical exclusion and spindle compaction. Video 3 shows biphasic migration of an intact spindle. Video 4 shows chromosome migration after spindle disassembly. Video 5 shows GV migration upon Fmn2-mCherry expression. Video 6 shows an oocyte undergoing spindle migration followed by first polar body extrusion. Video 7 shows an oocyte undergoing spindle migration followed by first polar body extrusion and MII spindle positioning. Video 8 shows chromosome movement when Arp2/3 was inhibited. A zip file is also provided that contains custom plugins written for ImageJ. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211068/DC1.
Supplementary Material
Acknowledgments
We thank Melainia McClain (Stowers Institute) for assistance in EM, Jenny Reynolds and the Laboratory Animal Facility of Stowers Institute for mice maintenance, Marie-Helene Verlhac for helpful discussion and the Fmn2 antibody, Philip Leder for Fmn2-null mice, William Bement for UtrCH-EGFP, Jan Ellenberg for Fmn2-mCherry, and Tom Rapoport for the Sec61-β plasmid.
This work is supported in part by National Institutes of Health grant P01 GM 066311.
Footnotes
Abbreviations used in this paper:
- AcGFP
- Aequorea coerulescens GFP
- GV
- germinal vesicle
- GVBD
- GV breakdown
- MSD
- mean square displacement
- STICS
- spatiotemporal image correlation spectroscopy
- UtrCH
- utrophin calponin homology
References
- Azoury J., Lee K.W., Georget V., Rassinier P., Leader B., Verlhac M.H. 2008. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr. Biol. 18:1514–1519 10.1016/j.cub.2008.08.044 [DOI] [PubMed] [Google Scholar]
- Azoury J., Lee K.W., Georget V., Hikal P., Verlhac M.H. 2011. Symmetry breaking in mouse oocytes requires transient F-actin meshwork destabilization. Development. 138:2903–2908 10.1242/dev.060269 [DOI] [PubMed] [Google Scholar]
- Brugués J., Nuzzo V., Mazur E., Needleman D.J. 2012. Nucleation and transport organize microtubules in metaphase spindles. Cell. 149:554–564 10.1016/j.cell.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Brunet S., Maro B. 2005. Cytoskeleton and cell cycle control during meiotic maturation of the mouse oocyte: integrating time and space. Reproduction. 130:801–811 10.1530/rep.1.00364 [DOI] [PubMed] [Google Scholar]
- Burkel B.M., von Dassow G., Bement W.M. 2007. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskeleton. 64:822–832 10.1002/cm.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco P.G. 1995. Polarization of mitochondria in the unfertilized mouse oocyte. Dev. Genet. 16:36–43 10.1002/dvg.1020160108 [DOI] [PubMed] [Google Scholar]
- Chesarone M.A., DuPage A.G., Goode B.L. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11:62–74 10.1038/nrm2816 [DOI] [PubMed] [Google Scholar]
- Deng M., Suraneni P., Schultz R.M., Li R. 2007. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell. 12:301–308 10.1016/j.devcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Dumont J., Million K., Sunderland K., Rassinier P., Lim H., Leader B., Verlhac M.H. 2007. Formin-2 is required for spindle migration and for the late steps of cytokinesis in mouse oocytes. Dev. Biol. 301:254–265 10.1016/j.ydbio.2006.08.044 [DOI] [PubMed] [Google Scholar]
- FitzHarris G., Marangos P., Carroll J. 2007. Changes in endoplasmic reticulum structure during mouse oocyte maturation are controlled by the cytoskeleton and cytoplasmic dynein. Dev. Biol. 305:133–144 10.1016/j.ydbio.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Higgs H.N. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30:342–353 10.1016/j.tibs.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Leader B., Lim H., Carabatsos M.J., Harrington A., Ecsedy J., Pellman D., Maas R., Leder P. 2002. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat. Cell Biol. 4:921–928 10.1038/ncb880 [DOI] [PubMed] [Google Scholar]
- Li H., Guo F., Rubinstein B., Li R. 2008. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat. Cell Biol. 10:1301–1308 10.1038/ncb1788 [DOI] [PubMed] [Google Scholar]
- Longo F.J., Chen D.Y. 1985. Development of cortical polarity in mouse eggs: involvement of the meiotic apparatus. Dev. Biol. 107:382–394 10.1016/0012-1606(85)90320-3 [DOI] [PubMed] [Google Scholar]
- Maro B., Verlhac M.H. 2002. Polar body formation: new rules for asymmetric divisions. Nat. Cell Biol. 4:E281–E283 10.1038/ncb1202-e281 [DOI] [PubMed] [Google Scholar]
- Mehlmann L.M., Terasaki M., Jaffe L.A., Kline D. 1995. Reorganization of the endoplasmic reticulum during meiotic maturation of the mouse oocyte. Dev. Biol. 170:607–615 10.1006/dbio.1995.1240 [DOI] [PubMed] [Google Scholar]
- Mendes Pinto I., Rubinstein B., Kucharavy A., Unruh J.R., Li R. 2012. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev. Cell. 22:1247–1260 10.1016/j.devcel.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mseka T., Cramer L.P. 2011. Actin depolymerization-based force retracts the cell rear in polarizing and migrating cells. Curr. Biol. 21:2085–2091 10.1016/j.cub.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Murrell M.P., Gardel M.L. 2012. F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc. Natl. Acad. Sci. USA. 109:20820–20825 10.1073/pnas.1214753109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., Pollard T.D. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A.C., Boujemaa-Paterski R., Martiel J.L., Guérin C., Cao W., Chin H.F., De La Cruz E.M., Théry M., Blanchoin L. 2012. Actin network architecture can determine myosin motor activity. Science. 336:1310–1314 10.1126/science.1221708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Ellenberg J. 2008. A new model for asymmetric spindle positioning in mouse oocytes. Curr. Biol. 18:1986–1992 10.1016/j.cub.2008.11.022 [DOI] [PubMed] [Google Scholar]
- Sun S.C., Wang Z.B., Xu Y.N., Lee S.E., Cui X.S., Kim N.H. 2011. Arp2/3 complex regulates asymmetric division and cytokinesis in mouse oocytes. PLoS ONE. 6:e18392 10.1371/journal.pone.0018392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., Unser M. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7:27–41 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- Van Blerkom J., Runner M.N. 1984. Mitochondrial reorganization during resumption of arrested meiosis in the mouse oocyte. Am. J. Anat. 171:335–355 10.1002/aja.1001710309 [DOI] [PubMed] [Google Scholar]
- van Steensel B., van Binnendijk E.P., Hornsby C.D., van der Voort H.T., Krozowski Z.S., de Kloet E.R., van Driel R. 1996. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 109:787–792 [DOI] [PubMed] [Google Scholar]
- Verlhac M.H., Lefebvre C., Guillaud P., Rassinier P., Maro B. 2000. Asymmetric division in mouse oocytes: with or without Mos. Curr. Biol. 10:1303–1306 10.1016/S0960-9822(00)00753-3 [DOI] [PubMed] [Google Scholar]
- Wilson C.A., Tsuchida M.A., Allen G.M., Barnhart E.L., Applegate K.T., Yam P.T., Ji L., Keren K., Danuser G., Theriot J.A. 2010. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 465:373–377 10.1038/nature08994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Unruh J.R., Deng M., Slaughter B.D., Rubinstein B., Li R. 2011. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol. 13:1252–1258 10.1038/ncb2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Graham O.S., Raposo A., St Johnston D. 2012. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science. 336:999–1003 10.1126/science.1219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.