ATP hydrolysis on both Arp2 and Arp3 influences dissociation of the Arp2/3 complex from the lamellipodial actin network but is not strictly necessary for network disassembly.
Abstract
We examined the role of ATP hydrolysis by the Arp2/3 complex in building the leading edge of a cell by studying the effects of hydrolysis defects on the behavior of the complex in the lamellipodial actin network of Drosophila S2 cells and in a reconstituted, in vitro, actin-based motility system. In S2 cells, nonhydrolyzing Arp2 and Arp3 subunits expanded and delayed disassembly of lamellipodial actin networks and the effect of mutant subunits was additive. Arp2 and Arp3 ATP hydrolysis mutants remained in lamellipodial networks longer and traveled greater distances from the plasma membrane, even in networks still containing wild-type Arp2/3 complex. In vitro, wild-type and ATP hydrolysis mutant Arp2/3 complexes each nucleated actin and built similar dendritic networks. However, networks constructed with Arp2/3 hydrolysis-defective mutants were more resistant to disassembly by cofilin. Our results indicate that ATP hydrolysis on both Arp2 and Arp3 contributes to dissociation of the complex from the actin network but is not strictly necessary for lamellipodial network disassembly.
Introduction
Actin-based cellular motility is critical for cell spreading, tissue formation, and in immune responses. Motility relies on the formation of a three-dimensional lamellipodial actin network, composed of actin, capping protein, Arp2/3 complex, and other factors. The Arp2/3 complex nucleates new (daughter) filaments from the sides of preexisting (mother) filaments to generate space-filling dendritic arrays, in vitro (Mullins et al., 1998; Blanchoin et al., 2000) and at the leading edge of migrating cells (Svitkina and Borisy, 1999). The architecture and assembly dynamics of this network are governed by the timing of Arp2/3 activation, and the disassembly of this network is critical for the recycling of its components and for sustained network growth (Cramer, 1999).
The Arp2/3 complex comprises seven subunits, two of which, Arp2 and Arp3, are actin-related proteins that contain actin-like, ATP-binding pockets. Yet, the role of ATP hydrolysis by the Arp2/3 complex is not well understood. Residues important for the catalytic mechanism of hydrolysis were elucidated by crystal structures of nonvertebrate actin (Vorobiev et al., 2003). Experiments in budding yeast, using mutant based on this crystal structure, indicate that ATP binding on Arp2 and Arp3 are required for function of the complex in endocytosis and in actin patch dynamics (Martin et al., 2005, 2006).
Although ATP is hydrolyzed on the Arp2 subunit at roughly the same time that the complex produces new filaments (Dayel and Mullins, 2004), hydrolysis on a single ATP-binding subunit (Arp2 or Arp3) does not appear to be required for nucleation (Martin, et al., 2006). However, nucleation by an Arp2/3 complex unable to hydrolyze ATP on both Arp2 and Arp3 has not been shown. Although conventional actin binds ATP with nanomolar affinity and hydrolyzes bound nucleotide soon after incorporating into a filament (Blanchoin and Pollard, 2002), Arp2 and Arp3 bind ATP with 1,000-fold weaker affinity (Dayel et al., 2001). The Arp2 subunit hydrolyzes bound ATP soon after creating a new filament or capping the pointed end of a preexisting one (Dayel and Mullins, 2004).
Actin nucleation by Arp2/3 is regulated by intracellular signals and requires the participation of a nucleation-promoting factor (NPF) such as N-WASP, WAVE, WHAMM, WASH, or JMY (Machesky et al., 1999; Rohatgi et al., 1999; Welch and Mullins, 2002; Campellone et al., 2008; Zuchero et al., 2009; Campellone and Welch, 2010; Duleh and Welch, 2010). The minimal NPF sequence able to activate Arp2/3 is a three-part motif, called a VCA domain. VCA consists of a verprolin homology (or WASP-homology 2, WH2) domain that binds monomeric actin (Higgs et al., 1999), an acidic region that binds to Arp2/3 complex (Marchand et al., 2001), and a central region that binds both actin and the Arp2/3 complex (Kelly et al., 2006). Nucleation of a new filament requires three factors: (1) binding of the Arp2/3 complex to the side of a preformed actin filament (Mullins et al., 1997, 1998; Machesky et al., 1999); (2) binding of two VCA domains to the Arp2/3 complex (Padrick et al., 2011); and (3) delivery of at least one actin monomer to the complex via the WH2 domain (Dayel and Mullins, 2004).
To examine the role of ATP hydrolysis by the Arp2/3 complex, in the context of building a cell’s leading edge, we used fluorescence speckle microscopy to follow the dynamics of Arp2/3 complexes containing nonhydrolyzing Arp2 and Arp3 subunits in the lamellipodia of Drosophila S2 cells. We also examined the structural properties of dendritic actin networks in an in vitro motility system reconstituted using purified components. Loss of ATPase activity on Arp2 and Arp3 has similar effects. Neither mutant inhibits cell spreading or assembly of the lamellipod. Rather, both mutant alleles prolong the association of the Arp2/3 complex with the lamellipodial actin network, promote expansion of the lamellipod, and prevent its disassembly. Likewise, in vitro, ATP hydrolysis mutants hinder disassembly of the actin network. Our results indicate that ATP hydrolysis on both Arp2 and Arp3 is not absolutely required for network disassembly, but does facilitate dissociation of the complex from lamellipodial actin networks.
Results
Mutations that abolish ATP hydrolysis in Arp2 and Arp3 have minimal effects on actin filament nucleation by the Arp2/3 complex
To study the role of ATP hydrolysis by the Arp2/3 complex, we mutated critical residues in the ATP-binding sites of Arp2 and Arp3. In actin, residue Q137 positions a water molecule near the gamma phosphate of the bound ATP, while residue H161 functions as a base catalyst, abstracting a proton from water, and priming it for nucleophilic attack on ATP (Vorobiev et al., 2003). We mutated Arp2 and Arp3 residues analogous to Q137 and H161 of actin (Fig. S1 A) and integrated these mutations into the yeast genome, under control of the endogenous gene promoters. We then purified five variants of the Arp2/3 complex from budding yeast: wild-type Arp2/3 complex and four ATP hydrolysis mutants, each containing a single mutation in Arp2 or Arp3 (Arp2Q137A, Arp2H161A, Arp3Q137A, or Arp3H161A).
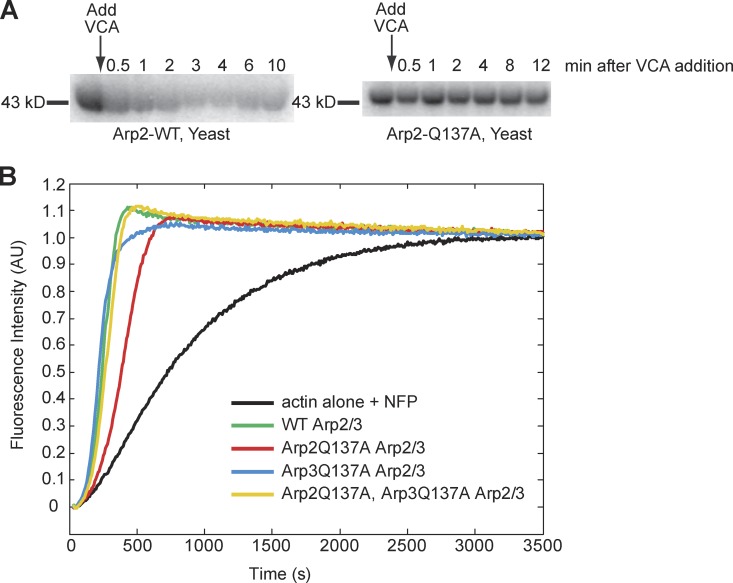
To confirm loss of ATPase activity in the Arp2/3 mutants, we performed ATP hydrolysis assays, using purified Arp2/3 complex cross-linked to 8-azido-γ-[32P]ATP. We stimulated hydrolysis of the radiolabeled ATP covalently cross-linked to the Arp2 and Arp3 subunits by adding actin and the purified VCA domain of yeast Arp2/3 activator, Las17. We observed rapid hydrolysis on the Arp2 subunit of the wild-type complex, but observed no detectable hydrolysis by Arp2Q137A or Arp2H161A subunits (Fig. 1 A; and unpublished data). This result is consistent with that of Martin, et al. (2006), who also found that Arp2H161A was unable to hydrolyze ATP. We could not accurately quantify ATP hydrolysis by Arp3 mutants due to poor cross-linking of azido-ATP to this subunit.
Figure 1.
Arp2/3 ATP hydrolysis mutants nucleate actin at levels near that of wild-type Arp2/3 complex. (A) Arp2Q137A does not hydrolyze ATP, in contrast to wild-type Arp2. We cross-linked azido-γ-[32P]ATP to WT and Arp2Q137A yeast Arp2/3. The VCA domain of the yeast NPF, Las17, was added to stimulate ATP hydrolysis by Arp2/3. Samples were removed from the reaction tubes at the indicated times, methanol precipitated, resuspended in sample buffer, and resolved by SDS-PAGE. Wild-type yeast Arp2/3 shows an appreciable loss of 32P signal over time, signifying hydrolysis and subsequent dissociation of the γ32-phosphate of cross-linked nucleotide from the ATP-binding pocket. (B) Fluorimetry of pyrene-labeled actin demonstrating Arp2/3-accelerated actin polymerization with WT and ATP hydrolysis mutant human Arp2/3 variants. Reactions were performed in 1× KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0) with 4 µM actin (5% pyrene labeled), 41 nM human Arp2/3, and 100 nM nWASP WWCA. Each pyrene reaction was performed two times. The data shown are from a single representative experiment.
When integrated into yeast under the native promoter, we found that, as previously described (Martin et al., 2006), arp2H161A exhibited growth defects on formamide and on 0.9 M NaCl, both stress-inducing conditions (Fig. S1 B). In contrast, yeast strains arp2Q137A and arp3Q137A exhibited normal growth under all conditions. This suggests that the H161A mutant complex may have additional defects unrelated to ATP hydrolysis, whereas the Q137A mutation likely does not. Thus, in our analysis of Arp2/3 ATPase activity, we focused on the Q137A mutation. We performed experiments in parallel, on wild-type (WT), Arp2 and Arp3 Q137A, and Arp2 and Arp3 H161A Arp2/3 complex. We include our results for the Arp2/3 H161A mutants, which are very similar to those obtained for the Q137A mutants, in Figs. S3 and S4. Surprisingly, despite the lack of phenotype displayed by the arp2- and arp3-Q137A alleles, even under stressful conditions, the two mutations together were synthetically lethal (Fig. S1 D). This result suggests that, despite our inability to detect it, ATP hydrolysis on the Arp3 subunit is important for the function of the complex. Nucleation activity of the four mutant yeast Arp2/3 complexes (Arp2Q137A, Arp3Q137A, Arp2H161A, and Arp3H161A) varied slightly from the activity of wild-type yeast Arp2/3, the H161A mutants being slightly more active than wild type and the Q137A mutants being slightly less active (Fig. S1 C).
Purification of Arp2/3 complex from yeast has some limitations: low quantities of purified protein and difficulty in obtaining a double ATP hydrolysis mutant (Arp2Q137A, Arp3Q137A) Arps2/3 complex, due to synthetic lethality. Thus, we used a baculovirus purification protocol, which allowed us to obtain a double ATP hydrolysis mutant human Arp2/3 complex, in addition to the same single mutant complexes that we had obtained previously from yeast. Using pyrenyl-actin assembly assays, we found that all four baculovirus-produced Arp2/3 variants (Arp2Q137A, Arp3Q137A, double mutant, and wild type) were capable of accelerating actin polymerization in an NPF-dependent manner (Fig. 1 B), indicating that nucleotide hydrolysis on either Arp2, Arp3, or both subunits is not specifically required for nucleation of actin filaments by the Arp2/3 complex.
Arp2/3 ATP hydrolysis mutants remain attached to the dendritic lamellipodial actin network longer than WT Arp2/3 in the leading edge of Drosophila S2 cells
To better understand the cellular and physiological role of ATP hydrolysis by the Arp2/3 complex, we examined the effects of ATPase mutants on the morphology of the cell’s leading edge. We created Drosophila S2 cell lines stably expressing fluorophore-labeled Arp2 and Arp3. We depleted endogenous Arp2 or Arp3 from the stable cell lines by RNAi, thus replacing endogenous protein with a stably expressed, fluorophore-tagged ATP hydrolysis-dead variant.
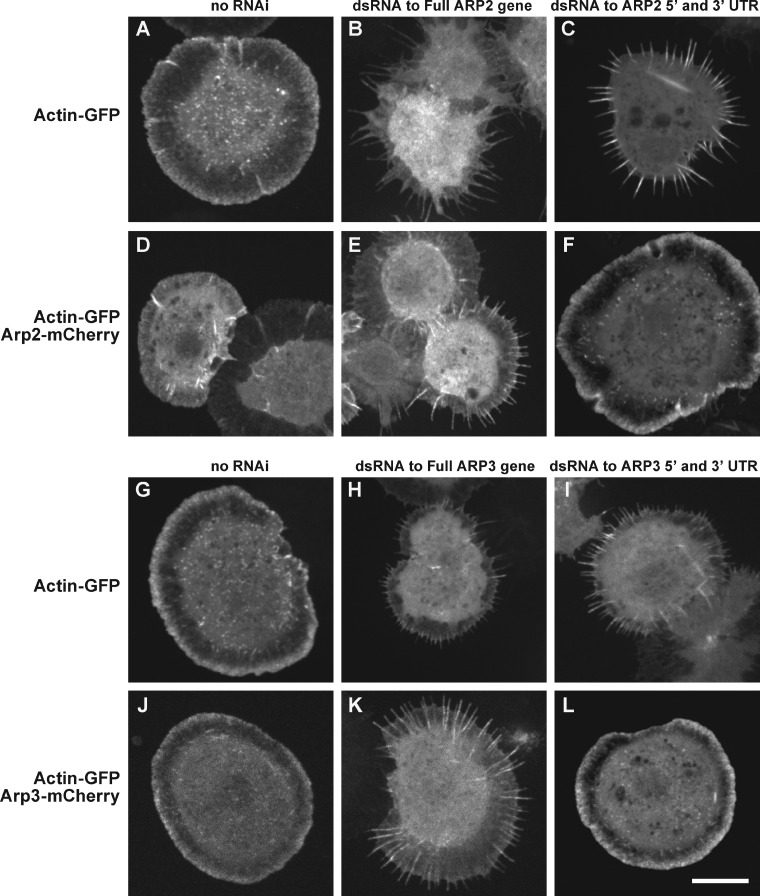
We verified the efficiency of our RNAi knockdown procedure using light microscopy and by immunoblotting cell extracts. Nucleation activity of the Arp2/3 complex, which helps form the dendritic actin network at the cell edge, is required for S2 cell spreading. When Arp2/3 complex subunits are depleted, cells fail to spread normally and exhibit a serrated morphology, rather than a smooth, circular boundary. We used stable S2 cell lines expressing actin-GFP alone (9), actin-GFP and Arp2-mCherry (9-1), or actin-GFP and Arp3-mCherry (9-2) (stable cell lines listed in Table 1). Consistent with previous studies, Arp2/3 complex activity was decreased by double-stranded RNA (dsRNA) targeting the full coding sequence of either ARP2 or ARP3, as judged by the fraction (90%) of cells with serrated phenotypes (Fig. 2, B, E, H, and K). We also observed serrated cells when we used dsRNA targeting the 5′ and 3′ untranslated regions (UTR) of ARP2 or ARP3 (Fig. 2, C and I). The serrated phenotype produced by dsRNA targeting the 5′ and 3′ UTRs of ARP2 or ARP3 was rescued by expression of mCherry-labeled Arp2 and Arp3 constructs, respectively (Fig. 2, F and L). We verified Arp2 knockdown by immunoblotting whole-cell Drosophila S2 extracts (Fig. S1 E) with Arp2 antibodies. Antibodies recognizing Drosophila Arp3 were not commercially available.
Table 1.
Stable S2 cell lines
| Cell line | Proteins expressed |
| 1-3 | pMT Arp2(WT) mCherry, pMT Arp2(WT) GFP |
| 1-4 | pMT Arp2(WT) mCherry, pMT Arp2Q137A GFP |
| 1-5 | pMT Arp2(WT) mCherry, pMT Arp2H161A GFP |
| 2-6 | pMT mCherry Arp3(WT), pMT GFP Arp3(WT) |
| 2-7 | pMT mCherry Arp3(WT), pMT GFP Arp3Q137A |
| 2-8 | pMT mCherry Arp3(WT), pMT GFP Arp3H161A |
| 9-1 | pMT GFP actin, pMT Arp2(WT) mCherry |
| 9-10 | pMT GFP actin, pMT Arp2Q137A mCherry |
| 9-12 | pMT GFP actin, pMT Arp2H161A mCherry |
| 9-2 | pMT GFP actin, pMT cherry Arp3(WT) |
| 9-13 | pMT GFP actin, pMT mCherry Arp3Q137A |
| 9-14 | pMT GFP actin, pMT mCherry Arp3H161A |
| 4-13 | pMT Arp2Q137A GFP, pMT Arp3Q137A mCherry |
| 10-7 | pMT Arp2Q137A mCherry, pMT Arp3Q137A GFP |
| 5-14 | pMT Arp2H161A GFP, pMT Arp3H161A mCherry |
| 12-8 | pMT Arp2H161A mCherry, pMT Arp3H161A GFP |
| 3-2 | pMT Arp2(WT) GFP, pMT Arp3(WT) mCherry |
| 1-6 | pMT Arp2(WT) mCherry, pMT Arp3(WT) GFP |
| 3 | pMT Arp2(WT) GFP |
| 4 | pMT Arp2Q137A GFP |
| 5 | pMT Arp2H161A GFP |
| 6 | pMT Arp3(WT) GFP |
| 7 | pMT Arp3Q137A GFP |
| 8 | pMT Arp3H161A GFP |
| 9 | pMT GFP actin |
Figure 2.
Double-stranded RNA (dsRNA) directed against the 5′ and 3′ untranslated regions (UTRs) of ARP2 or ARP3 depletes the targeted endogenous protein, producing the serrated phenotype. Arp2-GFP and Arp3-GFP each rescue cells depleted of endogenous Arp2 and Arp3, respectively. We created stable S2 cells lines that express actin-GFP, either alone or with Arp2-mCherry (or Arp3-mCherry). dsRNA targeting the coding sequence of ARP2 (or ARP3) depletes the corresponding protein, resulting in the serrated phenotype (B, E, H, and K). In contrast, cells transfected with dsRNA targeting the 5′ and 3′ UTRs of ARP2 or ARP3 are selectively depleted of endogenous Arp2 or Arp3 (C and I), and are rescued by exogenously expressed Arp2-mCherry (F) and Arp3-mCherry (L). Bar, 10 µm.
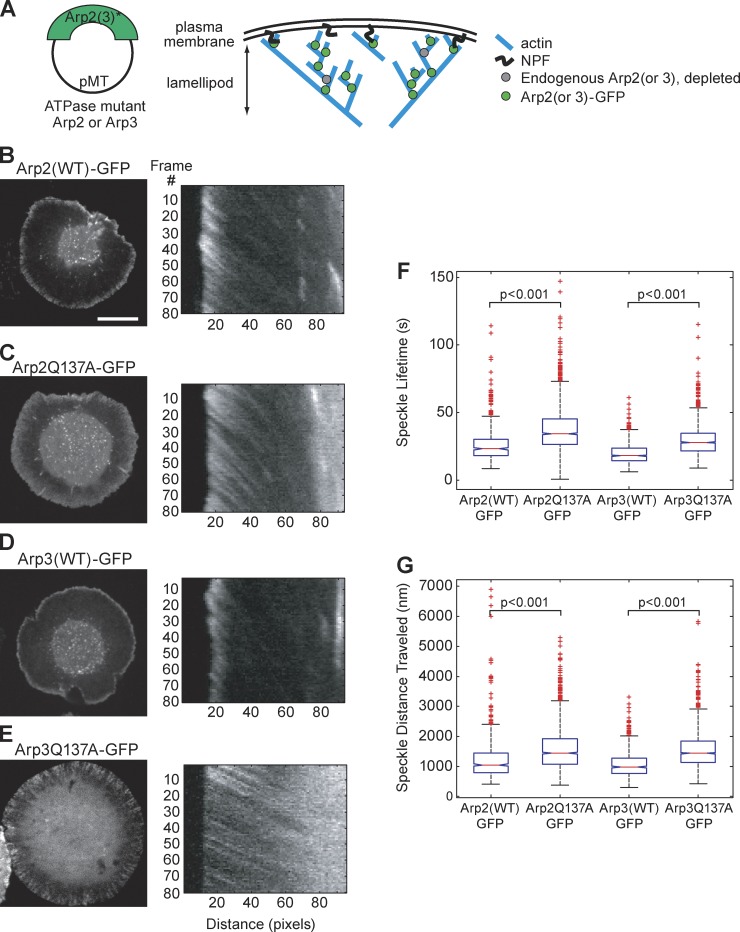
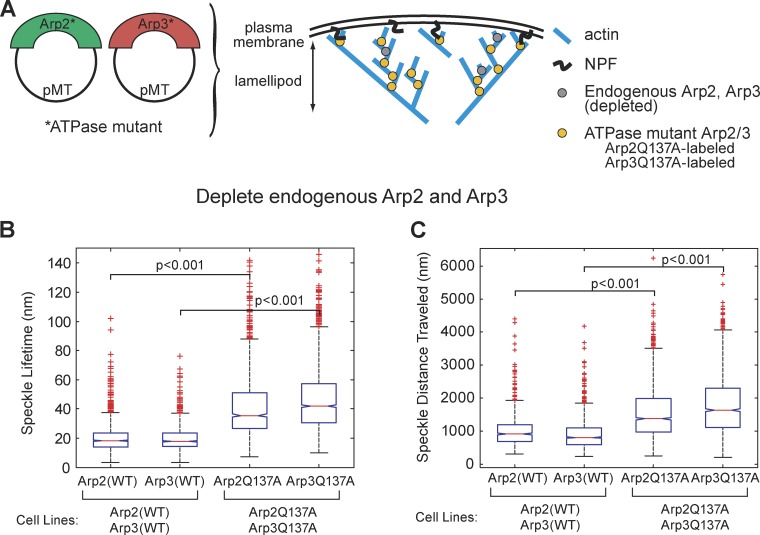
Confident that our dsRNA targeting the 5′ and 3′ UTR could deplete endogenous Arp2 and Arp3, while leaving exogenously expressed fluorophore-labeled Arp2 and Arp3 intact, we proceeded to examine the effects of Arp2 and Arp3 ATPase mutants on the behavior of Arp2/3 complex in the lamellipodial actin network. For these experiments, we created stable S2 cell lines expressing Arp2WT-GFP (3), Arp2Q137A-GFP (4), Arp3WT-GFP (6), and Arp3Q137A-GFP (7) (Table 1). We depleted endogenous Arp2 or Arp3 from cells, using dsRNA targeting the 5′ and 3′ UTRs, and replaced the depleted protein with the stably expressed GFP-labeled ATPase variant (Fig. 3 A). GFP-tagged proteins were expressed at low levels, allowing us to quantify the behaviors of individual Arp2 and Arp3 speckles. For movies of live cells, we used kymograph analysis to reconstruct speckle trajectories (Fig. 3, B–E; second column). Using speckle trajectories, we recorded distances traveled and speckle lifetimes for Arp2(WT)-GFP, Arp2Q137A-GFP, Arp3(WT)-GFP, and Arp3Q137A-GFP speckles (Fig. S2, A–D). We observed differences between the behaviors of speckles containing WT versus hydrolysis-dead variants of Arp2 and Arp3. Arp 2 and Arp3 ATP hydrolysis mutants had longer lifetimes and traveled greater distances than their wild-type counterparts (Fig. 3, F and G; Videos 1 and 2). For each cell line, we examined 20–50 cells, constructed at least 12 kymographs per cell from sections evenly distributed around the cell’s perimeter, and recorded metrics for at least 200 speckles.
Figure 3.
ATP hydrolysis mutant Arp2/3 complex produces dendritic actin network disassembly defects. This is demonstrated by the longer lifetimes and greater distances traveled of Arp2/3 speckles, as compared with WT Arp2/3. (A) We created the following Drosophila S2 stable cell lines: Arp2(WT)-GFP (3); Arp2Q137A-GFP (4); Arp3(WT)-GFP (6); and Arp3Q137A-GFP (7) (see Table 1 for complete list of cell lines). We imaged these stable cell lines and created kymographs from the movies (B–E, second column). We quantified the speckle distances traveled and speckle lifetimes. Endogenous Arp2 (B and C) and Arp3 (D and E) were depleted by dsRNA directed against the 5′ and 3′ UTRs, leaving Arp2-GFP (B and C) and Arp3-GFP (D and E) as the sole copy of Arp2 (B and C) and Arp3 (D and E) in the cell. Bar, 10 µm. (F) We replotted our speckle lifetime and distance-traveled measurements using a box-and-whisker plot. We used the nonparametric Kolmogorov-Smirnov test to compare ATP hydrolysis mutant data against WT data. We calculated p-values to be less than 0.001. The central mark in the box represents the median value; the edges of the box represent the 25th and 75th percentiles. Whiskers extend to the most extreme data points not considered outliers, and outliers are plotted individually, in red.
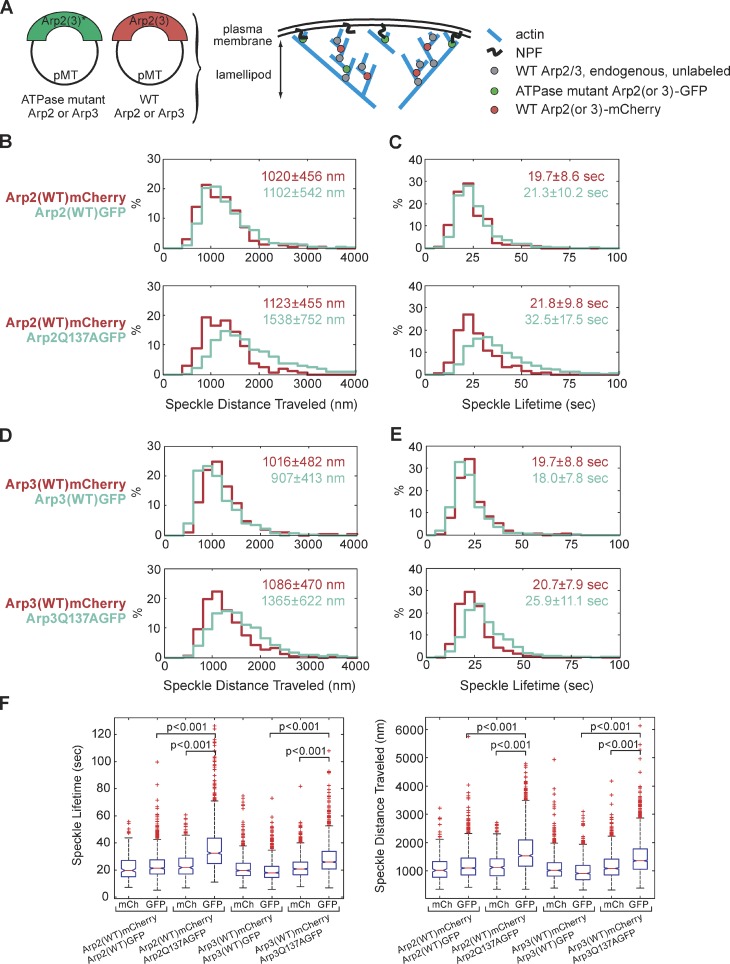
To further explore and contrast the behaviors of WT Arp2/3 versus ATPase mutant Arp2/3 in vivo, we created stable S2 cell lines that coexpress mCherry-labeled WT and GFP-labeled ATP hydrolysis mutant, as follows: (1-3): Arp2(WT)-mCherry, Arp2(WT)-GFP; (1-4): Arp2(WT)-mCherry, Arp2Q137A-GFP; (2-6): Arp3(WT)-mCherry, Arp3(WT)-GFP; and (2-7): Arp3(WT)-mCherry, Arp3Q137A-GFP (Table 1, Fig. 4 A). We quantified the behavior of Arp2/3 speckles, and could clearly distinguish behavior of WT Arp2/3 from that of hydrolysis-defective Arp2/3, within the same cell.
Figure 4.
ATP hydrolysis is necessary for timely detachment of Arp2/3 from the lamellipodial actin network, as seen from the ATP hydrolysis mutant’s longer lifetime and longer distance traveled. Arp2/3 containing a single ATP hydrolysis mutant subunit behaves differently than wild-type Arp2/3 within the same cell. (A) We expressed wild-type mCherry-tagged Arp2 (or Arp3) along with ATP hydrolysis mutant GFP-tagged Arp2 (or Arp3) in the same cells. (B and C) Imaging of stable S2 cell lines (1-3): Arp2(WT)-GFP, Arp2(WT)-mCherry (30 cells, 907 GFP speckles, 244 mCherry speckles); and (1-4): Arp2Q137A-GFP, Arp2(WT)-mCherry (34 cells, 972 GFP speckles, 321 mCherry speckles). (D and E) Imaging of stable S2 cell lines (2-6): Arp3(WT)-GFP, Arp3(WT)-mCherry (41 cells, 915 GFP speckles, 692 mCherry speckles.); and (2-7): Arp3Q137A-GFP, Arp3(WT)-mCherry (45 cells, 1,673 GFP speckles, 952 mCherry speckles). We created kymographs showing speckle trajectories, which we used to determine speckle distances traveled and speckle lifetimes. Median values for speckle distances traveled (B and D) or speckle lifetimes (C and E) are noted. Each imaging experiment was repeated on three separate occasions and speckle data were combined. (F) As in Fig. 3, F and G, we replotted our data in a box-and-whisker plot and used the nonparametric Kolmogorov-Smirnov test to compare ATP hydrolysis mutant data against WT data. We calculated p-values to be less than 0.001.
For each tracked GFP or mCherry speckle, we recorded its lifetime and distance traveled from the leading edge. Surprisingly, we found that, within single cells, ATP hydrolysis mutant Arp2Q137A-GFP speckles behave in a markedly differently manner than WT Arp2-mCherry speckles (Fig. 4, B, C, and F). Arp2Q137A-GFP speckles remain attached to the Arp2/3-actin network longer than WT Arp2/3 speckles, as indicated by their increased speckle lifetimes: 33 s for the Q137A Arp2 mutant versus 22 s for WT Arp2 (Fig. 4 C, bottom graph). Arp2 ATPase mutant speckles also traveled longer distances (1,500 nm) than WT speckles (1,100 nm) (Fig. 4 B, bottom graph) before disappearing. Remarkably, Arp3 ATPase mutants also had longer lifetimes and traveled greater distances than their WT counterparts (Fig. 4, D–F). This result is surprising, given the dearth of previous information about Arp3, but consistent with the synthetic lethality of the Arp2 ATP hydrolysis mutant allele with the Arp3 mutant allele in budding yeast (Fig. S1 D).
The synthetic lethality in yeast and the effect of the Q137A mutation on ARP3 prompted us to examine the effect of having both ATP hydrolysis–defective mutations simultaneously present in S2 cells. We created additional cell lines, this time placing fluorophore tags on both Arp2 and on Arp3: (3-2) Arp2(WT)-GFP, Arp3(WT)-mCherry; and (4-13) Arp2Q137A-GFP, Arp3Q137A-mCherry (Table1, Fig. 5 A). To avoid potential artifacts caused by differential photostability of GFP versus mCherry, we also created stable cells lines in which we switched the fluorophores used on Arp2 and Arp3: (1-6) Arp2(WT)-mCherry, Arp3(WT)-GFP; and (10-7) Arp2Q137A-mCherry, Arp3Q137A-GFP (Table 1). We used RNAi to simultaneously deplete both endogenous Arp2 and Arp3, and measured speckle lifetimes and distances traveled. RNAi depletes most, but not all, endogenous Arp2 and Arp3 in S2 cells. Thus, in knocking down and replacing both Arp2 and Arp3 with ATPase mutants, we also likely created complexes that contained one mutant and one wild-type subunit, along with the desired complexes, which contained two mutant subunits. This may explain the broadening of peaks in the histograms of mutant speckle lifetimes and distances traveled (Figs. S2 and S4). The effect of having two ATP hydrolysis mutations, one on Arp2 and the other on Arp3, was additive. WT Arp2 and Arp3 speckles have average lifetimes of 20.6 s (Table 2; Fig. S2, A and B). Double-mutant cells had greater speckle lifetimes (38.7 s) than cells in which a single subunit, either Arp2 or Arp3, was replaced with an ATP hydrolysis mutant (31 s) (Table 2; Fig. S2, E and F).
Figure 5.
Replacing both the Arp2 and the Arp3 subunits of the Arp2/3 complex with ATP hydrolysis mutant variants produces a more severe disassembly phenotype than replacing a single subunit. (A) We created stable cell lines that express mCherry-tagged Arp2 ATP hydrolysis mutant and GFP-tagged Arp3 ATP hydrolysis mutant. We also created stable cell lines in which we switched the fluorescent tags on Arp2 and Arp3; thus, cells express GFP-tagged Arp2 ATP hydrolysis mutant and mCherry-tagged Arp3 ATP hydrolysis mutant. In all cases, we combined speckle measurements from cells expressing Arp2-GFP and Arp3-mCherry with measurements from cells expressing Arp2-mCherry and Arp3-GFP. We plotted our speckle lifetimes (B) and speckle distances traveled (C) in a box-and-whisker plot and used the nonparametric Kolmogorov-Smirnov test to compare ATP hydrolysis mutant data against WT data. We calculated p-values to be less than 0.001.
Table 2.
Speckle properties
| S2 cell line | Speckle lifetime | Average | Speckle distance traveled | Average | |
| (s) | (s) | (nm) | (nm) | ||
| Single mutants | Arp2-WT, cell line (3) | 23.1 | 20.6 | 1,049 | 1,014.5 |
| Arp3-WT, cell line (6) | 18.0 | 20.6 | 980 | 1,014.5 | |
| Arp2Q137A, cell line (4) | 34.2 | 31.0 | 1,441 | 1,439.5 | |
| Arp3Q137A, cell line (7) | 27.8 | 31.0 | 1,438 | 1,439.5 | |
| Double mutants | Arp2-WT, cell lines (3-2;1-6) | 18.1 | 17.95 | 915 | 860.5 |
| Arp3-WT, cell lines (3-2;1-6) | 17.8 | 17.95 | 806 | 860.5 | |
| Arp2Q137A, cell lines (4-13;10-7) | 35.5 | 38.7 | 1,374 | 1,505 | |
| Arp3Q137A, cell lines (4-13;10-7) | 41.9 | 38.7 | 1,636 | 1,505 |
Median values, as noted in histograms shown in Fig. S2, are reprinted in this table for ease of comparison.
Arp2/3 ATP hydrolysis mutants are more resistant to cofilin-mediated disassembly in vitro
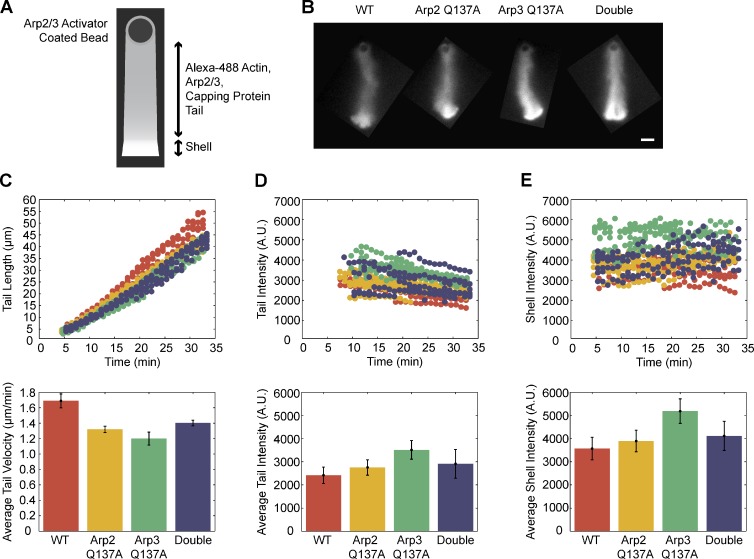
Our in vivo S2 cell observations led us to believe that ATP hydrolysis on both the Arp2 and Arp3 subunits of the complex aids in disassembly of the dendritic Arp2/3-actin network. We tested this hypothesis in a reconstituted in vitro motility system containing a limited number of purified components, as used by Akin and Mullins (2008). We used polystyrene beads with the VCA domain of the Arp2/3 activator, ActA, covalently linked to the bead surface. In the absence of “recycling” components, cofilin and profilin, a shell composed of actin, Arp2/3, and capping protein grew at the surface of the ActA-VCA–coated beads. After symmetry breaking of the shell, we observed sustained growth of an actin tail from the bead surface (Fig. 6, A and B). At 50-nM Arp2/3 concentrations, wild-type and ATP hydrolysis–defective Arp2/3 complexes supported growth of actin tails with similar lengths (Fig. 6 C), comparable tail intensities (Fig. 6 D), and similar shell intensities (Fig. 6 E; Videos 3 and 4). Actin tails constructed with higher Arp2/3 concentrations are shown in Fig. S5.
Figure 6.
Wild-type and ATP hydrolysis mutant variants of Arp2/3 complex build actin networks in a reconstituted actin-based motility system. (A) The VCA domain of ActA, an Arp2/3-activating protein from the pathogen Listeria monocytogenes, is covalently attached to polystyrene beads. In the presence of 3% Alexa Fluor 488–labeled cytoplasmic actin, Arp2/3 complex, and capping protein, a dendritic actin network grows in a spherical array from the bead surface, creating a shell of actin. After the actin shell breaks symmetry, a sustained tail of actin continues to grow from the bead surface. At 50 nM, wild-type Arp2/3, Arp2Q137A Arp2/3, Arp3Q137A Arp2/3, and Arp2Q137A/Arp3Q137A Arp2/3 all build (B) similar actin networks (C) with comparable actin tail lengths, and similar actin intensities in (C) the tail and (D) shell regions. Bar, 10 µm.
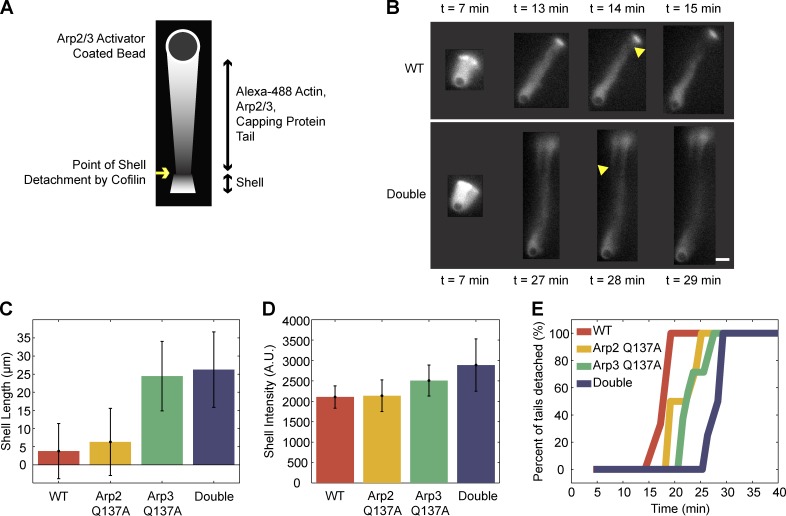
The addition of recycling components, profilin and cofilin, to the bead motility assay allowed us to observe differences between the behaviors of wild-type and ATP hydrolysis mutant Arp2/3 complexes during disassembly. Cofilin, which severs actin filaments, and profilin, which promotes nucleotide exchange by actin monomers, together promote turnover and recycling of actin monomers in the in vitro motility reaction. Inclusion of profilin and cofilin in the actin motility reaction leads to shell detachment from growing actin tails (Fig. 7, A and B). We measured shell length at time of detachment (Fig. 7 C), as well as shell intensity (Fig. 7 D). Despite having an actin nucleation rate comparable to wild-type Arp2/3 complex (Fig. 1 A) and building a shell and tail of similar intensity, double-ATP hydrolysis mutant Arp2/3 complex had a distinctly longer shell at its time of detachment and we observed a clear delay in time of shell detachment from the actin tail (Fig. 7 E; Videos 5 and 6). Based on these observations, we conclude that ATP hydrolysis by the Arp2 and Arp3 subunits contributes to Arp2/3 debranching and to actin network disassembly.
Figure 7.
Differences in actin disassembly between networks constructed with wild-type vs. ATP hydrolysis mutant Arp2/3 complex become apparent under recycling conditions. (A) The addition of cofilin and profilin to the in vitro actin-based motility system causes turnover and recycling of actin monomers by promoting disassembly of actin filaments. Cofilin severs the actin shell, detaching it from the tail. (B and E) Severing of shells from growing tails occurs 14 min earlier in networks built with wild-type Arp2/3 complex (top) than in networks built with double ATP hydrolysis mutant Arp2/3 complex (bottom). (C) Shells built with ATP hydrolysis mutant Arp2/3 grow to longer lengths than shells built with wild-type Arp2/3 complex. (D) Actin intensities of shells constructed with wild-type Arp2/3 are comparable or slightly lower than intensities of shells constructed with ATP hydrolysis mutant Arp2/3. (E) Quantification of the timing of tail severing for WT (13 tails), Arp2Q137A (9 tails), Arp3Q137A (9 tails), and double ATP hydrolysis mutant (8 tails). Arp2Q137A, Arp3Q137A, and double mutant tails result in shells severed later than wild-type tails by 3.5, 4.5, and 12 min, respectively. Bar, 10 µm.
Discussion
Arp2 and Arp3 are members of the actin superfamily, a group of proteins defined by a common fold that forms a nucleotide-binding pocket with a preference for ATP (Bork et al., 1992). The pocket consists of two globular domains separated by a flexible hinge region (Kabsch et al., 1990). Contacts between the globular domains and the bound ATP hold the pocket closed and hydrolysis of bound ATP weakens the contacts between the globular domains, opening the pocket. This simple conformational change has been adapted to many biochemical activities, including phosphorylation of sugar molecules (hexokinase), catalysis of protein folding (Hsp70), and destabilization of cytoskeletal filaments (actin and ParM).
A chemical mechanism of ATP hydrolysis by actin family proteins was suggested by high resolution atomic structures (Vorobiev et al., 2003). In this model, one critical residue in the nucleotide-binding pocket (Q137) positions a catalytic water molecule near the gamma phosphate of the bound ATP while another residue (H161) acts as a base catalyst to activate the water molecule, promoting hydrolysis of the γ-phosphate of ATP. This model was first tested experimentally in the bacterial actin-like protein ParM. The proposed catalytic residues of actin, Q137 and H161, were mapped onto the structure of ParM and mutated to alanines (Garner et al., 2004). ParM containing a mutation homologous to Q137A in actin lacked ATPase activity, but polymerized with essentially wild-type kinetics, whereas mutations spatially equivalent to H161A altered the ATP critical concentration and inhibited polymerization. These results verified the involvement of Q137 in ATP hydrolysis, but suggested that mutation of H161 produces additional defects unrelated to ATP hydrolysis.
Arp2 mutations equivalent to H161A and Q137A produced different growth phenotypes in budding yeast (Martin et al., 2006). Both mutations abolished ATP hydrolysis; H161A produced a growth defect, whereas Q137A did not. The authors focused on analysis of the H161A phenotype and, based on their analysis of actin patch dynamics in Arp2H161A cells, proposed that ATP hydrolysis on the Arp2 subunit promotes dissociation of the complex from actin and is required for disassembly of Arp2/3-generated networks (Martin et al., 2006).
Building on this work, we examined the dynamics of wild-type and ATPase mutant Arp2 and Arp3 in the context of a lamellipodial actin network. We focused primarily on mutations equivalent to Q137A because they completely abolish ATP hydrolysis, with minimal additional effects on protein structure and function. We used Drosophila S2 cells because they construct well-defined lamellipodial actin networks at the cell periphery, are flat and well suited to light microscopy, and are highly amenable to protein depletion by RNAi (Rogers et al., 2003; Iwasa and Mullins, 2007).
At the leading edge, the lamellipodial actin network enables cells to move, change shape, adhere to surfaces, and endocytose nutrients. This network is defined by the rapid assembly and retrograde flow of actin filaments (Salmon et al., 2002), as well as the presence of a characteristic set of actin regulatory proteins including the Arp2/3 complex, capping protein, and cofilin (Iwasa and Mullins, 2007). Filaments in the lamellipodial network are nucleated in a three-dimensional, dendritic array by the Arp2/3 complex; their rapid growth is terminated by capping protein, and they are disassembled by cofilin. Loss of either capping protein or the Arp2/3 complex abolishes the lamellipodial network entirely, whereas loss of cofilin leads to dramatic expansion of the lamellipod (Iwasa and Mullins, 2007). Additional factors, such as tropomyosin, also control the size of the lamellipod, probably by blocking access of the Arp2/3 complex to actin filaments. Similar to loss of cofilin, loss of tropomyosin causes dramatic expansion of the lamellipod (Gupton et al., 2005; Iwasa and Mullins, 2007).
If, as suggested by Martin et al. (2006), Arp2/3 complex dissociation from the actin network depends on its ability to hydrolyze ATP, we would expect nonhydrolyzing Arp2 mutants to mimic the loss of cofilin and to significantly expand the size of the lamellipodial network. We do, in fact, find that nonhydrolyzing Arp2 and Arp3 mutants widen the lamellipodial network, as judged from tracking both actin and Arp2/3 speckles. The effect, however, is mild compared with the loss of either cofilin or tropomyosin. The same effect is also produced by nonhydrolyzing Arp3 mutants. These results suggest that ATP hydrolysis occurs on both the Arp2 and Arp3 subunits and accelerates dissociation of the Arp2/3 complex, but is not strictly required for lamellipodial network disassembly.
In cells expressing both fluorophore-labeled WT and ATP hydrolysis mutant Arp2 or Arp3, we observed a clear disparity in the movements of ATPase mutant and WT Arp2 or Arp3 (Fig. 4). These observations concur with the in vitro experiments of Kawska et al. (2012). Our work similarly suggests that Arp2/3, along with other network components, builds up many entangled sub-networks, from which lamellipodial network properties emerge.
Previous studies of ATP hydrolysis by the Arp2/3 complex focused on the Arp2 subunit. Yet, it is not entirely surprising that ATP hydrolysis on the Arp3 subunit also plays a role in the activity cycle of the complex. Previously, it was reported that almost 100% of ATP bound to the Arp2 subunit of Acanthamoeba Arp2/3 complex is hydrolyzed during or soon after nucleation and that ∼15% of ATP bound to the Arp3 subunit is also hydrolyzed during the same time interval (Dayel et al., 2001). Likely, the covalent cross-linking of radiolabeled ATP into the active site of Arp3 inhibits efficient hydrolysis. Recent biochemical work (Padrick et al., 2011) has shown that the Arp2/3 complex binds two VCA domains, which deliver two actin monomers to the complex, one to each of the Arp subunits. In addition, high resolution structural analysis of the complex suggests that actin nucleation by the complex requires closure of the ATP-binding pocket of the Arp3 subunit (Xu et al., 2012). Our work concurs with the idea that ATP hydrolysis is important on both Arp subunits.
In S2 cells, Arp2 and Arp3 ATP hydrolysis mutants slow down, but do not completely inhibit, disassembly of the lamellipodial actin network. Our in vitro motility assay experiments show that Arp2/3 ATP hydrolysis mutants build actin tails very similar in structure to those built by WT Arp2/3. Cofilin’s known preference for ADP-actin (Blanchoin and Pollard, 1999) seemingly extends to the actin-related subunits of the Arp2/3 complex. Actin tails constructed with Arp2/3 ATP hydrolysis mutants have delayed network disassembly and delayed cofilin-mediated shell detachment from actin tails, supporting the hypothesis that ATP hydrolysis and/or phosphate release by Arp2 and Arp3 promotes more rapid disassembly, but is not absolutely required for network disassembly.
Similar to Akin and Mullins (2008) and Reymann et al. (2011), we found that cofilin preferentially targets regions of ADP-actin for disassembly. Our observation that dendritic actin networks constructed with ATP hydrolysis–deficient Arp2/3 complex are more resistant to cofilin-mediated disassembly echoes the idea of Reymann et al. (2011) that cofilin participate in a macroscopic network disassembly process, rather than in individual filament depolymerization. These results are consistent with the idea of “array treadmilling” proposed by Svitkina and Borisy (1999).
Recent work has shown that other proteins are involved in disassembly of the dendritic actin network. Cofilin destabilizes Arp2/3 branchpoints (Chan et al., 2009) and a related protein, GMF, which lacks actin severing and depolymerizing activity, has been shown to inhibit Arp2/3 nucleation activity (Gandhi et al., 2010). In addition, Coronin may serve to remodel Arp2/3-containing actin branchpoints (Cai et al., 2008). We propose that ATP hydrolysis on the Arp2 and Arp3 subunits of the complex contributes to the activities of other factors to induce actin network disassembly.
Materials and methods
Actin polymerization assays
Actin polymerization was measured by monitoring an increase in fluorescence (λex = 365 nm, λem = 407 nm) of 5% pyrene-iodoacetamide–labeled actin that was purified from Acanthamoeba castellanii. We mixed Arp2/3, Arp2/3 activator (VCA or WWCA) in KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0). Actin polymerization reactions contained 4 µM actin (5% pyrene labeled); all other components were as specified in figure legends. All reaction components, except for actin, were mixed in a single tube. Separately, actin was preincubated with ME (50 µM MgCl2 and 200 µM EGTA) for 2 min, to facilitate the exchange of Ca2+ ions for Mg2+ ions. Reactions were initiated by addition of actin to the other components.
Actin labeling
Actin was labeled on Cys-374 with pyrene iodoacetamide (Invitrogen) or Alexa Fluor 488–maleimide (Invitrogen), as described previously (Cooper et al., 1983). In brief, gel-filtered Acanthamoeba actin was diluted to around 1 mg/ml (27 µM) and polymerized by addition of KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0). Polymerized actin was dialyzed against L-KMEI (100 mM KCl, 2 mM MgCl2, 25 mM imidazole, pH 7.5, and 0.3 mM ATP). Actin was transferred to a small beaker and 4–7 mol of pyrene iodoacetamide (Invitrogen) per mole of actin were added while stirring. The reaction was covered with foil and allowed proceed overnight, with gentle stirring. The reaction was quenched with 10 mM DTT and precipitated dye was removed by low speed centrifugation. Labeled actin filaments were pelleted by centrifugation for 2 h at 38,000 rpm in a rotor (Type 45 Ti; Beckman Coulter). Filamentous, labeled actin was depolymerized by dialyzing into buffer A (2 mM Tris HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM TCEP, and 0.04% azide) and gel filtered on an S75 column (GE Healthcare). Labeled actin was stored at 4°C in the dark.
Actin was labeled with Alexa Fluor 488–maleimide (Invitrogen) while in the de-polymerized state. In brief, G-actin was dialyzed overnight into L-buffer A (5 mM Tris, pH 8.0, 0.2 mM ATP, and 0.1 mM CaCl2). 5 molar excess of Alexa Fluor 488 was added to the actin. The reaction proceeded on ice for 15 min and was quenched with 10 mM DTT. Actin was polymerized by addition of 1× KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0). Filamentous, labeled actin was collected by centrifugation, and both resuspended in and dialyzed against buffer A (2 mM Tris HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM TCEP, and 0.04% azide) to depolymerize. Labeled actin was gel filtered on an S75 column (GE Healthcare).
Yeast strain construction
We created all yeast strains in a haploid, wild-type S288c background strain. Plasmids pDD1697 and pDD1698, generously provided by the Drubin laboratory, were used to create ARP2 and ARP3 ATPase mutant yeast strains. Plasmid pDD1697 contains URA3-marked yeast ARP2; plasmid pDD1698 contains LEU2-marked ARP3. Using standard PCR and cloning techniques, ARP2 and ARP3 mutations analogous to yeast actin, act1-Q137A and act1-H161A, were made in plasmids pDD1697 and pDD1698, respectively. The plasmids were transformed into yeast using standard methods (Ito et al., 1983; Sherman et al., 1986). Yeast transformants were initially selected based on ability to growth on −leu or −ura plates. Presence of correctly integrated arp2-Q137A, arp2-H161A, arp3-Q137A, arp3-H161A mutations was verified by sequencing of yeast genomic DNA. Correctly integrated mutants were backcrossed to WT S288c, sporulated, and tetrads dissected, creating the final yeast strains.
Yeast growth assay
Yeast strains were grown overnight (in YPD or selective minimal media) at room temperature. In the morning, yeast density was measured spectrophotometrically, diluted back to OD600 = 0.1, and allowed to grow for two more doubling times. All yeast strains were diluted to exactly OD600 = 0.5, along with five 10-fold serial dilutions in a 96-well plate. A pin transfer device was used to transfer yeast to YPD, 3% formamide, 0.9 M NaCl plates. Plates were grown at room temperature and 37°C.
Protein purification
Cytoplasmic actin.
Actin was purified from A. castellanii by the method of Gordon et al. (1976). Approximately 15 liters of amoeba, OD600 = 4–5 were collected by centrifugation, rinsed with wash buffer (10 mM Tris, pH 8.0, and 40 mM NaCl), and resuspended in extraction buffer (10 mM Tris, pH 8.0, 1 mM EGTA, 11.6% sucrose, 1 mM ATP, 5 mM DTT, 15 µg/ml benzamidine, 5 µg/ml pepstatin, 40 µg/ml soybean trypsin inhibitor, 10 µg/ml leupeptin, 1 mM PMSF, and 5 µg/ml aprotinin). Amoeba were pressurized in a Parr Bomb to 400 psi with N2 gas and lysed by the slow release of pressure. Lysate was clarified by centrifugation at 167,000 g and batch bound to DEAE resin. Actin was eluted with a linear gradient, 0–600 mM KCl, in column buffer (10 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.5 mM ATP, 2 mM DTT, and 15 µg/ml benzamidine). Actin-containing fractions were pooled, polymerized by addition of 1 mM ATP and 2 mM MgCl2, centrifuged, and resuspended in buffer A (2 mM Tris HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM TCEP, and 0.04% azide). G-actin was gel filtered and stored in buffer A at 4°C.
Yeast Arp2/3.
Yeast Arp2/3 was purified, based on a modification of the A. castellanii Arp2/3 purification protocol (Dayel et al., 2001). Yeast of strain S288c were grown to OD600 = 1, harvested by centrifugation, washed once with ddH2O, and resuspended in a volume of ddH2O equal to 10% of the weight of yeast. Resuspended yeast was dripped into liquid nitrogen, forming small, frozen pellets, which were stored at −80°C. For the Arp2/3 purification, we weighed out 35–60 g of yeast pellets and ground them to a fine powder in a precooled coffee grinder (model BCG1000B1; KitchenAid). Yeast powder was resuspended in room-temperature 3× lysis buffer (60 mM Hepes, pH 7.1 at 4°C, 200 mM KCl, 3 mM EDTA, 3 mM DTT, supplemented with 1 µg/ml leupeptin, 5 µg/ml pepstatin, 1 µg/ml aprotinin, and 1 mM PMSF) equal to one half the weight of the yeast pellets. Yeast lysate was centrifuged at 4,000 g to pellet unlysed cells. Cleared lysate was then centrifuged at 100,000 g for 2 h, resulting in high-speed supernatant (HSS). HSS was passed over a 15–20-ml bed volume DEAE column that had been equilibrated with DEAE buffer (20 mM Hepes, pH 7.1 at 4°C, 67 mM KCl, and 1 mM DTT). We collected the DEAE column flow-through, along with one bed volume wash of the DEAE resin, measured the conductivity of the DEAE flow-through, and added enough dilution buffer (20 mM Hepes, pH 7.1 at 4°C, 100 µM ATP, 300 mM sucrose, and 1 mM DTT) to obtain a final conductivity of 3 mS. Diluted DEAE flow-through was passed over a 4.5-ml CH-Sepharose (activated CH Sepharose 4B; GE Healthcare) affinity column to which we had conjugated nWASP-WWCA(398–501). The nWASP-WWCA affinity column was washed with 20 column volumes of VCA column buffer (20 mM Hepes, pH 7.1 at 4°C, 5 mM KCl, and 1 mM DTT), followed by 10 column volumes of VCA wash buffer (20 mM Hepes, pH 7.1 at 4°C, 80 mM KCl, and 1 mM DTT). Yeast Arp2/3 was eluted from the column with VCA elution buffer (20 mM Hepes, pH 7.1 at 4°C, 1 M KCl, and 1 mM DTT). The first elution fraction volume was ∼3 ml; subsequent elution fraction volumes were ∼4.5–5 ml. Elution fractions 2 and 3 were combined and dialyzed overnight into MonoQ low salt buffer (20 mM Hepes, pH 7.1 at 4°C, 25 mM NaCl, and 1 mM DTT). After dialysis, Arp2/3 was filtered and loaded onto a MonoQ column 5/50GL column, connected to an ÄKTA Purifier system (GE Healthcare). The column was washed with 10 column volumes of 10% MonoQ high salt buffer (20 mM Hepes, pH 7.1 at 4°C, 1 M NaCl, and 1 mM DTT) and eluted with a linear gradient from 10–35% MonoQ buffer B, over 60 column volumes. Fractions containing Arp2/3, as verified by Coomassie-stained SDS-PAGE gel, were combined and dialyzed overnight into Arp2/3 freezing buffer (20 mM Hepes, pH 7.1 at 4°C, 50 mM KCl, 10% glycerol, and 1 mM TCEP). Dialyzed Arp2/3 was concentrated in a Vivaspin6 5-kD MWCO spin column (GE Healthcare), flash frozen in liquid nitrogen, and stored at −80°C.
Human Arp2/3.
Baculovirus expression vectors encoding the seven subunits of the human Arp2/3 complex were obtained from the Welch Laboratory at UC Berkeley (Berkeley, CA). The four vectors were: (1) pFastBac Dual Arp3/p21; (2) pFastBac Dual Arp2/p16; (3) pFastBac Dual p34/p20; and (4) pFastBac1 p40. The pFastbac1 p40 vector also contained a 6× His tag at the C terminus. We used QuikChange site-directed mutagenesis to mutate the Arp2 and Arp3-encoding vectors such that they would contain the Arp2Q137A and Arp3Q137A mutations, respectively. Viral stocks were made using standard baculovirus protocols (Invitrogen). We infected Sf9 cells with four viruses at a time, producing WT Arp2/3 complex and with the Q137A mutation on either Arp2 or Arp3, or on both Arp2 and Arp3.
All Arp2/3 complexes were purified as follows. Sf9 cells were infected for 72 h, collected by centrifugation, and flash frozen in liquid nitrogen. Cell pellets were thawed and resuspended in lysis buffer (20 mM Hepes, pH 7.4 at 4°C, 300 mM NaCl, 20 mM imidazole, 100 µM ATP, 1 mM MgCl2, 1 mM DTT, and 0.5% NP-40) that was supplemented with 2 µg/ml leupeptin, 5 µg/ml pepstatin, 2 µg/ml aprotinin, and 1 mM PMSF. Resuspended cells were lysed by passing several times through a 27-gauge, 1/2-inch syringe needle. Cell lysates were cleared by centrifugation at 150,000 g. The cleared lysate was batch bound to Nickel NTA resin (QIAGEN) at 4°C, nutating for 1 h. The lysate–resin slurry was poured into a column, washed with 20 column volumes of wash buffer (20 mM Hepes, pH 7.4 at 4°C, 300 mM NaCl, 30 mM imidazole, 100 µM ATP, 1 mM MgCl2, and 1 mM DTT), and protein was eluted with two column volumes of elution buffer (20 mM Hepes, pH 7.4 at 4°C, 300 mM NaCl, 500 mM imidazole, 100 µM ATP, 1 mM MgCl2, and 1 mM DTT). In preparation for cation exchange, the Nickel NTA column eluate was passed over a HiPrep 26/10 desalting column equilibrated with MonoS low salt buffer (20 mM MES, pH 6.1 at 4°C, 15 mM KCl, 100 µM ATP, 1 mM MgCl2, and 1 mM DTT). The buffer-exchanged Nickel NTA eluate was loaded onto a MonoS 5/50GL column connected to an ÄKTA Purifier system (GE Healthcare). The column was washed with 10 column volumes of MonoS low salt buffer and eluted with a linear gradient from 0–60% MonoS high salt buffer (20 mM MES, pH 6.1 at 4°C, 1 M KCl, 100 µM ATP, 1 mM MgCl2, and 1 mM DTT) over 30 column volumes. Fractions containing Arp2/3, as verified by Coomassie-stained SDS-PAGE gel, were combined and dialyzed overnight into Arp2/3 freezing buffer (20 mM Hepes, pH 7.1 at 4°C, 50 mM KCl, 100 µM ATP, 1 mM MgCl2, 10% glycerol, and 1 mM TCEP). Dialyzed Arp2/3 was flash frozen in liquid nitrogen, and stored at −80°C. Baculovirus-infected Sf9 cells express recombinant human Arp2/3 at levels far exceeding those of endogenous insect cell Arp2/3. Gournier et al. (2001), in systematically purifying hArp2/3 complex with missing subunits, demonstrated that insect cell Arp2/3 complex does not copurify with exogenously expressed human Arp2/3 complex. We also observed that we could not purify any Arp2/3 complex when infecting Sf9 cells with very low-titer or nonfunctional baculovirus. Thus, we are confident that no mixing of subunits occurs between insect cell and human Arp2/3 complex.
nWASP-WWCA398-501 domain.
Recombinant rat N-WASP-WWCA398-501 containing an N-terminal 6×His tag was purified by metal-affinity chromatography, using Nickel NTA resin (QIAGEN) and standard methods.
Las17-VCA domain.
A single colony from a fresh transformation of pGEX-6P-2-Las17534-634 into BL21 Rosetta bacterial cells (EMD Millipore) was used to start an overnight 5-ml culture. The 5-ml culture was sub-cultured to OD600 = 0.015 into two 1-liter cultures, grown to OD600 = 0.4 at 37°C, and induced with 0.5 mM IPTG for 3 h. Bacteria were harvested, rinsed with cold PBS (2.9 mM NaH2PO4, 7.1 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KCl), frozen in liquid nitrogen, and stored at −80°C. Frozen bacteria were thawed in PBSED (PBS supplemented with 1 mM EDTA and 2 mM DTT) in the presence of 1 mM PMSF. Bacteria were lysed using a microfluidizer (Microfluidics) and lysate was clarified by centrifugation at 30,000 rpm for 30 min in a rotor (Type 45-Ti; Beckman Coulter) at 4°C. Cleared lysate was incubated on a nutator for 1 h with 2 ml of glutathione-4B Sepharose resin that had been equilibrated with PBSED. Resin was transferred to a glass column washed with 20 column volumes of PBSED. Protein was eluted from the column with elution buffer (20 mM Tris, pH 8.0, and 25 mM pH-adjusted glutathione) and dialyzed overnight into PBSED. During the last hour of dialysis, the GST tag was cleaved from Las17-VCA using PreScission Protease (GE Healthcare). Las17-VCA was cleared of cleaved GST tag by passing over a second column of glutathione-4B Sepharose that had been preequilibrated with PBSED. After overnight dialysis into MonoS buffer A (25 mM Hepes, pH 7.4, and 2 mM DTT), cleared Las17-VCA was loaded onto a MonoS 5/50GL column connected to an ÄKTA Purifier system (GE Healthcare). The column was washed with 15 column volumes of MonoS buffer A and Las17-VCA was eluted with a linear, 0–50% gradient of MonoQ buffer B (25 mM Hepes, pH 7.4, and 1 M NaCl) over 20 column volumes. Fractions containing Las17-VCA, as verified by Coomassie-stained SDS-PAGE gel, were combined, dialyzed into 20 mM Hepes, pH 7.4, and 0.5 mM TCEP, flash frozen in liquid nitrogen, and stored at −80°C.
Capping protein.
Recombinant mouse capping protein CPα1β2 was purified as described previously (Palmgren et al., 2001). In brief, the protein construct, in a pET3d vector, was expressed in BL21 E. coli cells. Bacterial pellets were resuspended in a 3× volume of lysis buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF) and lysed by passage through a microfluidizer and cleared by centrifugation. A 50–70% ammonium sulfate cut was taken from the cleared lysate and centrifuged at 32,000 g for 20 min at 4°C. The pellet was resuspended in 20 ml of HA dialysis buffer (10 mM NaH2PO4, pH 7.0, and 1 mM DTT) and dialyzed overnight into the same buffer with at least three changes. The dialysate was loaded onto an HA column (GE Healthcare) with 10 mM NaH2PO4, pH 7.0, and then eluted with a gradient from 10–250 mM NaH2PO4. Peak fractions were pooled and dialyzed against QA dialysis buffer (10 mM Tris, pH 8.0, 10 mM KCl, 0.5 mM EDTA, and 0.5 mM TCEP). The sample was loaded onto a MonoQ column (GE Healthcare) and eluted with a 10–400 mM KCl gradient. Selected fractions were dialyzed into SA dialysis buffer (10 mM MES, pH 5.8, 0.5 mM EDTA, and 1 mM DTT). The sample was loaded onto a MonoS column (GE Healthcare) and eluted with a 0–350 mM NaCl gradient. Selected fractions were dialyzed into storage buffer (10 mM Tris, pH 8.0, 40 mM KCl, 0.5 mM TCEP, and 50% glycerol), flash frozen, and stored at −80°C.
ActA30-612-KCK-6XHis.
ActA30-612-KCK-6XHis was purified as described previously (Akin and Mullins, 2008). In brief, the construct, cloned in a pET 17b vector, was expressed in BL21 E. coli cells. Bacterial pellets were resuspended in 4–5× volume of lysis buffer (100 mM NaH2PO4, pH 8.0, 10 mM Tris base, 6 M guanidium HCl, 10 mM 2-mercaptoethanol, and 1 mM PMSF) and lysed by nutating overnight at room temperature. Clarified lysate was batch bound to Ni-NTA resin (1–2 ml of resin per liter of culture) for 30 min. The resin was washed with wash buffer (100 mM NaH2PO4, pH 7.8, 10 mM Tris base, 10 mM imidazole, 6 M guanidium HCl, and 10 mM 2-mercaptoethanol) and eluted with elution buffer (100 mM NaH2PO4, pH 7.5, 10 mM Tris base, 250 mM imidazole, 6 M guanidium HCl, and 10 mM 2-mercaptoethanol). The eluate was dialyzed into buffer QA (10 mM bistrispropane, pH 7.0, 1 mM EDTA, and 0.5 mM TCEP), loaded onto a MonoQ column (GE Healthcare), and eluted with a 100–300-mM KCl gradient. Selected fractions were dialyzed into storage buffer (10 mM Hepes, pH 7.0, 50 mM KCl, and 1 mM TCEP), flash frozen, and stored at −80°C.
Profilin 1.
Recombinant human Profilin1 was purified as described previously (Reichstein and Korn, 1979), with the following modifications. The Profilin1, in vector pET16B, was expressed in E. coli BL21 pLysS. Bacterial cell pellets were resuspended in lysis buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 2 mM DTT, and 1 mM PMSF) and lysed using an Emulsiflex. Ammonium sulfate was added to the clarified lysate to a concentration of 35%. After centrifugation, an additional 26% ammonium sulfate to a final concentration of 61% was added to the supernatant. After centrifugation, the pellet was resuspended in lysis buffer and dialyzed overnight against more lysis buffer. Protein was applied to a DEAE column and the flow-through and wash fractions were collected. These fractions were dialyzed against HA buffer (5 mM K2HPO4, pH 7.5, and 1 mM DTT) and applied to a hydroxyapatite column. Flow-through and wash fractions were pooled, and gel filtered (10 mM Tris, pH 7.5, 50 mM NaCl, and 0.5 mM TCEP) using a Superdex 200 column connected to an ÄKTA Purifier system. Glycerol was added to 20% final concentration to the peak fractions. Protein was flash frozen in liquid nitrogen and stored at −80°C.
hCofilin 1.
Recombinant human Cofilin1 was purified as described previously (Carlier et al., 1997), with the following modifications. The construct, cloned into a pET16b vector, was expressed in BL21 Rosetta cells (EMD Millipore). E. coli lysate was dialyzed into DEAE buffer (10 mM Tris, pH 7.8, 50 mM NaCl, 0.2 mM EDTA, 2 mM DTT, and 1 mM PMSF). The flow-through and wash from the DEAE column was dialyzed into 10 mM Pipes, pH 6.5, 15 mM NaCl, 2 mM DTT, and 0.2 mM EDTA and loaded onto a MonoS 5/50 GL column connected to an ÄKTA Purifier system (GE Healthcare). Protein was gel filtered using a Superdex 200 column into 10 mM Tris, pH 7.5, 50 mM NaCl, and 0.5 mM TCEP; glycerol was added to 20% to peak fractions and protein was flash frozen and stored at −80°C.
ATP binding and hydrolysis
130 nM freshly thawed yeast Arp2/3 was equilibrated with 6 µM 8-azido γ-[32P]ATP (Affinity Photoprobes, LLC) in KMEH (10 mM Hepes, pH 7.0, 50 mM KCl, 1 mM MgCl2, and 1 mM EGTA). The mixture was cross-linked with a hand-held UV lamp and the reaction was quenched by addition of an equivalent volume of 2 mM ATP and 2 mM DTT in KMEH. To the quenched reaction, we added 2 µM actin that had been preincubated with 50 µM MgCl2 and 200 µM EGTA. A sample of this mixture was removed and Las17 VCA was added to a final concentration of 50 nM to commence Arp2/3-mediated actin nucleation. 175-µl reaction volumes were removed at 0.5, 1, 2, 3, 4, 6, and 10 min after addition of VCA. Each time point was immediately precipitated by addition of 400 µl of methanol and 100 µl of chloroform. Samples were methanol precipitated, dried, resuspended in sample buffer, and resolved on SDS-PAGE. Gels were dried and exposed a phosphorimaging for autoradiography by unhydrolyzed γ-[32P]ATP.
Plasmid construction
The region coding for the VCA domain of LAS17 was amplified from yeast genomic DNA using the following primers: 5′CGCGGATCCAAATGCAAATCCGGAGGCGGTTCATTCGCTGAAACTAC-3′ and 5′-CCGCTCGAGTTACCAATCATCACCATTGTCCATATCGTCATG-3′. The PCR product was ligated into vector pGEX-6P-2 (GE Healthcare) using restriction enzyme sites BamHI and XhoI.
Baculovirus vectors generously provided by the Welch laboratory (UC Berkeley, Berkeley, CA) were mutated by standard QuikChange site-directed mutagenesis techniques (Agilent Technologies) in order to make the Arp2 and Arp3 Q137A mutations. The following primers were used to mutagenize the pFastBac Dual Arp2/p16 plasmid: 5′-GTAGCCATCGCCGCAGTTCTGACTTTGTACGCTCAAGGTTTATTG-3′ and 5′-CAGAACTGCggcGATGGCTACATATACACCGGAAAACTGGTAAGT-3′. The following primers were used to mutagenize the pFastBac Dual Arp3/p21 plasmid: 5′-CATTGCTGTGGCCGCTGTTCTTGCCTTAGCTGCATCTTGGACCTCAAG-3′ and 5′-CAAGAACAGCGGCCACAGCAATGTACAAGCCTGGAACATTGAAGGACT-3′.
Culture of S2 cells, construction of stable cell lines
S2 cells were cultured in Schneider’s media (Invitrogen), supplemented with 10% fetal bovine serum and 1× antibiotic/antimycotic, as described previously (Rogers et al., 2003). Stable cell lines were created by transfecting plasmids containing chimeric, fluorophore-labeled protein of interest under the copper-inducible pMT promoter. Transfections were performed using Effectene transfection reagent (QIAGEN). A confluent T25 flask of S2 cells was resuspended in its own media and 200 µl of cells were plated per well of a 6-well plate for use in a transfection the next day. On the day of transfection, media was removed from cells and replaced with 3 ml fresh media. A total of 1 µg of DNA, in a volume of not more than 10 µl, was diluted with buffer EC to a final volume of 150 µl. To this, we added 8 µl of Enhancer, mixed well, and incubated at room temperature for 2–5 min. After incubation, we added 25 µl of Effectene reagent to the DNA-Enhancer mixture, mixed well, and incubated at room temperature for 10 min. We added 1 ml of complete media to the transfection mixture, pipetted up and down gently to mix, and added the mixture drop-wise to cells.
For single plasmid transfections, we used 800 ng of plasmid plus 200 ng of the selection plasmid, pCoHygro (Invitrogen). For double plasmid transfections, we used 450 ng of each plasmid plus 100 ng of pCoHygro. 2 d after transfection, cells were passaged 1:2 to media containing 9 µl/ml hygromycin (Invitrogen). Cells were passaged for ∼4 wk before freezing.
RNAi of S2 cells
RNAi of S2 cells slated for microscopy was performed in 96-well plate format, as described previously (Goshima et al., 2007). 2 × 106 cells were plated per well of a 96-well plate in serum-free Schneider’s media. Cells were incubated with 3 µg of dsRNA for 45 min. After incubation, serum-containing media was added to the cells, bringing the serum concentration to 7%. Cells were treated with dsRNA for 7 d.
Microscopy of S2 cells
The day before microscopy, a 25-µM final concentration of CuSO4 was added to cells to induce expression of genes under control the metallothionein promoter. The day of imaging, one well of a 96-well plate of cells was resuspended in ∼100 µl of media. 8–12 µl of resuspended cells were plated onto a concanavalin A–coated well of a 96-well Matrical plate. Additional fresh S2 media was added and cells were allowed to adhere for 2 h before imaging. All S2 cells imaged were expressing either eGFP, mCherry, or both.
Images were acquired in the Nikon Imaging Center (UCSF, San Francisco, CA) using a 100×, 1.40 NA oil Plan Apo objective on a motorized inverted microscope (Eclipse Ti-E; Nikon) equipped with a spinning disk (model CSU22, Yokogawa Corporation of America; Solamere Technology Group) and a cooled CCD camera (Evolve; Photometrics) running MicroManager open source software. Pixel size was 91 nm. All movies were taken under speckling conditions at room temperature.
Image analysis of S2 cells
Movies of S2 cells were analyzed with MATLAB (Mathworks), using code written in house. Speckle trajectories were followed and each speckle’s lifetime and distance traveled from the cell cortex was recorded. The number of speckles analyzed is noted the figure legends.
Actin motility assays and microscopy
Carboxylated, polystyrene 5-µm-diameter beads (Bangs Laboratories) were coated with ActA30-612-KCK-6XHis using EDC-SulfoNHS chemistry, as described previously (Akin and Mullins, 2008). Coated beads were stored in 1 mg/ml BSA, 2 mM Tris, pH 8.0 and used at 1:120 dilution in motility assays.
For motility assays without recycling agents, we mixed the ActA-coated beads with 7.4 µM 3% Alexa Fluor 488–labeled actin, 100 nM capping protein, and varying concentrations (50 nM, 75 nM, 100 nM) of wild-type, Arp2Q137A, Arp3Q137A, and double mutant Arp2/3. All assays were performed in 0.2% methyl cellulose (400 cP), 2.5 mg/ml BSA, 1 mM MgCl2, 1 mM EGTA, 20 mM Hepes, pH 7.0, 50 mM KOH (pH 7.0 with 15 mM TCEP HCl), and 250 µM ATP. For motility assays with recycling agents, we included 500 nM hCofilin1 and 500 nM hProfilin1 in the reactions.
All images were acquired at room temperature on a software-controlled (MicroManager) inverted epifluorescence microscope (Eclipse TE2000-E; Nikon) equipped with an EM CCD camera (iXon; Andor Technology) and a 60×, 1.2 NA water immersion objective (Nikon). To minimize nonspecific protein absorption, glass slides were washed in ethanol and KOH. Glass coverslips were washed in ethanol and KOH, and additionally treated with 2% solution of diethyldichlorosilane (Gelest) in 5% water, 95% isopropanol, pH 4.5 (with acetic acid). Microscopy chambers were prepared with double-stick tape between the slide and coverslip. After flowing in 10 µl of the motility mix, chambers were sealed with VALAP (molten mixture of Vaseline, lanolin, and paraffin at 1:1:1 mass ratio) and imaged immediately. Images were taken at 60-s intervals.
Motility assay image analysis
All image analysis was performed using MATLAB code written by the authors. Intensity values were taken from the original images, where the intensity values had not been adjusted. For nonrecycling assays, the lengths of the motile actin comet tails were determined by hand-drawn line scans through the middle of the long axis of the tail in each image. The actin intensity profile along this line was recorded. The maximum intensity of the shell and the average intensity of the tail (without the shell) were plotted over time, as well as time averaged in the bar graphs. For assays with recycling components, the length of the shell (defined to be from the point of detachment to the end of the shell) as well as the maximum intensity of the shell was measured by hand-drawn line scans. The number of tails detached at a particular time was counted versus the total number of tails present and plotted value over time.
Online supplemental material
Fig. S1 shows protein alignments of yeast actin with yeast and Drosophila Arp2 and Arp3, yeast growth assays, and depletion of Drosophila Arp2 by RNAi. Figs. S2–S4 show histograms of speckle distances traveled and speckle lifetimes. Fig. S5 shows additional bead motility experiments, at varying concentrations of Arp2/3 complex. Video 1 shows movement of WT Arp2-GFP speckles in a Drosophila S2 cell. Video 2 shows movement of Arp2Q137A-GFP speckles in a Drosophila S2 cell. Video 3 shows bead motility of network with WT Arp2/3 complex, no recycling. Video 4 shows bead motility of network with double ATP hydrolysis mutant Arp2/3 complex, no recycling. Video 5 shows bead motility and severing of shell built with WT Arp2/3 complex, with recycling. Video 6 shows bead motility and severing of shell built with double ATP hydrolysis mutant Arp2/3 complex, with recycling. Bars, 5 µm. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211069/DC1.
Supplementary Material
Acknowledgments
We are grateful to D. Drubin for generously sharing yeast strains and plasmids and to M. Welch for generously sharing Arp2/3 baculovirus constructs. We thank E.A. Ingerman for advice and assistance with MATLAB. We thank P. Bieling for sharing his hCofilin1 and hProfilin1 plasmids. We are grateful to L. LeClaire, O. Akin, and M. Quinlan for sharing reagents and for much helpful advice. We especially thank C. Rivera for his unflagging enthusiasm, countless thoughtful discussions regarding experimental design and data presentation, and for carefully proofreading and editing the manuscript. We thank L. Goins and other members of the Mullins Laboratory for their insights and comments. We are grateful to E. Griffis and M. D’Ambrosio for advice on S2 cell culture and to other members of the Vale Laboratory for useful discussions and for use of the actin-GFP plasmid. We would like to thank J. Schaefer and other members of the Morgan Laboratory for their helpful advice about yeast. We are very grateful to K. Thorn and the UCSF Nikon Imaging Center, where all live-cell imaging was performed.
This work was supported, in part, by National Institutes of Health grant 2R01GM061010 to R.D. Mullins and a National Institutes of Health post-doctoral fellowship to E. Ingerman.
E. Ingerman would like to dedicate this paper to teachers of Lowell High School: Ms. A. Puretz, Ms. O. Tilton, Mr. R. Shapiro, Mr. P. Luk, Mr. S. Granucci, and Dr. T. Briggs.
J.Y. Hsiao would like to dedicate this work to her coach and lifelong mentor, Joanna Santarsiere.
Footnotes
Abbreviations used in this paper:
- ARP
- actin-related protein
- dsRNA
- double-stranded RNA
- NPF
- nucleation-promoting factor
- VCA
- verprolin homology/central/acidic
- WT
- wild type
References
- Akin O., Mullins R.D. 2008. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 133:841–851 10.1016/j.cell.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D. 1999. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274:15538–15546 10.1074/jbc.274.22.15538 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D. 2002. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 41:597–602 10.1021/bi011214b [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K.J., Higgs H.N., Marchand J.B., Kaiser D.A., Pollard T.D. 2000. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 404:1007–1011 10.1038/35010008 [DOI] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA. 89:7290–7294 10.1073/pnas.89.16.7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Makhov A.M., Schafer D.A., Bear J.E. 2008. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 134:828–842 10.1016/j.cell.2008.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K.G., Welch M.D. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11:237–251 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K.G., Webb N.J., Znameroski E.A., Welch M.D. 2008. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 134:148–161 10.1016/j.cell.2008.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., Laurent V., Santolini J., Melki R., Didry D., Xia G.X., Hong Y., Chua N.H., Pantaloni D. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322 10.1083/jcb.136.6.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Beltzner C.C., Pollard T.D. 2009. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19:537–545 10.1016/j.cub.2009.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.A., Walker S.B., Pollard T.D. 1983. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4:253–262 10.1007/BF00712034 [DOI] [PubMed] [Google Scholar]
- Cramer L.P. 1999. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr. Biol. 9:1095–1105 10.1016/S0960-9822(99)80478-3 [DOI] [PubMed] [Google Scholar]
- Dayel M.J., Mullins R.D. 2004. Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2:E91 10.1371/journal.pbio.0020091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel M.J., Holleran E.A., Mullins R.D. 2001. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. USA. 98:14871–14876 10.1073/pnas.261419298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh S.N., Welch M.D. 2010. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken). 67:193–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M., Smith B.A., Bovellan M., Paavilainen V., Daugherty-Clarke K., Gelles J., Lappalainen P., Goode B.L. 2010. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 20:861–867 10.1016/j.cub.2010.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E.C., Campbell C.S., Mullins R.D. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 306:1021–1025 10.1126/science.1101313 [DOI] [PubMed] [Google Scholar]
- Gordon D.J., Eisenberg E., Korn E.D. 1976. Characterization of cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J. Biol. Chem. 251:4778–4786 [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier H., Goley E.D., Niederstrasser H., Trinh T., Welch M.D. 2001. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell. 8:1041–1052 10.1016/S1097-2765(01)00393-8 [DOI] [PubMed] [Google Scholar]
- Gupton S.L., Anderson K.L., Kole T.P., Fischer R.S., Ponti A., Hitchcock-DeGregori S.E., Danuser G., Fowler V.M., Wirtz D., Hanein D., Waterman-Storer C.M. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619–631 10.1083/jcb.200406063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H.N., Blanchoin L., Pollard T.D. 1999. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 38:15212–15222 10.1021/bi991843+ [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa J.H., Mullins R.D. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17:395–406 10.1016/j.cub.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H.G., Suck D., Pai E.F., Holmes K.C. 1990. Atomic structure of the actin:DNase I complex. Nature. 347:37–44 10.1038/347037a0 [DOI] [PubMed] [Google Scholar]
- Kawska A., Carvalho K., Manzi J., Boujemaa-Paterski R., Blanchoin L., Martiel J.L., Sykes C. 2012. How actin network dynamics control the onset of actin-based motility. Proc. Natl. Acad. Sci. USA. 109:14440–14445 10.1073/pnas.1117096109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Kranitz H., Dötsch V., Mullins R.D. 2006. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J. Biol. Chem. 281:10589–10597 10.1074/jbc.M507470200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M., Mullins R.D., Higgs H.N., Kaiser D.A., Blanchoin L., May R.C., Hall M.E., Pollard T.D. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 96:3739–3744 10.1073/pnas.96.7.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand J.B., Kaiser D.A., Pollard T.D., Higgs H.N. 2001. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3:76–82 10.1038/35050590 [DOI] [PubMed] [Google Scholar]
- Martin A.C., Xu X.P., Rouiller I., Kaksonen M., Sun Y., Belmont L., Volkmann N., Hanein D., Welch M., Drubin D.G. 2005. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J. Cell Biol. 168:315–328 10.1083/jcb.200408177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Welch M.D., Drubin D.G. 2006. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 8:826–833 10.1038/ncb1443 [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Stafford W.F., Pollard T.D. 1997. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J. Cell Biol. 136:331–343 10.1083/jcb.136.2.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R.D., Heuser J.A., Pollard T.D. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 95:6181–6186 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick S.B., Doolittle L.K., Brautigam C.A., King D.S., Rosen M.K. 2011. Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. USA. 108:E472–E479 10.1073/pnas.1100236108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren S., Ojala P.J., Wear M.A., Cooper J.A., Lappalainen P. 2001. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 155:251–260 10.1083/jcb.200106157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichstein E., Korn E.D. 1979. Acanthamoeba profilin. A protein of low molecular weight from Acanpthamoeba castellanii that inhibits actin nucleation. J. Biol. Chem. 254:6174–6179 [PubMed] [Google Scholar]
- Reymann A.C., Suarez C., Guérin C., Martiel J.L., Staiger C.J., Blanchoin L., Boujemaa-Paterski R. 2011. Turnover of branched actin filament networks by stochastic fragmentation with ADF/cofilin. Mol. Biol. Cell. 22:2541–2550 10.1091/mbc.E11-01-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Wiedemann U., Stuurman N., Vale R.D. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M.W. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 97:221–231 10.1016/S0092-8674(00)80732-1 [DOI] [PubMed] [Google Scholar]
- Salmon W.C., Adams M.C., Waterman-Storer C.M. 2002. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 158:31–37 10.1083/jcb.200203022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: 120 pp [Google Scholar]
- Svitkina T.M., Borisy G.G. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009–1026 10.1083/jcb.145.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobiev S., Strokopytov B., Drubin D.G., Frieden C., Ono S., Condeelis J., Rubenstein P.A., Almo S.C. 2003. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. USA. 100:5760–5765 10.1073/pnas.0832273100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.D., Mullins R.D. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247–288 10.1146/annurev.cellbio.18.040202.112133 [DOI] [PubMed] [Google Scholar]
- Xu X.P., Rouiller I., Slaughter B.D., Egile C., Kim E., Unruh J.R., Fan X., Pollard T.D., Li R., Hanein D., Volkmann N. 2012. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 31:236–247 10.1038/emboj.2011.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero J.B., Coutts A.S., Quinlan M.E., Thangue N.B., Mullins R.D. 2009. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11:451–459 10.1038/ncb1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.