The hsp-16.2 promoter is sufficient for recruitment of hsp-16.2 to nuclear pore complexes in a manner dependent on RNA pol II and ENY-2, but not on full-length mRNA production.
Abstract
Some inducible yeast genes relocate to nuclear pores upon activation, but the general relevance of this phenomenon has remained largely unexplored. Here we show that the bidirectional hsp-16.2/41 promoter interacts with the nuclear pore complex upon activation by heat shock in the nematode Caenorhabditis elegans. Direct pore association was confirmed by both super-resolution microscopy and chromatin immunoprecipitation. The hsp-16.2 promoter was sufficient to mediate perinuclear positioning under basal level conditions of expression, both in integrated transgenes carrying from 1 to 74 copies of the promoter and in a single-copy genomic insertion. Perinuclear localization of the uninduced gene depended on promoter elements essential for induction and required the heat-shock transcription factor HSF-1, RNA polymerase II, and ENY-2, a factor that binds both SAGA and the THO/TREX mRNA export complex. After induction, colocalization with nuclear pores increased significantly at the promoter and along the coding sequence, dependent on the same promoter-associated factors, including active RNA polymerase II, and correlated with nascent transcripts.
Introduction
Increasing evidence argues that the organization of the genome within the nucleus depends on sites of chromosomal anchorage at the inner face of the nuclear envelope (NE; Akhtar and Gasser, 2007; Kind and van Steensel, 2010). This occurs through heterochromatin binding to the stabilizing meshwork of nuclear lamin and associated proteins, or in some cases, such as stress-induced genes in yeast, with nuclear pores (for reviews see Dieppois and Stutz, 2010; Taddei et al., 2010; Egecioglu and Brickner, 2011). Indeed, the visualization of chromatin within the nucleus by electron microscopy has revealed a nonhomogeneous distribution of chromatin (Heitz, 1928). Generally, dark staining heterochromatin that fails to incorporate labeled UTP clusters at the NE, whereas light-staining, transcriptionally competent chromatin is internal (Visser et al., 2000; Rouquette et al., 2009). Closer observation showed, however, that the silent heterochromatic domains are excluded from nuclear pores, which suggests that active chromatin might bind the nuclear pore complex (NPC). Consistently, screens for yeast genes recovered with inner nuclear pore basket components detected both stress-induced genes and ribosomal protein genes (Brickner and Walter, 2004; Casolari et al., 2004; Cabal et al., 2006; Dieppois et al., 2006; Taddei et al., 2006; Yoshida et al., 2010). Furthermore, in both budding yeast and Drosophila melanogaster, the boundary or insulator elements that separate active from inactive chromatin were associated with NPC proteins (Ishii et al., 2002; Kalverda and Fornerod, 2010).
The pore association of activated yeast genes contrasts with the positioning of developmentally regulated genes in worms, flies, and human cells. The latter shift to internal sites upon induction or shift to the nuclear lamina upon tissue-specific repression (for review see Meister et al., 2011). In mammalian cultured cells, the heat shock (HS) gene Hsp70 localized preferentially to internal nuclear speckles upon activation (Hu et al., 2010), whereas the Hsp70 gene in cultured Drosophila Schneider 2 (S2) cells was perinuclear (Kurshakova et al., 2007). Similarly, the up-regulated X chromosome in male flies is pore associated (for review see Akhtar and Gasser, 2007). Given the diversity of these results, it has remained unclear whether mechanisms that tether expressed genes at nuclear pores are conserved.
One common feature of NPC-bound genes in yeast is their response to stressful conditions (Brickner and Walter, 2004; Casolari et al., 2004; Cabal et al., 2006; Dieppois et al., 2006; Taddei et al., 2006). On a molecular level, it appears that Sus1, a protein present both in the histone acetylation and de-ubiquitination complex called SAGA (Spt-Ada-Gcn-Acetyltransferase) and in the TREX-2 (transcription and mRNA export linking) complex, is implicated in pore association (García-Oliver et al., 2012). Sus1 binds directly the nuclear pore protein Nup1 and anchors TREX-2 to the NPC. This mediates gene recruitment upon transcriptional activation and facilitates mRNA processing (García-Oliver et al., 2012). The peripheral anchoring of Hsp70 in Drosophila S2 cells also requires E(y)2/ENY-2, the Sus1 homologue, which colocalizes weakly with nucleoporins, and Xmas2, the homologue of TREX-2 subunit Sac3 (Kurshakova et al., 2007). This suggested, but did not prove, that pore association is linked to mRNA processing or export. Whether the NPC regulates either mRNA synthesis or maturation remained unclear.
Here we explore the link between stress-induced gene activation and subnuclear gene positioning in an intact organism by tracking the essential heat-responsive locus hsp-16.2 in Caenorhabditis elegans. The hsp-16.2 gene is one of four related HS genes found in two clusters of two and four genes on chromosome V (chr V). The smaller cluster contains divergently transcribed hsp-16.41 and hsp-16.2, whose expression level is 14-fold that of the homologous gene in the larger cluster (Stringham et al., 1992). The common promoter region of 394 bp contains two HS elements (HSEs; Fernandes et al., 1994; Trinklein et al., 2004; Guertin and Lis, 2010), which bind the conserved HS transcription factor 1 (HSF-1). HSF-1 is essential for hsp-16.2 activation (Hajdu-Cronin et al., 2004). The hsp-16.2 promoter contains a second HS-associated site (HSAS), with no known ligand, which improves expression in transgenic arrays if the distal HSE is absent (GuhaThakurta et al., 2002).
The promoter-associated changes that correlate with Hsp70 activation are well characterized, particularly in Drosophila. Upon HS, HSF-1 trimerizes and binds in a cooperative manner to HSEs in the promoter (Xiao et al., 1991). Binding of HSF-1 affects the chromatin structure and composition by recruiting coactivators, elongation factors, histone modifying enzymes, and chaperones (for review see Guertin et al., 2010). Even without induction, the promoter is held in an open state and harbors a paused RNA polymerase II (pol II), which produces a short RNA of ∼25 nucleotides (Rougvie and Lis, 1988). Gene activation coincides with the recruitment of positive transcription elongation factor b (P-TEFb), a serine/threonine kinase that phosphorylates the carboxy-terminal domain of the pol II catalytic subunit. This phosphorylation releases paused pol II, which then productively transcribes the gene (Guertin et al., 2010). Activation leads to a rapid eviction of nucleosomes after ∼30 s in a transcription-independent manner, an event proceeded by dTip60-mediated histone H2A acetylation (Petesch and Lis, 2012).
Spatial organization analysis shows that the endogenous hsp-16.2 locus in C. elegans, like transgenes that carry only the hsp-16.2 promoter, are at or near the NE. Super-resolution microscopy shows that upon activation, transgenes bearing the hsp-16.2 promoter colocalize efficiently with the NPC. Thus, peripheral positioning requires both HSE and HSAS elements before induction, but either is sufficient after HS. Importantly, both HSF-1 and active pol II are essential for perinuclear anchoring both before and after induction. We propose that this stress-activated promoter autonomously directs chromatin to the nuclear pore, where continued association correlates with the abundance of engaged RNA polymerases.
Results
Repetitive hsp-16.2 promoter arrays are peripherally retained upon transcriptional induction
By studying arrays bearing developmentally regulated promoters in C. elegans, we have shown that the activation of tissue-specific promoters leads to their relocation toward the nuclear interior (Meister et al., 2010b). We asked whether a stress-induced promoter does the same, using a worm strain homozygous for a large integrated gene array that expresses cytoplasmic GFP under control of the hsp-16.2 promoter (Link et al., 1999). The strain of interest carries a second large gene array expressing GFP-lacI from the constitutive baf-1 promoter (Meister et al., 2010b). GFP-lacI binds to lacO sites on both integrated constructs, allowing live imaging of array position. Given that baf-1::gfp-lacI expression is insensitive to temperature shift (Fig. S1), it serves as an internal control for quantitation.
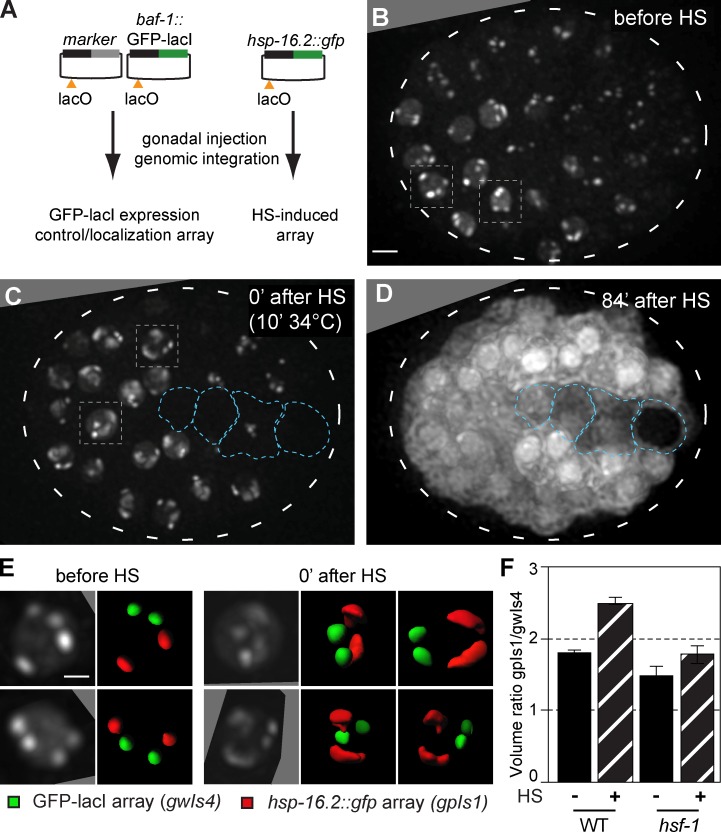
Before HS (10 min at 34°C), the four integrated gene arrays were localized at the NE (Meister et al., 2010b; Towbin et al., 2010; Fig. 1, B and E, before HS). The hsp-16.2 array, which is slightly larger than the control array in the absence of HS (1.8-fold volume; Fig. 1, E and F), expanded visibly and immediately upon temperature shift to occupy 2.6-fold of the control array volume (Fig. 1, C, E, and F). We confirmed that the expanded array contained the hsp-16.2 promoter by repeating this in worms in which only the hsp-16.2 promoter array carries lacO sites (Fig. S1 A). The observed array unfolding resembles that reported during induced expression of array-borne genes in cultured mammalian cells (Tumbar et al., 1999; Hu et al., 2009). However, we note that the expanded HS-induced array did not shift inwards, but rather unfolded along the inner NE (Fig. 1 D). Gene induction and array decondensation were correlated, as both were ablated in a temperature-sensitive hsf-1 mutant (hsf-1(sy441); Hajdu-Cronin et al., 2004; Figs. 1 F and S1 B).
Figure 1.
HS-induced transcription leads to peripheral decondensation of large, HS-inducible arrays. (A) Plasmids used to create large arrays by gonadal injection, bearing either baf-1::GFP-lacI with a muscle-specific marker (gwIs4) or the hsp-16.2 promoter driving GFP (gpIs1), and a single lacO site per plasmid. Arrays contain 250–500 plasmid copies. (B) Fluorescence from an embryo of a strain carrying both gwIs4 and gpIs1 is shown, before HS induction. Four spots reflecting the large integrated gene arrays are located at the nuclear periphery (Meister et al., 2010b; Towbin et al., 2012). Boxed nuclei are enlarged in E. See Fig. S1. Bar, 5 µm. (C) Fluorescence from the same embryo in B after 10 min at 34°C. Boxed nuclei, enlarged in E, show decondensation of two of the arrays. Blue encircled nuclei do not respond to HS. (D) Pervasive cytoplasmic fluorescence from the same embryo in B and C captured 84 min after HS, showing hsp-16.2::gfp induction from the gpIs1 array. Entire embryos are encircled by broken lines. (E) Enlarged nuclei from the embryo in B and C, with two 90° rotations for C. Array volume rendering shows no shift in subnuclear localization of gwIs4 (green) or gpIs1 (red), decondensed at 0 min after HS. Bar, 2 µm. (F) Array volume was quantified using the GFP-lacI signals in WT and hsf-1 mutant worms, presented as the ratio of gwIs4 and HS-activated gpIs1 before and 0 min after HS (WT; n = 100, 11; hsf-1 mutant; n = 3, 6). Error bars indicate mean ± SEM.
The endogenous hsp-16.2 locus is efficiently recruited to the NE upon activation
The fact that HS-responsive arrays remain peripheral after induction could mean either that activation enhances the association or that hsp-16.2 induction cannot overcome heterochromatin tethering (Meister et al., 2010b; Towbin et al., 2012). We therefore analyzed the positioning of the endogenous hsp-16.2 locus in wild-type (WT) C. elegans embryos before and after HS by FISH. The endogenous hsp-16.2 gene is found next to hsp-16.41 on the left arm of chr V, 1.8 Mb from the left telomere. Genome-wide lamin-DAM-ID (Towbin et al., 2012; Fig. 2 A) and LEM-2 chromatin immunoprecipitation (ChIP) analysis (Gerstein et al., 2010; Ikegami et al., 2010) both showed an enrichment of the terminal 4–5 Mb of chr V at the nuclear lamina. To determine the subnuclear position of the hsp-16.2 locus more precisely, we turned to quantitative 3D microscopy.
Figure 2.
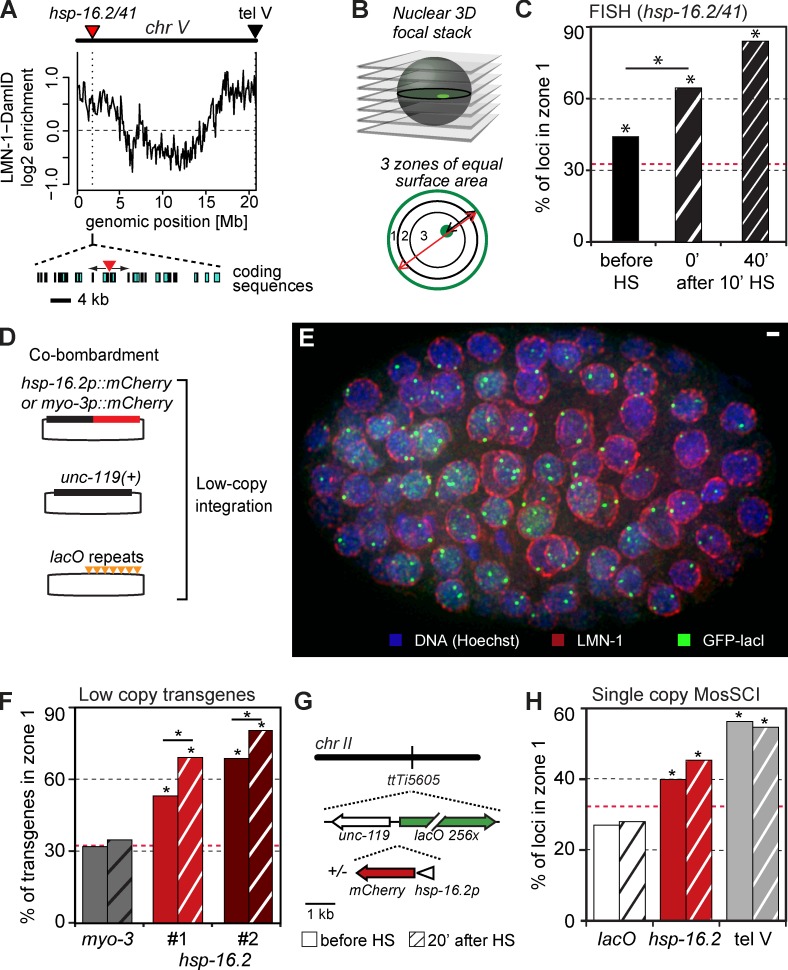
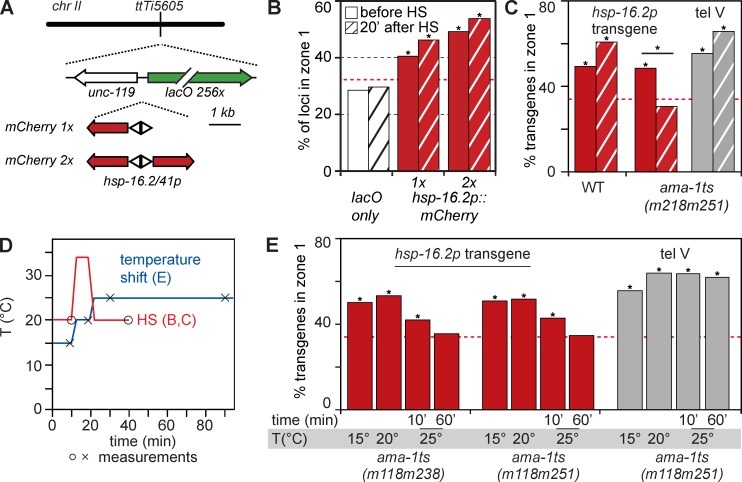
The hsp-16.2 promoter is sufficient to anchor chromatin at the nuclear periphery. (A) Sketch shows the hsp-16.2/41 locus on chr V left arm (red triangle), 1.8 Mb from the telomere, and the MosSCI-inserted lacO repeat (tel V; black triangle), at ttTi9115, 170 kb from TG repeats (Towbin et al., 2012). Below is LMN-1-Dm-ID data showing terminal 4–5 Mb enriched for lamin association (Towbin et al., 2012). An expanded view of coding sequences shows the divergent hsp-16.2 and hsp-16.41 genes. (B) Quantitation of radial positioning of endogenous loci and GFP-lacI–tagged transgenes, as described in Materials and methods. Random localization = 33% in each zone. (C) The endogenous hsp-16.2/41 locus is enriched in zone 1 in early embryos and becomes more enriched after HS. FISH signal positions were quantified for the hsp-16.2 locus as in B. Only zone 1 values are shown (n = 229, 293, 281; P < 0.01 vs. random distribution for all). Red broken lines indicate a 33% or random distribution of the foci against which all values are compared. Asterisks indicate distributions significantly different from random, or different between indicated conditions. (D) Plasmids used to create small bombarded transgenes (tg). mCherry is driven by either the hsp-16.2 or the muscle-specific myo-3 promoter, co-bombarded with an array of 256 lacO sites and the unc-119+ marker. The copy number of the hsp-16.2 promoter is 1 (tg #1) or 74 (tg #2; Fig. S2). (E) Maximal Z projection of a partial 3D reconstruction of a 200-cell-stage embryo (GW421, expressing GFP-lacI) carrying a lacO-tagged transgene gwIs28[hsp-16.2::mCherry; 256xlacO; unc-119+]. The embryo is stained for GFP (anti-GFP, green), nuclear lamina (anti–LMN-1, red), and DNA (Hoechst, blue). Bar, 1 µm. (F) Quantitation of the GFP-lacI signal position for either the myo-3 transgene, or the two hsp-16.2 promoter–containing transgenes, as in B. Zone 1 values and asterisks are defined as in C. Hatched bars, after HS. Loci scored were (left to right) n = 275, 183, 503, 188, 226, and 200; p-values versus random = 0.47, 0.6 for myo-3; and P < 10−10 for hsp-16.2–containing transgenes. A Fisher’s exact test for significance before versus after HS for hsp-16.2 transgenes yielded P = 7.2 × 10−6, 7.5 × 10−3). (G) Sketch of DNA used for MosSCI insertion at ttTi5605 in mid–chr II: lacO sites and unc-119+ were integrated with or without the hsp-16.2 promoter driving mCherry. (H) Quantitation of the GFP-lacI signal for lacO only insertion, the ectopic hsp-16.2::mCherry construct at ttTi5605, and the MosSCI lacO insertion at tel V (see A). Method, bars, and asterisks are defined as in B and C. Numbers scored were (left to right) n = 117, 95, 309, 346, 213, and 204; p-values versus random = 0.17, 0.31, 0.02, 3 × 10−5, 3 × 10−12, and 2 × 10−10, respectively.
We scored the position of the endogenous hsp-16.2 locus before and after HS (10 min at 34°C), relative to the edge of the DAPI-stained nucleus, in embryos of 50–150 cells. Within 3D confocal stacks of images, we located optical sections with the strongest hsp-16.2 FISH signals, divided each focal plane into three zones of equal surface, and scored the FISH signals relative to these zones (Fig. 2 B; Meister et al., 2010a). Even before HS, the hsp-16.2 locus showed a significant enrichment in outermost zone 1 (Fig. 2 C). After induction, the hsp-16.2 locus became more peripheral: zone 1 values increased from 44% to 65% immediately after HS, and to 84% by 40 min (Fig. 2 C). Thus, the endogenous hsp-16.2 gene was enriched at the nuclear rim before exposure to HS, and peripheral positioning was significantly enhanced by HS induction at 34°C.
Promoter sequences are sufficient to mediate perinuclear positioning
To distinguish the relative contributions of chromosomal context, coding, and promoter sequences to gene localization, we generated small transgene arrays by microparticle bombardment that contained only the 394-bp hsp-16.2 promoter driving the mCherry coding sequence, and lacO repeats. The mCherry gene contains two synthetic introns and was flanked by the unc-54 3′ UTR, allowing us to quantify nascent and processed transcripts, and to monitor induction by mCherry fluorescence (Fig. 2 D). We obtained two independent transgenic inserts that contained either 1 (GW421, tg #1) or 74 (GW391, tg #2) copies of the promoter, integrated among 24–88 copies of plasmid, respectively (Fig. S2). Similar small transgenes, e.g., one containing the myo-3 promoter, were shown previously to be randomly positioned in embryonic nuclei, independent of their transcriptional status (Meister et al., 2010b). For embryos with the hsp-16.2 promoter transgenes, we observed a significant enrichment of the array at the NE during growth at normal temperatures (Fig. 2, E and F; hsp-16.2 tg #1 zone 1 = 50% and hsp-16.2 tg #2 zone 1 = 69%), unlike the randomly distributed myo-3 promoter transgene (Fig. 2, E and F). Peripheral enrichment correlated with the promoter copy number, as higher values were scored for tg #2.
After HS, the lacO-tagged hsp-16.2 promoter transgenes were even more peripheral (Fig. 2 F; tg #1 = 69% and #2 = 81%; P vs. non-HS = 7 × 10−6 and 0.008, respectively), whereas the myo-3 transgene retained its random distribution (Fig. 2 F), which is consistent with earlier results (Meister et al., 2010b). Indeed, among all transgenes tested, only ones containing the hsp-16.2 promoter were peripherally enriched in early embryos, and this was the case both before and after HS.
Because it is not possible to control copy number or insertion site of such transgenes, we next used the MosSCI insertion system to integrate desired sequences at a specific target site by homologous recombination, namely at the ttTi5605 locus on chr II (Fig. 2 G; Materials and methods; Frøkjaer-Jensen et al., 2008). This target site (ttTi5605) is randomly localized in embryonic nuclei when tagged with lacO sites only (Fig. 2 H, lacO). To test the effect of a single hsp-16.2 promoter on localization, we integrated a single copy of the hsp-16.2 promoter driving mCherry, along with the lacO repeats. The hsp-16.2::mCherry construct was now significantly enriched at the nuclear rim (39%, P = 0.02 vs. 27%, P = 0.17 for lacO only). After 10 min at 34°C, the proportion of hsp-16.2–containing loci at the nuclear periphery was 44% (before vs. after HS, P = 0.27), whereas the lacO alone single-site insertion remained randomly distributed (28% in zone 1). A MosSCI insertion of lacO repeats into a subtelomeric region on chr V (tel V, Fig. 2 A; Towbin et al., 2012) showed strong association with the nuclear periphery that was unchanged upon HS (Fig. 2 H).
In summary, the endogenous hsp-16.2 locus is distributed nonrandomly with respect to the NE, even under noninducing conditions. Importantly, the hsp-16.2 promoter alone is significantly peripheral when integrated randomly as a transgene, or as a targeted single-copy integration in the middle of chr II (ttTi5605). This argues that the promoter sequence itself drives peripheral localization, even when transcription levels are low. Promoter induction significantly increased enrichment at the nuclear rim at the endogenous locus and in the case of small transgenes (P < 0.7 × 10−5). Chromosomal context could well influence the efficiency of perinuclear localization: the endogenous hsp-16.2 locus is on a distal chromosome arm that is more likely to contact the NE than the mid-chromosomal ttTi5605 MosSCI insert. We note, however, that the hsp-16.2 promoter drives a single transcript at the MosSCI insert, whereas at the endogenous locus there are two divergent transcripts.
Super-resolution microscopy shows colocalization of induced hsp-16.2 transgene with NPC
In yeast, inactive genes at telomeres and silent HM loci contact nonpore sites at the NE, whereas stress-induced genes are enriched at NPCs (Taddei et al., 2010). To examine which NE structure binds the hsp-16.2 promoter either before or after HS in worms, we exploited super-resolution structured illumination microscopy (SR-SIM). With its 100-nm resolving power, we could distinguish individual fluorescently tagged nuclear pores and differentiate them from the lamina (Fig. 3 A and Video 1). We scored the positioning of the GFP-lacI–bound hsp-16.2 promoter tg #1 relative to pore and lamina staining. SR-SIM measurements yielded a mean diameter of 190 nm for an immunostained NPC, which agrees perfectly with measurements derived from structural studies, given a 15 nm size for the 1° and 2° antibodies (Strambio-De-Castillia et al., 2010). Mean NPC density was ∼3 per µm2 or 115 per embryonic nucleus, with a GFP-LacI spot diameter of ∼300 nm, providing sufficient resolution to accurately map locus position relative to pores and lamina.
Figure 3.
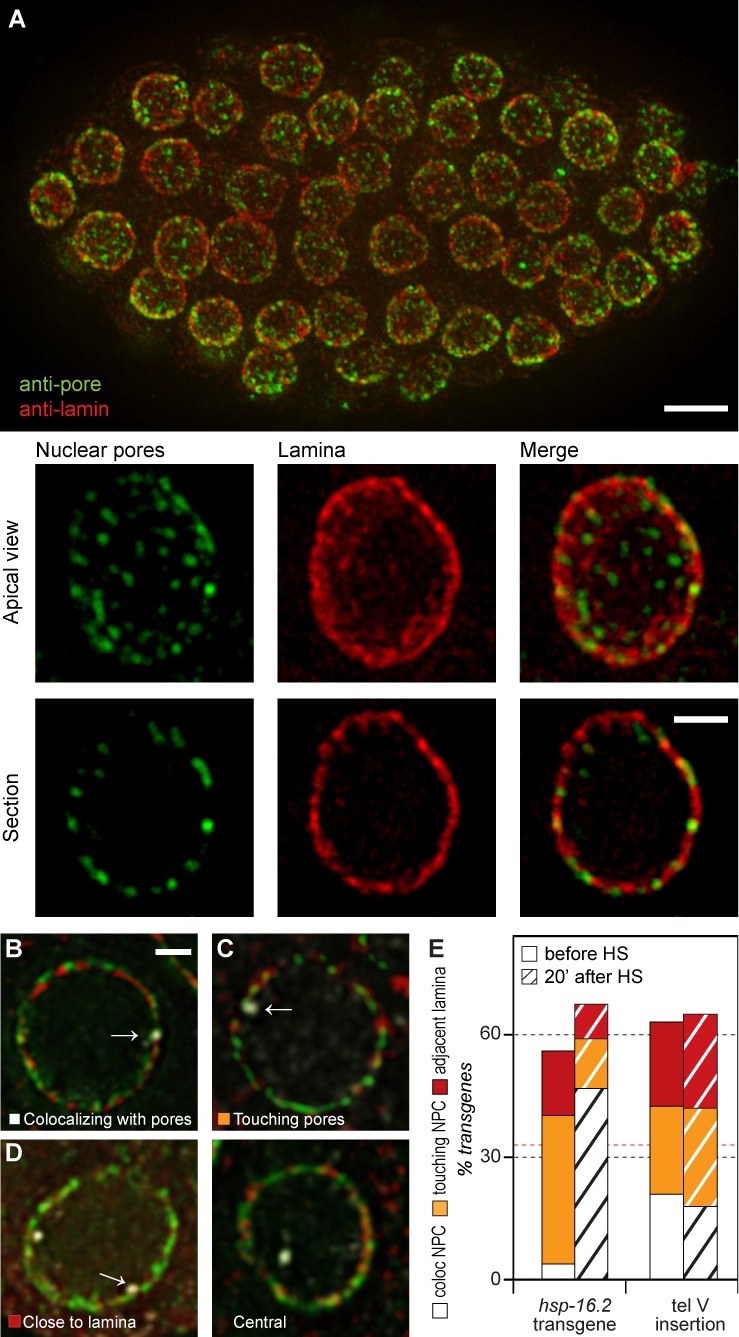
hsp-16.2 transgenes colocalize with pores after HS, and are near pores before. (A) SR-SIM of WT worm embryo immunostained with rabbit anti–LMN-1 (Ce lamin; red) and anti-nucleoporin FG repeat (mAb414; green). See Video 1 for 3D imaging. Below are higher magnifications of an apical and a mid-nuclear view with separated channels. (B–D) Single mid-nucleus focal planes from SR-SIM, showing hsp-16.2 tg1 localization by GFP-lacI signal (white) relative to LMN-1 (red) or NPC (green). Bar, 1 µm. (E) Scoring of hsp-16.2 tg #1 and MosSCI insertion tel V before and after HS (hatched), as in B–D. White, colocalizing with NPC (pore); orange, touching NPC; red, adjacent to lamina. Non-peripheral transgenes are not indicated. The number of hsp-16.2 tg counted, n colocalizing with NPC, n touching NPC, and n adjacent to LMN-1 were as follows. no HS, n = 197, 10, 68, and 35; and after HS, n = 199, 106, 23, and 18. For tel V, numbers were as follows. no HS, n = 215, 45, 46, 44; and after HS (n = 184, 33, 44, 42. The red line is defined as in Fig. 2. Bars: (A, top) 10 µm; (A, bottom) 3 µm; (B–D) 1 µm.
We classified the position of the peripheral hsp-16.2 transgenes as NPC colocalizing (<50 nm between the centers of mass of nearest pore and GFP-lacI spot), touching NPC (>50 nm between centers of mass), or away from NPC and adjacent to lamina (Fig. 3, B–D). The MosSCI lacO insertion at the chr V telomere (Fig. 2 A, tel V) served as a control.
Before HS, 35% of all hsp-16.2 foci were touching the NPC, while a minority were either colocalized with pores (5%) or adjacent to the lamina (18%; Fig. 3 E). After HS, 74% of hsp-16.2 transgene foci were peripheral, of which 53% were colocalized with the NPC and 12% were touching the NPC, whereas a small fraction remained adjacent to lamins (Fig. 3 E, after HS). In contrast, the tel V control locus, despite being strongly perinuclear, showed no favored distribution either before or after HS (Fig. 3 E). Thus, induced hsp-16.2 promoter within the transgene is preferentially associated with nuclear pores.
ChIP confirms preferential hsp-16.2 promoter association with nuclear pores
To confirm that the endogenous hsp-16.2 locus and the MosSCI hsp-16.2 promoter insertion also prefer a pore over the nuclear lamina, we performed ChIP with antibodies against NPP-13, a pore protein located in the intramembrane domain of the NPC (Nic96 in yeast, Nup93 in mammals), and LEM-2, a lamin-associated protein (Ikegami et al., 2010). Quantitative PCR (qPCR) confirms the association of these proteins with the endogenous hsp-16.2 locus (Fig. 4 A, en), as well as the single-copy hsp-16.2 promoter insertion at ttTi5605 (Fig. 2 A, ec; Fig. 2 G and Fig. 4 A). Unrelated loci were used to normalize PCR values among the biological ChIP replicates (ctrl1) and to provide a non–HS-responsive control (ctrl2). These two loci were previously characterized as not interacting with the NPC (unpublished data; Fig. 4 C).
Figure 4.
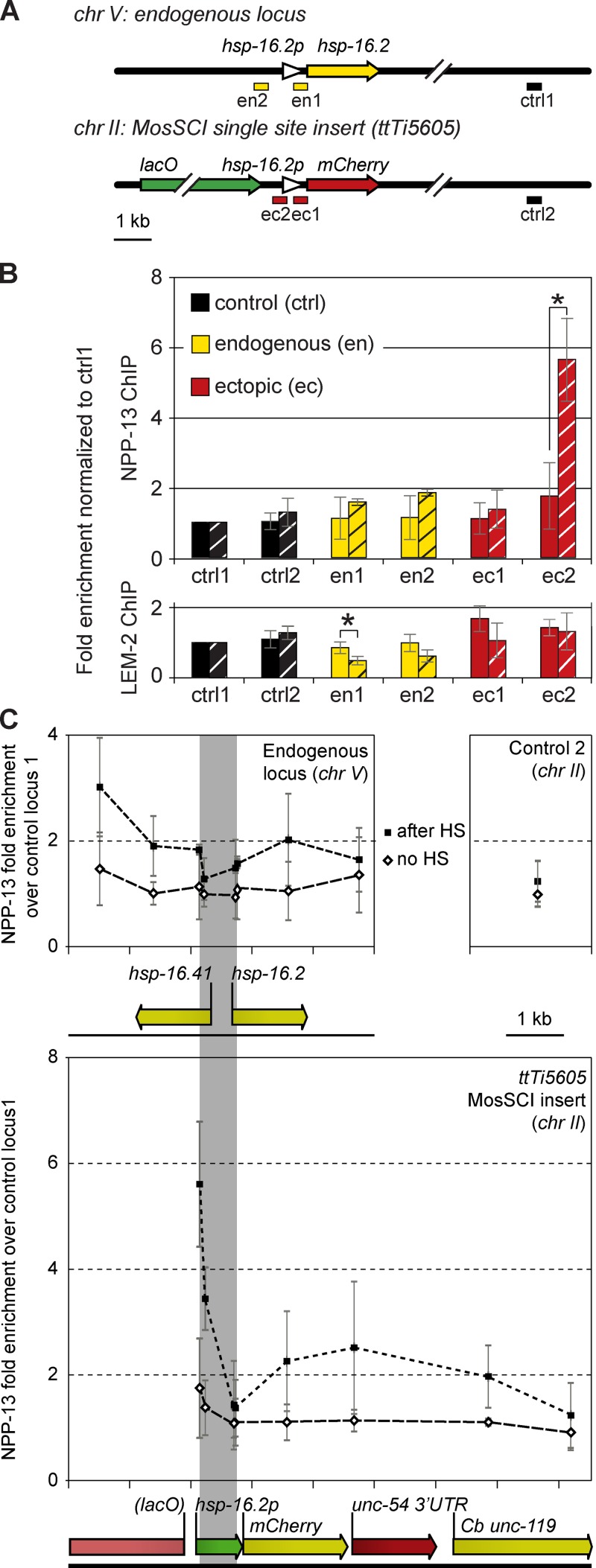
NPC interaction at endogenous hsp-16.2 and at the MosSCI–hsp-16.2 promoter insert increases after HS. (A) Sketch of the endogenous hsp-16.2 locus (top, chr V) and the ectopic MosSCI insert (bottom, chr II) of hsp-16.2::mCherry at ttTi5605, showing probes used for qPCR (en1 and -2, endogenous; ec1 and -2, MosSCI insert). Probes en1, en2, and ec1 are near TSS; control loci ctrl1 and ctrl2 do not to interact with NPC or LMN-1. crtl1 is used for normalization and ctrl2 is not HS-inducible. All ChIP was performed in triplicate on a single strain; see Materials and methods for quantitation. (B) NPP-13 (top) and LEM-2 (bottom) ChIP enrichment of the probes in A, normalized to ctrl1. Values are shown before (unhatched) and 10 min after (hatched) HS. Asterisks indicate statistically significant change. (C) Additional qPCR probes were used to monitor ChIP enrichment at the endogenous hsp-16.2 and MosSCI hsp-16.2 promoter integration, before (Δ) and after HS (▪). Loci are aligned (gray column). ctrl2 normalized to ctrl1 reflects no HS response. Error bars indicate mean ± SEM.
We note that the probes nearest the transcription start sites (TSSs) at the ectopic MosSCI locus (ec1), the endogenous hsp-16.2 (en1), or the hsp-16.41 promoter (en2) failed to cross-link efficiently to NPP-13, either before or after HS. This may be caused by interference in DNA–pore contact by holo–pol II binding. However, the promoter probe further upstream (ec2) yielded a strong enrichment for NPP-13 after HS within the hsp-16.2 MosSCI insertion (Fig. 4 B, compare ec1 with ec2). At the endogenous locus, the association of both en1 and en2 with LEM-2 dropped after HS (Fig. 4 B), which is consistent with the SR-SIM localization data presented in Fig. 3.
Given the poor ChIP efficiency for NPP-13 with the promoter probes used in Fig. 4 A, we extended ChIP analysis along the coding sequence, and to additional sites in the promoters of both the endogenous locus and the hsp-16.2::mCherry MosSCI insertion. In Fig. 4 C, we compared NPP-13 recovery before HS (open diamond) and 10 min after HS (closed squares) and normalized enrichment to the ctrl1 sequence. At both sites we observed a small but significant enrichment for NPP-13 after HS, and the association spread along the coding sequence (Fig. 4 C). The hsp-16.2 MosSCI insertion has particularly strong NPP-13 binding at the distal end of the promoter after HS, confirming the results shown in the preceding paragraph, whereas NPP-13 enrichment was low at the TSS of all constructs analyzed. The presence of NPP-13 along the gene body after HS suggests that the transcribing gene perhaps becomes exposed to contact the NPC. In summary, SR-SIM and ChIP results indicate that before HS, the hsp-16.2 promoter is near, but does not colocalize with the NPC, whereas after HS, the promoter directly interacts with the NPC, as does the 3′ end of each transcribed gene.
Endogenous hsp-16.2 and the MosSCI hsp-16.2 insertion show similar induction kinetics
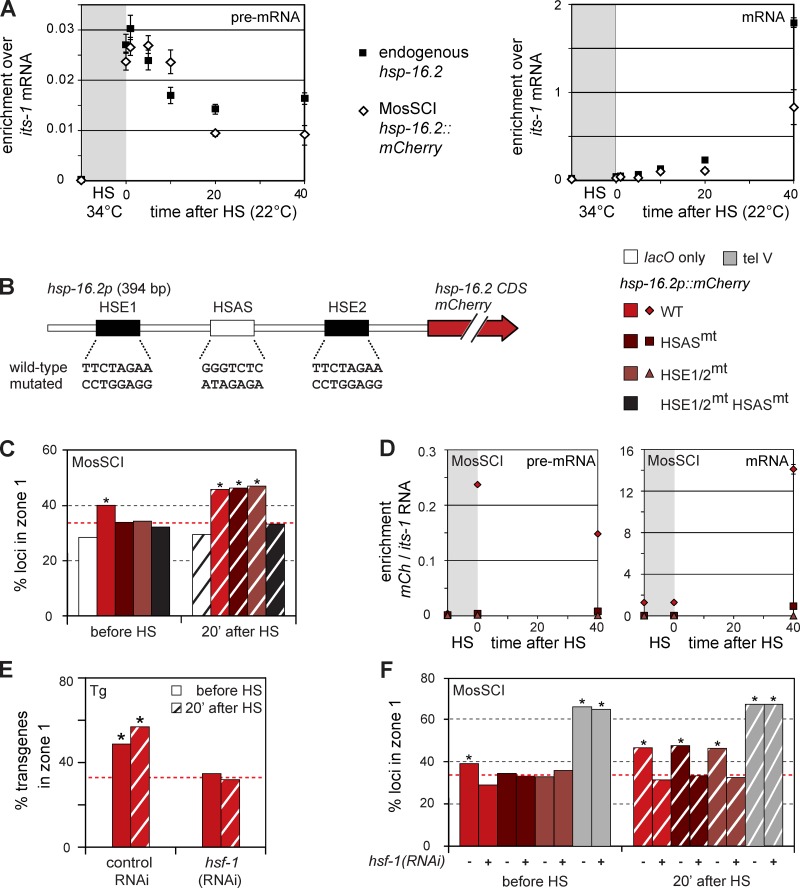
To validate the use of the MosSCI-integrated hsp-16.2 promoter, we analyzed its kinetics of induction alongside those of the endogenous locus by analyzing levels of either nascent (pre-mRNA) or spliced transcripts during HS and for 40 min after return to 22°C. Unspliced (pre-mRNA) was monitored by real-time PCR, using primers that generate a unique intron–exon junction-spanning product. Values are plotted relative to its-1, a transcribed spacer in ribosomal DNA (rDNA), which does not fluctuate upon HS. Both unspliced hsp-16.2 and mCherry transcripts (hsp-16.2 promoter-driven) showed an immediate and proportionate increase after HS (Fig. 5 A). Nascent transcripts peaked 1 min after return to 22°C, which suggested induced rapid transcriptional shutoff. Yet significant amounts of unprocessed transcript persisted for 20–40 min after induction, which is consistent with the persistent NPC anchorage observed at both endogenous and transgenic loci after HS (Figs. 2 and 3).
Figure 5.
HSE and HSAS sequences are necessary to anchor hsp-16.2 at the nuclear rim. (A) Nascent (left) and spliced (right) RNA levels of hsp-16.2 (◇) and mCherry (▪) before and at six time points after HS for the indicated loci. Values are normalized to its-1, a transcribed rDNA spacer. (B) Scheme of the hsp-16.2/41 promoter, containing two HSF-1 binding sites, HSE1 and HSE2, on either side of the HSAS sequence, with hsp-16.2 or mCherry transcription driven toward the right. Changes in mutated versions of HSE1, HSAS, or HSE2 are shown below the sketch. (C) Quantitation of subnuclear positions of the GFP-lacI signal for the MosSCI insertion of WT or mutated hsp-16.2 promoters; color-coded as in B. Zone 1 values, bar labels, and asterisks are defined as in Fig. 2 (B and C). Cell numbers were (left to right) n = 117, 309, 367, 339, 306, 95, 346, 329, 347, and 249. P-values versus random = 0.17, 0.02, 0.80, 0.73, 0.33, 0.31, 3 × 10−5, 1 × 10−6, 1 × 10−6, and 0.59, respectively. (D) Nascent and spliced RNA levels of mCherry driven from WT or mutated hsp-16.2 promoters at MosSCI insertion, normalized to its-1. Values are shown for HSE1/2mt, HSASmt, and WT, color-coded as in B. (E) HSF-1 is essential for peripheral targeting of the hsp-16.2 tg #1 both before and after HS. Quantification of positions of the GFP-lacI signal for progeny from control RNAi or hsf-1 RNAi fed adult progeny as in Fig. 2 B. Zone 1 scoring, asterisks, and hatching are defined the same as in Fig. 2 (B and C). Numbers counted were (left to right) n = 681, 704, 281, and 244. P-values versus random = 3 × 10−4, 3 × 10−7, 0.16, and 0.07, respectively. (F) Quantitation of GFP-lacI focus position for WT or mutated hsp-16.2 promoter MosSCI inserts (coded as in B) and the lacO-tagged insert tel V in progeny of hsf-1 RNAi fed adults. Scoring, asterisks, and hatching are as in Fig. 2 (B and C). Loci counted were (left to right) n = 309, 209, 367, 209, 339, 227, 95, 198, 346, 210, 329, 242, 347, 204, 91, and 209. P-values versus random = 0.02, 0.16, 0.80, 0.54, 0.73, 0.92, <0.01, <0.01, 3 × 10−5, 0.56, 1 × 10−6, 0.88, 1 × 10−6, 0.86, <0.01, and <0.01, respectively.
The appearance and accumulation of spliced mRNA occurred 10–20 min after HS for both loci, with a slight delay for the mCherry mRNA, which has two introns instead of one. Spliced products continue to accumulate for 40 min after HS-induced transcription for both transcripts. We conclude that induction kinetics and processing are very similar for the endogenous gene and the MosSCI insertion (Fig. 5 A), and that NPC association correlates with the timing of transcript elongation and processing.
Two different pathways recruit the hsp-16.2 promoter at the nuclear periphery
Having shown that the hsp-16.2 promoter is sufficient for both rapid induction and relocation of the locus to the NPC, we checked whether both are controlled by the same promoter elements. The bidirectional endogenous hsp-16.2 promoter drives transcription of both hsp-16.2 and hsp-16.41. The promoter contains two HSEs, flanking the HSAS, which has no known ligand (Fig. 5 B). Deletion analysis and induction of high copy number ectopic arrays indicated that the proximal HSE site was most important for transcription, whereas loss of both HSE sites abolished transcription (GuhaThakurta et al., 2002). In addition, mutation of HSAS decreased reporter gene transcription supported by the distal HSE site.
We asked whether mutation of the HSE or HSAS sites would influence hsp-16.2 promoter-driven subnuclear localization. lacO-tagged single-copy reporters with modified hsp-16.2 promoters were introduced into an otherwise identical chromatin environment by MosSCI-mediated recombination at the ttTi5605 locus. Confirming the observations in Figs. 2–4, the MosSCI hsp-16.2 promoter shifted the ttTi5605 locus to the nuclear periphery in 40% of the cells before HS (Fig. 5 C). After HS, NE association increased to 44%. Mutation of either HSAS or HSE elements (Fig. 5, B and C), or both, led to a random distribution before HS (Fig. 5 C; zone 1 = 33% for HSASmt, 32% for HSE1/2mt, and 31% for HSASmt HSE1/2mt). Intriguingly, when the same constructs were monitored 20 min after HS, the locus was significantly peripheral despite loss of HSAS or HSE consensuses (Fig. 5 C; zone 1 = 45% for HSASmt and 46% for HSE1/2mt). Loss of all three sites, however, ablated NE association (Fig. 5 E; 32% for HSASmt HSE1/2mt). In conclusion, before HS, both HSAS and HSE sites are used to position the hsp-16.2 promoter near the periphery, whereas after HS either the HSAS or HSE sequence suffices.
To test whether relocation correlates with expression efficiency, we performed real-time PCR on reverse-transcribed RNA, extracted from the strains bearing mutant and WT hsp-16.2 promoters, before and after HS. Real-time PCR values are expressed relative to those of its-1 (Fig. 5 D; for an agarose gel of amplicons, see Fig. S3). HS robustly induces the WT promoter-driven mCherry, with the same kinetics as the endogenous hsp-16.2 mRNA (Fig. 5 A), although mCherry protein is only visible 80 min after HS (Fig. S4). Mutation of the HSE sites compromises mCherry expression (Fig. 5 D), whether or not the HSAS is intact. There is, nonetheless, a very low level of nascent and spliced transcript detectable in the HSE mutant (Figs. S3 and S4 A), even though induction is compromised and mCherry fluorescence is not detected.
When the HSAS site alone is mutated, leaving functional HSE sites, the expression level of mCherry is reduced ∼10-fold (Fig. 5 D), yet spliced transcript could be detected after 40 min (Fig. S3) and mCherry fluorescence could be detected by 120 min (Fig. S4 A). This argues that HSAS contributes to full activation of hsp-16.2, even in the presence of HSE sequences. Given that HSAS is needed for peripheral positioning before induction, NE positioning before HS correlates with maximal activation.
After HS, the NPC localization of the hsp-16.2 promoter does not require induced RNA levels because promoters that lack the HSE sites (no increased mRNA) or the HSAS site (increased mRNA) support similar recruitment to the NE upon HS (Fig. 5, C and D). The HSAS element may contribute to NPC binding through a factor that contributes to activation, but which is insufficient for induced gene expression. In contrast, HSE elements support both HS-induced transcription and relocation in the absence of HSAS, most probably through HSF-1 and pol II recruitment.
Peripheral anchoring of the hsp-16.2 promoter depends on HSF-1
We next tested whether HSF-1 is needed for peripheral positioning (Figs. 1 and 5 E). We used hsf-1(RNAi) on worms bearing the lacO-tagged hsp-16.2 transgene. Upon depletion of HSF-1 in embryos of RNAi-treated worms, both before and after HS, the hsp-16.2 transgene was released from the nuclear rim (Fig. 5 E; 35% and 32% for hsf-1 RNAi, vs. 50% and 55% control RNAi; n and p-values are given in the figure legend). A myo-3 transgene showed no effect of hsf-1 RNAi (unpublished data). We extended the analysis of hsf-1(RNAi) effects to the MosSCI-integrated hsp-16.2::mCherry construct. For the MosSCI insert with the hsp-16.2 promoter intact or lacking either HSAS or HSE sites, HSF-1 was necessary for the peripheral enrichment scored either before or after HS (Fig. 5 F). As expected, hsf-1(RNAi) had no effect on the tagged tel V locus (Fig. 5 F). We conclude that HSF-1 binding to the hsp-16.2 promoter is necessary for perinuclear localization, both before and after HS. This argues that NPC anchoring through the HSAS site after HS also depends on HSF-1, even though HSF-1 does not bind HSAS directly.
RNA pol II is essential for peripheral anchoring by the hsp-16.2 promoter
Given that HSF-1 is necessary for pol II–dependent elongation, we next tested the role of pol II itself in NPC anchoring of the hsp-16.2 promoter. To see if bidirectional transcription contributes to peripheral anchoring, we added a second mCherry gene between the promoter and the lacO sites at the MosSCI ttTi5605 locus (Fig. 6 A). Peripheral localization increased significantly both before and after HS due to insertion of a second ORF (39% vs. 49% before HS and 44% vs. 53% after HS; Fig. 6 B). Thus, by doubling the amount of pol II bound (or the number of transcripts possible), we increased peripheral anchoring, both before and after HS.
Figure 6.
Active RNA pol II is necessary for peripheral anchoring of the hsp-16.2 promoter. (A) MosSCI insertion at ttTi5605 of the hsp-16.2 promoter driving either one or two mCherry genes as indicated. The top shows control lacking the hsp-16.2 promoter. (B) Quantitation of the GFP-lacI position for lacO-only control and uni- or bidirectional hsp-16.2 promoter constructs (A). Scoring, zone 1 plotting, asterisks, and hatching are the same as in Fig. 2 (B and C). Loci counted were (left to right) n = 117, 95, 309, 346, 232, and 246. P-values versus random = 0.17, 0.31, 0.02, 3 × 10−5, 7 × 10−7, and 3 × 10−11, respectively; for 1× versus 2× genes, both with and without HS, P < 0.05. (C) A thermosensitive (ts) mutation in RNA pol II AMA-1 impairs anchoring of the hsp-16.2 tg #1. GFP-lacI signal positions in WT or ama-1(m218m251) embryos were scored and zone 1 values were plotted before and 20 min after HS (hatched), as in Fig. 2 (B and C). The lacO-tagged tel V insert in the same ama-1ts background shows the opposite effect upon AMA-1 inactivation (HS). Loci counted were (left to right) n = 309, 346, 179, 96, 155, and 155. P-values versus random = 0.02, 3 × 10−5, 6 × 10−5, 0.39, 2 × 10−9, and <10−10, respectively. (D) Scheme of HS kinetics (red indicates samples in B and C) showing gradual temperature increase (E) testing ama-1ts mutants for the hsp-16.2 tg1 position. At × and ○, images were taken and quantified. (E) Progressive temperature increase in the indicated ama-1 mutants releases hsp-16.2 tg #1 without HS. Locus scoring, zone 1, and asterisks are the same as in Fig. 2 (B and C). Loci counted were (left to right) n = 186, 217, 208, 167, 199, 210, 256, 274, 85, 123, 76, and 39. P-values versus random = 6 × 10−6, 1 × 10−8, 0.02, 0.70, 4 × 10−6, 3 × 10−7, 0.01, 0.93, 2 × 10−5, 1 × 10−12, 3 × 10−8, and 2 × 10−4, respectively.
To see if pol II itself was involved in the anchorage, we used two previously characterized thermo-sensitive alleles of the pol II large subunit, ama-1(m118m238) and ama-1(m118m251). These mutations are in the catalytic subunit and appear to trigger pol II dissociation either because of loss of template binding (m118m238 [C777Y, G1406R]) or to the disruption of contacts between AMA-1 and RPB-2, two core subunits of pol II (m118m251 [C777Y, A364V]; Bowman et al., 2011). Animals homozygous for either mutation develop normally at 15–20°C, but arrest early in development at 25°C. At permissive temperature, ama-1 mutants containing the hsp-16.2 tg #1 support NE association, as in WT animals (Fig. 6 C, ama-1ts). However, after 10 min at 34°C, the hsp-16.2 transgene became randomly distributed within the nucleus of mutant embryos (Fig. 6 C, WT vs. ama-1ts). This was not caused by a general release of chromatin from the NE, as the control tel V locus remains enriched in zone 1 upon pol II inactivation (Fig. 6 C, tel V). Engaged and active pol II is specifically involved in hsp-16.2 transgene anchoring. Strikingly, even transient inactivation of pol II leads to release from the NPC.
Given that there is basal transcription of hsp-16.2 under noninducing conditions (Fig. S3) and that the transgenes are peripherally enriched before HS induction, we examined whether progressive inactivation of pol II (AMA-1) by a step-wise temperature increase would ablate hsp-16.2 transgene association with the NE. To test this, we took embryos from two ama-1ts mutant strains at a permissive temperature and increased the temperature while imaging (Fig. 6 D). Results are similar for both mutants: whereas hsp-16.2 transgenes are enriched at the NE at a permissive temperature (15° or 20°C; Fig. 6 E), transgenes progressively lose their peripheral attachment after 10 min at 25°C, and are randomly distributed by 60 min (Fig. 6 E). In contrast, the peripherally located tel V insert was unaffected by temperature shift (Fig. 6 E). We conclude that enzymatically active pol II, either paused or actively transcribing, is necessary to anchor noninduced or induced hsp-16.2 promoters to the NPC.
THO/TREX and anchoring and mRNA export complex factor ENY-2 bridges hsp16.2 to NPC
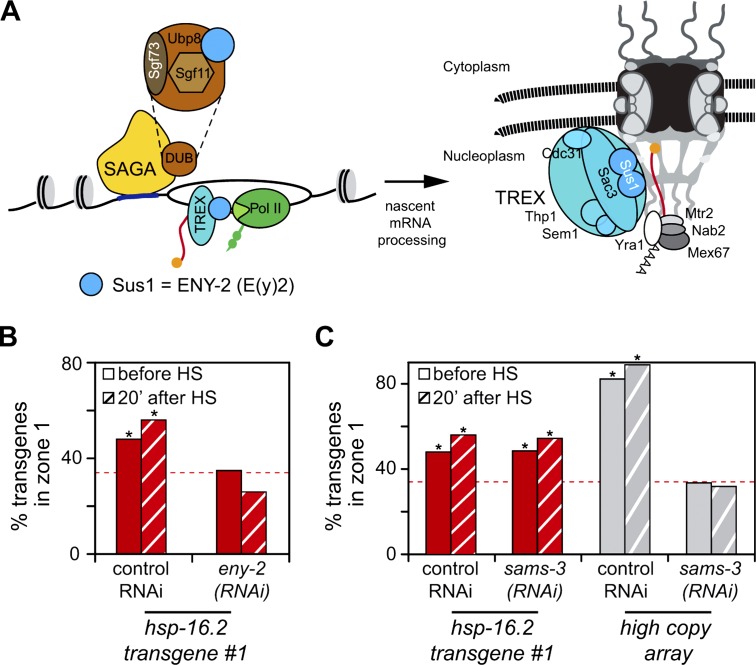
Several complexes that stimulate both elongation and mRNA packaging and export are associated with engaged pol II. One such is the THO/TREX complex (García-Oliver et al., 2012; Fig. 7 A), which is implicated in the NPC anchoring of several yeast inducible genes and the Drosophila Hsp70 cluster. Anchoring requires the S. cerevisiae Sus1 or Drosophila E(y)2/ENY2 subunit (Rodríguez-Navarro et al., 2004; Kurshakova et al., 2007; Kopytova et al., 2010), which has affinity for proteins of the inner nuclear pore basket (Kurshakova et al., 2007).
Figure 7.
The SAGA and THO/TREX component ENY-2 is required for peripheral positioning of hsp-16.2 promoter. (A) Sketch of budding yeast SAGA and THO/TREX complexes with yeast names (García-Oliver et al., 2012). The yeast subunit Sus1 (ENY-2, formerly e(y)2 in flies and T01C3.2 in worms) is present in both complexes (blue circle) and binds the nuclear basket. Other equivalents are Xmas-2 (ScSac3) and DSS1 (ScSem1). (B) Quantitation of hsp-16.2 transgene 1 position in progeny from control RNAi or eny-2(RNAi) fed adults, scored before and after HS. Scoring, zone 1, asterisks, and hatched bars are as in Fig. 2 (B and C). Loci counted were (left to right) n = 681, 704, 157, and 157. P-values versus random = 3 × 10−4, 3 × 10−7, 0.65, and 0.55, respectively. (C) Peripheral position of hsp-16.2 promoter tg #1 is unaffected by sams-3 RNAi, unlike the heterochromatic myo-3 array, GW76. GFP-lacI signals were scored in progeny of control RNAi or sams-3 RNAi fed worms. Scoring, zone 1 plotting, and hatching are as in Fig. 2 (B and C). Loci counted were (left to right) n = 681, 704, 298, and 270. P-values versus random = 3 × 10−4, 3 × 10−7, 2 × 10−8, and 2 × 10−13, respectively.
We examined the peripheral localization of the hsp-16.2 transgenes before and after HS after eny-2(RNAi). Indeed, hsp-16.2 transgene enrichment in zone 1 was lost without ENY-2 (Fig. 7 B, before HS, 35% vs. 44% for control RNAi), and HS treatment did not restore or improve NPC association (26% in zone 1 for eny-2(RNAi) vs. 56% for control RNAi; Fig. 7 B). In contrast, eny-2(RNAi) had no significant effect on myo-3 transgene localization (unpublished data). Furthermore, eny-2(RNAi) delayed the appearance of mCherry fluorescence from 80 to 120 min after HS (Fig. S4), which is consistent with a profound effect on HS gene induction, processing, or export. We conclude that ENY-2 is essential both for anchoring the hsp-16.2 transgene to NPCs and for maximum induction from the hsp-16.2 promoter.
The hsp-16.2–NPC anchoring is distinct from the histone H3K9 methylation-dependent anchoring of heterochromatin, which is ablated by sams-3(RNAi) (Towbin et al., 2012). Down-regulation of S-adenosyl methyltransferases releases large heterochromatic arrays from the NE, but had no effect on the subnuclear distribution of the hsp-16.2 transgene (Fig. 7 C).
Discussion
Several hypotheses have attempted to explain the logic of tethering highly transcribed genes at nuclear pores. Initially, Blobel proposed that targeting genes to pores would spatially facilitate mRNA export to distinct cytoplasmic domains in asymmetric cells, the so-called “gene gating” hypothesis (Blobel, 1985). Alternatively, pore-proximal localization was proposed to establish an epigenetic state that confers a transcriptional memory, facilitating reactivation of stress-inducible genes (Brickner et al., 2007). Other results suggested that the NPC could influence the fine-tuning of mRNA levels (for review see Akhtar and Gasser, 2007) or the degradation of nascent transcripts (Woolcock et al., 2012). Here we implicate specific promoter sequences and the trans-activator HSF-1 in the positioning of both the uninduced and induced C. elegans hsp-16.2 gene at nuclear pores. We rule out chromosomal context and gene-specific intron or 3′ UTR sequences as major determinants of NE binding, and show instead that promoter-bound pol II, but not abundant mRNA, is essential for gene positioning.
The physiological significance of pore-proximal gene positioning has been tested in yeast, yet no function has been shown to be universally relevant (for reviews see Akhtar and Gasser, 2007; Dieppois and Stutz, 2010; Kind and van Steensel, 2010; Egecioglu and Brickner, 2011). In budding yeast, interaction with the NE was essential for maximal transcriptional activity of an inducible subtelomeric gene (Taddei et al., 2006), and the NPC tethering of nontelomeric genes facilitated derepression in the absence of activating factors (Brickner and Walter, 2004). However, in other cases such as ribosomal protein genes, HSP104, and some GAL loci, steady-state levels of mRNA were higher when genes were released from pores (Dieppois et al., 2006; Yoshida et al., 2010; Green et al., 2012). Similarly, transcripts of heat-induced fission yeast genes were kept low by the action of pore-associated Dicer (Woolcock et al., 2012). This diversity of phenotypes linked to gene–pore interaction most likely reflects locus-specific differences in their modes of activation, or in pathways of processing and export.
In metazoans, the association of the X chromosome in male flies contributes to the up-regulation of mRNAs (for review see Akhtar and Gasser, 2007), whereas the Drosophila HSP70 locus was found near the NE both before and after HS in cultured S2 cells (Kurshakova et al., 2007). The down-regulation of E(y)2/ENY2 led to the release of HSP70 from the NE of S2 cells, and reduced mRNA by ∼50% (Kurshakova et al., 2007), although in fly imaginal discs the same locus was not uniformly peripheral (Yao et al., 2007). These results suggest, but do not prove, that NPC association correlates with control over mature mRNA levels in metazoan cells.
Intriguingly, transcription of yeast INO1 and GAL1 was not required either for the establishment or maintenance of their perinuclear positioning (Schmid et al., 2006; Brickner et al., 2007). Analogously, the induced ectopic C. elegans hsp-16.2 promoter was strongly enriched at the NE in the absence of HSF-1 binding sites, although the production of full-length mRNA was significantly impaired (Fig. 5, C and D). The perinuclear positioning of the hsp-16.2 promoter is nonetheless sensitive to the loss of functional pol II both before and after HS. Reconciling these results, we propose that either a paused polymerase, or short pol II–dependent transcripts, drive association of C. elegans genes with the NPC.
The hsp-16.2 promoter autonomously determines perinuclear localization
By inserting a 394-bp hsp-16.2 promoter, either as a low-copy number array or as a single-copy MosSCI insert, we found that perinuclear positioning is intrinsic to the promoter sequence. No sequence was shown to drive positioning of the Drosophila Hsp70 locus (Kurshakova et al., 2007), whereas in yeast, a short “DNA zip code” called gene recruitment sequence I (GRS-I) led to NPC association when inserted ectopically (Ahmed et al., 2010). Intriguingly, at the endogenous yeast INO1 locus, pore targeting required an inositol-dependent event, which suggests that a change in either transcription factor affinity or in surrounding sequences can alter the positioning of the uninduced locus (Ahmed et al., 2010).
In our case, HSF-1 down-regulation, as well as conditional mutations that trigger pol II release from the template, impaired NE localization both before and after HS (Figs. 5 and 6). We propose that the recruitment of additional factors upon HS allows a HSE-deficient promoter to confer perinuclear localization but not to support induced gene expression. A similar situation may occur in budding yeast, where relocalization of GRS-I–containing promoters required transcriptional induction and binding of Put3, an activating transcription factor (Ahmed et al., 2010; Brickner et al., 2012). At hsp.16.2, the relevant factor is likely to be the NPC-binding Sus1 homologue, ENY-2, part of SAGA and THO/TREX (Fig. 8).
Figure 8.
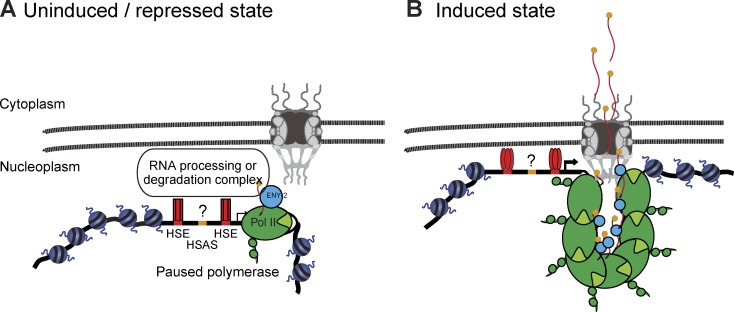
Model of hsp-16.2 positioning in relation to NPC before and after stress induction. Model based on the results presented, proposing that hsp-16.2 binds nuclear pores before and after HS in a similar manner, except that before HS nascent RNA is degraded and after HS it is efficiently spliced and exported. See text for details.
Our ability to exploit well-characterized ts mutants of the large pol II RPB1 subunit (AMA-1 in worms) allowed us to show that functional pol II is essential for NE positioning of hsp-16.2. Loss of template binding (m118m238 [C777Y, G1406R]) or loss of interaction between the two core subunits AMA-1 and RPB-2 (m118m251 [C777Y, A364V]; Bowman et al., 2011) compromise the perinuclear enrichment of the hsp-16.2 promoter both before and after HS. Consistent with the notion that pol II is engaged at an uninduced hsp-16.2 promoter, genome-wide mapping revealed endogenous hsp-16.2 transcripts under normal embryonic growth conditions (www.modencode.org), which we confirmed by RT-PCR of hsp-16.2 promoter-driven mCherry transcripts (Fig. S3). ModEncode ChIP data (www.modencode.org) suggest the presence of pol II along the entire hsp-16.2 gene before HS, with enrichment of its unphosphorylated form in the promoter. Small RNAs were detected at the 5′ end of the gene, arguing that pol II may be stalled or nonproductively engaged at the hsp-16.2 promoter. These results suggest the presence of engaged pol II before HS. However, the hsp-16.2p::mCherry reporter is peripheral before HS, with extremely low levels of spliced mCherry mRNA (Fig. S3), and some mutant promoters supported relocation (HSEmt) without supporting induced transcription (Fig. 5). Thus, processed mRNA (as opposed to engaged pol II) is unlikely to be the critical link to the NE.
In Drosophila and other organisms, paused pol II is found 30–50 bp away from the HSP70 TSS (Levine, 2011). Two complexes, negative elongation factor (NELF) and DRB sensitivity–inducing factor (DSIF) bind the nascent transcript, and DSIF is phosphorylated by P-TEFb, a cyclin/Cdk complex, to release paused pol II (Levine, 2011). There is no reported homologue of NELF in C. elegans (Baugh et al., 2009), but the C. elegans DSIF homologue has been shown to repress transcription of hsp-16.2 (Shim et al., 2002). From our studies, we conclude that perinuclear anchoring of the uninduced hsp-16.2 is mediated by a complex based either on engaged pol II or on the unprocessed mRNA itself, and that factors associated with an engaged pol II, such as the ENY-2–containing THO/TREX complex or SAGA-associated DUB, are likely tethers (Fig. 8). Consistently, loss of ENY-2 correlated with loss of peripheral enrichment of the hsp-16.2 transgene.
Strong colocalization of the hsp-16.2 promoter with NPC after induction
Our study demonstrates by super-resolution microscopy that after HS induction, the majority of hsp-16.2 transgenes colocalize with the NPC, whereas before HS the same transgene is near but not overlapping with nuclear pores (Fig. 3). Similarly, ChIP results show enriched pore association, and a drop in LEM-2 association, after HS. Thus, gene induction coincides with a shift of the hsp-16.2 promoter from a site near pores to direct NPC colocalization, where it remains even after a return to 22°C. This is consistent with two related modes of association, both dependent on pol II, but one being enhanced by nascent transcript.
Rapid induction and rapid repression: the dual function of nuclear pores?
Locus retention at the pore after HS, despite a return to 22°C, could also reflect a process that either degrades or delays processing of the induced mRNA, triggered by the return to non-HS conditions. Indeed, NPC association may promote both message processing and export, as well as degradation, depending on the transcription factors bound to the promoter, and perhaps their phosphorylation status. It is noteworthy that in mammalian cells, c-Myc appears to regulate not only induction of c-Myc regulated genes, but also the stability of the resulting mRNA in the cytoplasm (Rounbehler et al., 2012).
Based on recent results in fission yeast (Woolcock et al., 2012), we propose that pol II– and mRNA-mediated association of stress-inducible genes with nuclear pores has evolved to control genes that require rapid induction, rapid repression, and efficient clearance of unwanted mRNA once conditions change. In S. pombe, a set of divergently transcribed heat-induced genes were seen to be repressed cotranscriptionally by the RNAi machinery (Woolcock et al., 2012). Intriguingly, at least some of these genes are targeted to the NPC for induction, and remain there after the shift back to noninducing conditions (Woolcock et al., 2012). RNA degradation, or simply impaired processing of unspliced mRNAs, may be used to down-regulate temperature-induced genes once the stress is no longer present. It will be interesting to test whether enzymes that control splicing or cotranscriptional RNA degradation also contribute to the persistent NPC localization of HS-induced genes in C. elegans.
Materials and methods
Transgenic strains and molecular biology
Standard C. elegans culture conditions and crossing procedures were used. Unless otherwise stated, worms were maintained at 22.5°C. GFP-lacI was expressed from gwIs39[baf-1::GFP-lacI; vit-5::GFP]. For strains bearing more than one large array, integrations were performed separately and backcrossed to WT worms before crossing strains. A full strains list is available in Table S1. Note that although a blastP of T01C3.2 against fly or human protein databases does not detect ENY2, the Brugia malayi homologue of T01C3.2 detects ENY2 in both flies and mammals (WormBase).
Cloning of hsp-16.2p::mCherry constructs
The hsp-16.2 WT promoter construct driving mCherry expression was obtained by replacing GFP in pPD49.78 by WmCherry. hsp-16.2 promoter constructs containing mutations in HSE or HSAS sites were created by replacing the promoter in pPD49.78 with the mutated promoters described in GuhaThakurta et al. (2002). Specifically, the original sequence of HSE (5′-TTCTAGAA-3′) was replaced by 5′-CCTGGAGG-3′, and the original HSAS sequence (5′-GGGTCTC-3′) was replaced by 5′-ATAGAGA-3′. Insertion of lacO repeats at ttTi5605 (middle of chr II) was achieved by cloning the repeats from pSR1 (Rohner et al., 2008) into pCJ151 (Frøkjaer-Jensen et al., 2008). To insert hsp-16.2::mCherry constructs at ttTi5605, hsp-16.2::mCherry fusions (WT or mutated promoter versions) were inserted in pCFJ151-lacO. For the bidirectional promoter construct, the mCherry coding sequence was obtained by PCR and was inserted in appropriately digested pPD49.78-mCherry. Insertion of lacO repeats on tel V was achieved using Mos insertion ttTi9115 (right arm of chr V). The myo-3::mCherry promoter fusion used is described in Meister et al. (2010b).
Small transgenes were obtained by microparticle co-bombardment of hsp-16.2::mCherry with a lacO repeat construct (pSR1) (Rohner et al., 2008) and the unc-119 rescuing construct (Fig. 2 D). In brief, worms were bombarded with micrometer-sized gold beads loaded with DNA. Bombardment leads to the formation of small-sized transgenes containing 10–100 cointegrated plasmids (Praitis, 2006). Strains were backcrossed to unc-119(ed3) III parents after integration. MosSCI strains were obtained according to the method of Frøkjaer-Jensen et al. (2008), a method based on homologous recombination targeted to transposon sites, through either a direct or indirect method.
FISH
For the single-gene FISH, cosmid F36H9 and a plasmid covering hsp-16.2 were labeled with Alexa Fluor 555 and Alexa Fluor 647, respectively, using the FISHTag kit (Invitrogen). FISH was performed as follows: embryos from bleached worms were fixed for 5 min in 2% PFA and spread on poly-l-lysine–coated slides. They were freeze-cracked on dry ice before 2 min of dehydration in 100% ethanol. Slides were washed in SSC, treated with RNase, and dehydrated progressively in 70%, 85%, 95%, and 100% ethanol, then air-dried for 5 min. The sample was denatured with heat after probe addition. Probe and samples were incubated overnight at 37°C before stringent washes in SSC buffers. Samples were DAPI-stained quickly before mounting in ProLong Gold antifade (Invitrogen). Image acquisition was performed at room-temperature on a wide-field microscope (DeltaVision; Olympus IX70) with a UPlan-SApochromat 100×/1.4 NA UIS2 oil objective lens, a charge-coupled device (CCD) camera (HQ CoolSNAP; Photometrics), and SoftWoRx software (Fig. 2 C). Samples were deconvolved using the multidimensional deconvolution software Huygens (Scientific Volume Imaging), and position scoring relative to nuclear periphery was determined with ImageJ software (National Institutes of Health) using the Point Picker plugin (http://bigwww.epfl.ch/thevenaz/pointpicker). This experiment was performed twice.
Immunofluorescence staining for SIM
For immunostaining, embryos were fixed for 5 min in 2% PFA before freeze-cracking, followed by dehydration in −20°C 100% ethanol. After three washes in PBS and 0.25% Triton X-100 (PBS-T), slides were blocked in PBS-T 0.5% BSA before 1 h of incubation with primary antibody (mAB414 [ab24609; Abcam]; anti–LMN-1 [a gift of Y. Gruenbaum, The Hebrew University of Jerusalem, Jerusalem, Israel]; or anti-GFP [D153_3; MBL]) at RT. After three washes with PBS-T, samples were incubated for 1 h with secondary antibodies (Alexa Fluor 488 anti–rabbit and Alexa Fluor 555 anti–mouse [Fig. 3 A], or Alexa Fluor 488 anti–rat, Alexa Fluor 555 anti–mouse, and Alexa Fluor 647 anti–rabbit [Fig. 3, B–D]; Invitrogen) at RT before final washes and DNA staining with Hoechst 33258.
Microscopy and quantitation
Live microscopy was performed on 2% agarose pads at room temperature. For microscopy of embryos, a spinning disk multipoint confocal microscope (AxioImager M1 [Carl Zeiss] + Yokogawa CSU-22 scan head, plan-Neo-Fluar 100×/1.45 NA oil objective lens, an EM-CCD camera [Cascade II; Photometrics], and MetaMorph 7.7.2 software [Molecular Devices]) was used (Figs. 1, 2 [E, F, and H], 5 [C, E, and F], 6 [B, C, and E], and 7 [B and C]). For each picture, a stack with a z spacing of 0.2 µm was taken and deconvolved using the multidimensional deconvolution software Huygens (Scientific Volume Imaging). 3D reconstructions used Imaris software (Bitplane). For quantitative analysis of arrays, transgenes, and locus position, measurements were made with ImageJ using Point Picker, and scoring of radial positioning of endogenous loci and GFP-lacI–tagged transgenes were performed as described in Meister et al. (2010a). In brief, through-focus stacks of images are acquired at 200-nm intervals, and in the plane of the fluorescent locus, the nuclear cross section is divided in three concentric zones of equal surface area. To score spot position, the ratio of the distance from spot center to periphery (black line) over the nuclear radius (red line/2) is determined for many foci, which are binned into zones 1–3, such that a random localization would imply 33% in each zone. All scoring was performed on at least two biologically independent experiments. Significance of locus distribution relative to a random distribution was performed using a χ2 test with a degree of freedom of 2, when an entire distribution is compared with a random distribution, whereas the significance of changes in two test conditions (e.g., zone 1 in ±HS) was determined using the Fisher’s exact test. For position scoring on SR-SIM microscopy, single sections with lacO/GFP-lacI spots were scored relative to the NPC and lamina. The transgene position analysis (Fig. 3 E) is based on two independent experiments, scored by two independent researchers.
Z projections of embryos were done in ImageJ using maximal intensity projection. High-resolution imaging was done with a super-resolution structured illumination microscope (Elyra S.1 [Carl Zeiss], Plan-Apochromat 63×/1.4 NA objective lens, EM-CCD camera [iXon 885; Andor Technology], and ZEN Blue 2010 D software [Carl Zeiss]; Fig. 3) at RT. Processing was performed with Zen software (Carl Zeiss) and 3D reconstruction and analysis was performed with Imaris software.
The Lamin Dam-ID study shown in Fig. 2 A was performed as described previously (Towbin et al., 2012) using LMN-1-Dam-ID data from three biological replicates of WT C. elegans embryos.
Temperature shift experiments
For HS, the embryos or worms were shifted to 34°C for 10 min either in a slide incubator for a PCR machine or in a water bath. For gradual increase of temperature on the microscope stage, embryos from adults grown at 15°C were transferred on 2% agarose pads on a mini-stage temperature controller (CB164-V1; EMBL). Embryos were imaged at 15°C, temperature was shifted at 20°C, then at 25°C. Pictures were taken always after a 10-min incubation, and, while at 25°C, images were taken again after 1 h.
RNAi experiments
RNAi was performed by feeding on plates as described previously with minor adaptations (Timmons et al., 2001). Worms were put on the feeding plates either as L4 and left for 24 h (hsf-1, eny-2) or left for two generations on RNAi plates starting with synchronized L1s (sams-3). An EcoRV fragment containing 25 bp of perfect identity to GFP-LacI was removed from vector L4440 (Fire vector library) and used as a mock RNAi control.
RNA extraction and qPCR
Extraction of RNA was performed on embryos according to the WormBook protocol (Stiernagle, 2006). The RNA was purified using the RNeasy kit (catalog no. 74104; QIAGEN) including DNase treatment. Reverse transcription PCR was done with ProtoScript AMV First Strand cDNA Synthesis kit (E6550S; New England Biolabs, Inc.). Real-time qPCR was done with GoTaq qPCR Master Mix with an ABI 7500 Fast qPCR machine. Primer sequences can be found in Table S2. The data shown in Fig. 5 (A and D) were obtained by RNA extraction of two independent experiments.
ChIP-qPCR
Standard worm culture techniques were used to obtain embryos from GW615 strain in S liquid media (Stiernagle, 2006). Embryos in M9 buffer were divided into two aliquots: one aliquot was subsequently incubated in 34°C M9 buffer for 10 min (HS), while the other aliquot was incubated in 20°C M9 buffer for 10 min (non-HS). Embryos were further incubated in buffer at 25°C for 10 min and then cross-linked in 2% formaldehyde solution. Chromatin extracts were prepared by sonicating the cross-linked embryos in FA buffer (50 mM Hepes/KOH, pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and 150 mM NaCl). ChIP was performed by incubating the chromatin extract with anti–NPP-13 or anti–LEM-2 antibodies immobilized onto Protein A–conjugated Sepharose beads (Ikegami et al., 2010). Affinity-purified rabbit polyclonal antibodies for NPP-13 (aa 667–766; SDQ4094) and for LEM-2 (aa 1–100; SDQ4051) were produced by genetic immunization at SDIX. After 12 cycles of ligation-mediated PCR amplification, the ChIP DNA was quantified using real-time PCR amplification monitored by a SYBR green dye in a PCR instrument (7900HT; Applied Biosystems). ChIP enrichment represented by an amount of ChIP DNA ([ChIP]) over an amount of input DNA ([Input]) at a given locus was calculated by: [ChIP]/[Input] = 2(CTinput − CTchip)/PCR efficiency, where CTinput and CTchip are Ct (threshold cycle) values for input and ChIP DNA, respectively. PCR efficiencies for primer pairs were: ctrl1, 0.998; ctrl2, 0.971; en1, 1.034; en2, 1.044; ec1, 0.951; and ec2, 1.002.
Online supplemental material
Fig. S1 shows controls for the HS induced integrated array of hsp-16.2 promoters (Fig. 1). Fig. S2 shows quantification by real-time PCR of the number of plasmids present in the small arrays carrying hsp-16.2::mCherry (Fig. 2). Fig. S3 shows that PCR amplification detects mCherry mRNA in a reverse transcription–dependent manner. Fig. S4 shows mCherry protein detection after HS induction. Video 1 shows 3D reconstruction of WT C. elegans embryos stained for nuclear pores and nuclear lamina (Fig. 3). Table S1 gives an overview of C. elegans strains used including genetic details. Table S2 gives an overview of primers used. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207024/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201207024.dv.
Supplementary Material
Acknowledgments
We thank Y. Gruenbaum for antibodies; M. Thomas, R. Arpagaus, and I. Katic for assistance; I. Katic, the Caenorhabditis Genetics Center, D. Riddle, and P. Sternberg for strains; C. Link for plasmids; and R. Thierry, J. Pielage, L. Gelman, and S. Bourke for microscopy support.
This work was supported by the Novartis Research Foundation and the “Fondation Suisse de Recherche sur les Maladies Musculaires.”
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- chr
- chromosome
- DSIF
- DRB sensitivity–inducing factor
- HS
- heat shock
- HSAS
- HS-associated site
- HSE
- HS element
- HSF-1
- HS transcription factor 1
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- pol
- polymerase
- qPCR
- quantitative PCR
- rDNA
- ribosomal DNA
- SAGA
- Spt-Ada-Gcn-Acetyltransferase
- SR-SIM
- super-resolution structured illumination microscopy
- tel
- telomere
- TSS
- transcription start site
- WT
- wild type
References
- Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., Brickner J.H. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A., Gasser S.M. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Baugh L.R., Demodena J., Sternberg P.W. 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 324:92–94 10.1126/science.1169628 [DOI] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 82:8527–8529 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman E.A., Riddle D.L., Kelly W. 2011. Amino acid substitutions in the Caenorhabditis elegans RNA polymerase II large subunit AMA-1/RPB-1 that result in α-amanitin resistance and/or reduced function. G3 (Bethesda). 1:411–416 10.1534/g3.111.000968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J.H., Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., Brickner J.H. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Ahmed S., Meldi L., Thompson A., Light W., Young M., Hickman T.L., Chu F., Fabre E., Brickner J.H. 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev. Cell. 22:1234–1246 10.1016/j.devcel.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., Nehrbass U. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Dieppois G., Stutz F. 2010. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J. Cell Sci. 123:1989–1999 10.1242/jcs.053694 [DOI] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26:7858–7870 10.1128/MCB.00870-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D., Brickner J.H. 2011. Gene positioning and expression. Curr. Opin. Cell Biol. 23:338–345 10.1016/j.ceb.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M., Xiao H., Lis J.T. 1994. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor—heat shock element interactions. Nucleic Acids Res. 22:167–173 10.1093/nar/22.2.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Oliver E., García-Molinero V., Rodríguez-Navarro S. 2012. mRNA export and gene expression: The SAGA-TREX-2 connection. Biochim. Biophys. Acta. 1819:555–565 10.1016/j.bbagrm.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Gerstein M.B., Lu Z.J., Van Nostrand E.L., Cheng C., Arshinoff B.I., Liu T., Yip K.Y., Robilotto R., Rechtsteiner A., Ikegami K., et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 330:1775–1787 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E.M., Jiang Y., Joyner R., Weis K. 2012. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol. Biol. Cell. 23:1367–1375 10.1091/mbc.E11-06-0547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin M.J., Lis J.T. 2010. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 6:e1001114 10.1371/journal.pgen.1001114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin M.J., Petesch S.J., Zobeck K.L., Min I.M., Lis J.T. 2010. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 75:1–9 10.1101/sqb.2010.75.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuhaThakurta D., Palomar L., Stormo G.D., Tedesco P., Johnson T.E., Walker D.W., Lithgow G., Kim S., Link C.D. 2002. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 12:701–712 10.1101/gr.228902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin Y.M., Chen W.J., Sternberg P.W. 2004. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 168:1937–1949 10.1534/genetics.104.028423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. 1928. Das Heterochromatin der Moose. Jahrbücher für wissenschaftliche Botanik. 69:762–818 [Google Scholar]
- Hu Y., Kireev I., Plutz M., Ashourian N., Belmont A.S. 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J. Cell Biol. 185:87–100 10.1083/jcb.200809196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Plutz M., Belmont A.S. 2010. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J. Cell Biol. 191:711–719 10.1083/jcb.201004041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Egelhofer T.A., Strome S., Lieb J.D. 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11:R120 10.1186/gb-2010-11-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U.K. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 109:551–562 10.1016/S0092-8674(02)00756-0 [DOI] [PubMed] [Google Scholar]
- Kalverda B., Fornerod M. 2010. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 9:4812–4817 10.4161/cc.9.24.14328 [DOI] [PubMed] [Google Scholar]
- Kind J., van Steensel B. 2010. Genome-nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol. 22:320–325 10.1016/j.ceb.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Kopytova D.V., Orlova A.V., Krasnov A.N., Gurskiy D.Y., Nikolenko J.V., Nabirochkina E.N., Shidlovskii Y.V., Georgieva S.G. 2010. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 24:86–96 10.1101/gad.550010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova M.M., Krasnov A.N., Kopytova D.V., Shidlovskii Y.V., Nikolenko J.V., Nabirochkina E.N., Spehner D., Schultz P., Tora L., Georgieva S.G. 2007. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 26:4956–4965 10.1038/sj.emboj.7601901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. 2011. Paused RNA polymerase II as a developmental checkpoint. Cell. 145:502–511 10.1016/j.cell.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C.D., Cypser J.R., Johnson C.J., Johnson T.E. 1999. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 4:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Gehlen L.R., Varela E., Kalck V., Gasser S.M. 2010a. Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol. 470:535–567 10.1016/S0076-6879(10)70021-5 [DOI] [PubMed] [Google Scholar]
- Meister P., Towbin B.D., Pike B.L., Ponti A., Gasser S.M. 2010b. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 24:766–782 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Mango S., Gasser S.M. 2011. Locking the genome: nuclear organization and cell fate. Curr. Opin. Genet. Dev. 21:167–174 10.1016/j.gde.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch S.J., Lis J.T. 2012. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol. Cell. 45:64–74 10.1016/j.molcel.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V. 2006. Creation of transgenic lines using microparticle bombardment methods. Methods Mol. Biol. 351:93–107 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro S., Fischer T., Luo M.J., Antúnez O., Brettschneider S., Lechner J., Pérez-Ortín J.E., Reed R., Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 116:75–86 10.1016/S0092-8674(03)01025-0 [DOI] [PubMed] [Google Scholar]
- Rohner S., Gasser S.M., Meister P. 2008. Modules for cloning-free chromatin tagging in Saccharomyces cerevisae. Yeast. 25:235–239 10.1002/yea.1580 [DOI] [PubMed] [Google Scholar]
- Rougvie A.E., Lis J.T. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 54:795–804 10.1016/S0092-8674(88)91087-2 [DOI] [PubMed] [Google Scholar]
- Rounbehler R.J., Fallahi M., Yang C., Steeves M.A., Li W., Doherty J.R., Schaub F.X., Sanduja S., Dixon D.A., Blackshear P.J., Cleveland J.L. 2012. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 150:563–574 10.1016/j.cell.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J., Genoud C., Vazquez-Nin G.H., Kraus B., Cremer T., Fakan S. 2009. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 17:801–810 10.1007/s10577-009-9070-x [DOI] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U.K. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21:379–391 10.1016/j.molcel.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Shim E.Y., Walker A.K., Shi Y., Blackwell T.K. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135–2146 10.1101/gad.999002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. WormBook. 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Stringham E.G., Dixon D.K., Jones D., Candido E.P. 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell. 3:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S.M. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 441:774–778 10.1038/nature04845 [DOI] [PubMed] [Google Scholar]
- Taddei A., Schober H., Gasser S.M. 2010. The budding yeast nucleus. Cold Spring Harb. Perspect. Biol. 2:a000612 10.1101/cshperspect.a000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D.L., Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Towbin B.D., Meister P., Pike B.L., Gasser S.M. 2010. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb. Symp. Quant. Biol. 75:555–565 10.1101/sqb.2010.75.041 [DOI] [PubMed] [Google Scholar]
- Towbin B.D., González-Aguilera C., Sack R., Gaidatzis D., Kalck V., Meister P., Askjaer P., Gasser S.M. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 150:934–947 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Trinklein N.D., Murray J.I., Hartman S.J., Botstein D., Myers R.M. 2004. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol. Biol. Cell. 15:1254–1261 10.1091/mbc.E03-10-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., Belmont A.S. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354 10.1083/jcb.145.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser A.E., Jaunin F., Fakan S., Aten J.A. 2000. High resolution analysis of interphase chromosome domains. J. Cell Sci. 113:2585–2593 [DOI] [PubMed] [Google Scholar]
- Woolcock K.J., Stunnenberg R., Gaidatzis D., Hotz H.R., Emmerth S., Barraud P., Bühler M. 2012. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 26:683–692 10.1101/gad.186866.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Perisic O., Lis J.T. 1991. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 64:585–593 10.1016/0092-8674(91)90242-Q [DOI] [PubMed] [Google Scholar]
- Yao J., Ardehali M.B., Fecko C.J., Webb W.W., Lis J.T. 2007. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell. 28:978–990 10.1016/j.molcel.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Shimada K., Oma Y., Kalck V., Akimura K., Taddei A., Iwahashi H., Kugou K., Ohta K., Gasser S.M., Harata M. 2010. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 6:e1000910 10.1371/journal.pgen.1000910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.