Abstract
In embryonic stem cells, removal of oxidatively damaged proteins is triggered upon the first signs of cell fate specification but the underlying mechanism is not known. Here, we report that this phase of differentiation encompasses an unexpected induction of genes encoding the proteasome activator PA28αβ (11S), subunits of the immunoproteasome (20Si), and the 20Si regulator TNFα. This induction is accompanied by assembly of mature PA28-20S(i) proteasomes and elevated proteasome activity. Inhibiting accumulation of PA28α using miRNA counteracted the removal of damaged proteins demonstrating that PA28αβ has a hitherto unidentified role required for resetting the levels of protein damage at the transition from self-renewal to cell differentiation.
Protein carbonylation, resulting from a metal catalysed oxidation involving hydrogen peroxide (H2O2) as the oxidant1,2, has become one of the most commonly used biomarkers of severe oxidative damage to proteins. Protein carbonylation increases during aging and this oxidative modification has been linked to the development of age-related neurological disorders and an age-dependent decline in the activity of many proteins, including the proteasome1,3,4,5,6. Therefore, it has been hypothesized that the protein carbonyl load of young individuals must be sufficiently low to avoid detrimental effects on fitness1,3, a notion which is true for the progeny of many aging species1,7,8. However, undifferentiated mouse embryonic stem (ES) cells were found to contain relatively high levels of carbonylated proteins and advanced glycation end products but upon differentiation such damage was efficiently eliminated9. In this work, we have investigated the mechanisms by which damage removal is accomplished during the onset of ES cell differentiation and report on an unexpected induction and requirement of the proteasome activator PA28, normally associated with the immunoproteasome and processing of antigens10.
Results
Differentiation-induced elimination of protein damage in ES cells requires active proteasomes
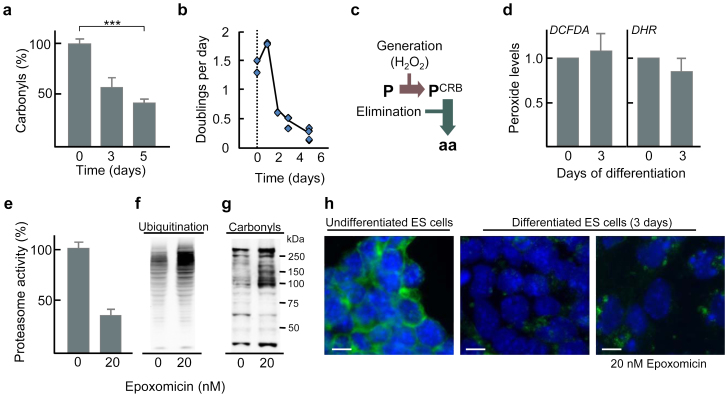
The decrease in the levels of proteins carbonyls observed upon ES cell differentiation (Fig 1a)9 could be a result of dilution of damage by an increased growth rate. This does not appear to be the case, however, since the number of doublings per day decreased rather than increased during differentiation (Fig. 1b). Instead, damage elimination could be due to a reduced rate of damage formation and/or an enhanced rate of damage removal (Fig. 1c). In order to approach these possibilities, we first determined if differentiation resulted in a lowered cellular concentration of hydrogen peroxide since protein carbonylation in biological samples is mainly formed by a metal catalysed oxidation that involves a reaction with this oxidant2. However, there were no statistically significant differences in peroxide levels between undifferentiated and differentiated cells (Fig 1d).
Figure 1. Proteasome activity is essential for limiting protein damage during early differentiation of ES cells.
(a) Levels of protein carbonyls during differentiation of ES cells. The mean value of undifferentiated cells was set to 100% in each run and error bars represent SEM (n = 3), p*** = 8.8E-04 (one-way ANOVA followed by Tukey's post hoc test, see Supplementary Fig. S5). (b) Doubling time of ES cells during differentiation. (c) Illustration of different means by which protein carbonylation may be reduced; P denotes a native protein, PCRB the carbonylated form of the protein, and aa amino acids. (d) Peroxide levels determined by DCFDA (left) and DHR (right) staining and FACS analysis. The value for undifferentiated cells was set to 1.0 and error bars represent SEM (n = 3). (e) Reduction of proteasome activity of ES cells during early differentiation by 20 nM epoxomicin. The mean value of the undifferentiated cells was set to 100%, and error bars represent SD (n = 2). (f) Ubiquitinated proteins and (g) protein carbonyls in ES cells after 3 days of differentiation with or without epoxomicin as indicated (n ≥ 2). (h) SSEA-1 detection (green) and DAPI (blue) staining of undifferentiated cells (left) and differentiated (3 days) cells with (right) and without (middle) epoxomicin treatment. (Images have been cropped; scale bar, 10 μm.)
Focusing instead on damage elimination, we treated differentiating ES cells with the proteasome-specific inhibitor epoxomicin (at a concentration causing a modest inhibition of activity) to test if proteasome activity affects protein carbonyl levels. As shown in figure 1e–g, 20 nM epoxomicin led to a 67% inhibition of the proteasome (Fig. 1e) and an increased level of both ubiquitinated (Fig. 1f) and carbonylated proteins (Fig. 1g). This elevation of carbonylated proteins upon proteasome inhibition occurred without inducing apoptosis (cleaved caspase-3 did not localize to the nucleus; Supplementary Fig. S1), affecting viability (Supplementary Fig. S1), or blocking differentiation (the undifferentiation marker SSEA-1 did not remain localized to membrane as it would have been should the cells have remained undifferentiated; Fig. 1h). These data suggest that proteasome activity during early ES cell differentiation is required to keep protein carbonyl levels at bay.
Differentiation of ES cells triggers production and assembly of the PA28-20S proteasome complex
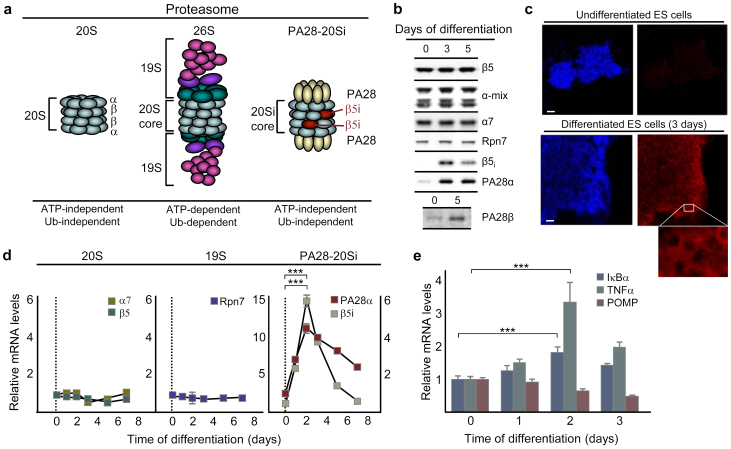
To elucidate the mechanism behind the differentiation-induced boost in proteasome activity demonstrated previously9, we quantified the absolute levels of proteasome subunits. Since the protein carbonyls in ES cells are mainly cytosolic9 we focused on the 20S core and the two known cytosolic regulators of proteasome activity, 19S and PA28 (see schematic representation in Fig. 2a)6,11. We found that the levels of subunits of the 20S core (β5 and a mixture of α-subunits) and 19S (Rpn7) were similar in undifferentiated and differentiated cells (Fig. 2b). However, the levels of the PA28 subunits PA28α and β and the 20Si immunoproteasome subunit β5i become markedly elevated upon differentiation (Fig. 2b and immunocytochemical detection of PA28α in Fig. 2c). Both the β5 and the β5i proteins were found to be processed (around 23 kDa rather than 28–30 kDa) suggesting that the β5i produced is incorporated into the 20S (Supplementary Fig. S2).
Figure 2. Subunits of PA28-20S(i) are elevated upon differentiation of ES cells.
(a) Schematic presentation of the 20S and 20Si proteasomes and the two cytosolic regulators, 19S and PA28. There are also hybrid proteasomes which contain both 19S and PA28 activator complexes38,39 which are not shown here. (b) Representative Western blots of β5, α-subunits, α7 (20S core), Rpn7 (19S subunit), β5i (20Si subunit), PA28α, and PA28β (PA28 subunit) in undifferentiated ES cells (0) and cells that have been differentiating for 3 and 5 days. (c) PA28α (red) and nuclei (blue) in undifferentiated and differentiated ES cells, inset displays a zoomed in box of the PA28α image. (Scale bar, 25 μm.) (d) Relative levels of mRNA encoding 20S (β5 and α7), 19S (Rpn7), 20Si (β5i; y-axis to the left), and PA28 (PA28α; y-axis to the right) subunits with HPRT as a reference gene. Values were normalized to that of undifferentiated cells and error bars represent SEM (n ≥ 3), p*** < 0.0000001 (one-way ANOVA followed by Tukey's post hoc test, see Supplementary Fig. S6). (e) Relative levels of mRNA encoding TNFα (green), IκBα (blue), and POMP (red) with HPRT as a reference gene. Values were normalized to that of undifferentiated cells and error bars represent SEM (n ≥ 3); p***TNFα = 0.000011 and p***IκBα = 0.00018 (one-way ANOVA followed by Tukey's post hoc test, see Supplementary Fig. S7).
Real-time qPCR established that the increase in PA28α and β5i levels is accompanied by a 4-fold elevation in PA28α transcripts and a 15-fold induction of β5i mRNA during early differentiation (Fig. 2d). In contrast, the transcripts of the 19S subunit Rpn7 and 20S subunits α7 and β5 did not change (Fig. 2d). The cytokine TNFα12 and the transcription factor NF-κB13 are regulators of βi gene expression and we found that TNFα expression was elevated upon differentiation (Fig. 2e). In addition, IκBα, a known transcriptional target of NF-κB14 was modestly induced upon differentiation suggesting that the TNFα and, possibly, the NF-κB pathways are both activated (though NF-κB activation may be a result of an active TNFα pathway). In contrast, the levels of the third regulator of βi and PA28α transcription, the immunomodulatory cytokine interferon-γ (IFN-γ)15, were below the detection limit in both undifferentiated and differentiated ES cells. In addition, mRNA levels of the transcriptional target of IFN-γ, the proteasome maturation protein (POMP)16, was not elevated upon differentiation (Fig. 2e). Hence, IFN-γ is unlikely to be the upstream regulator for PA28α and β5i induction during ES cell differentiation.
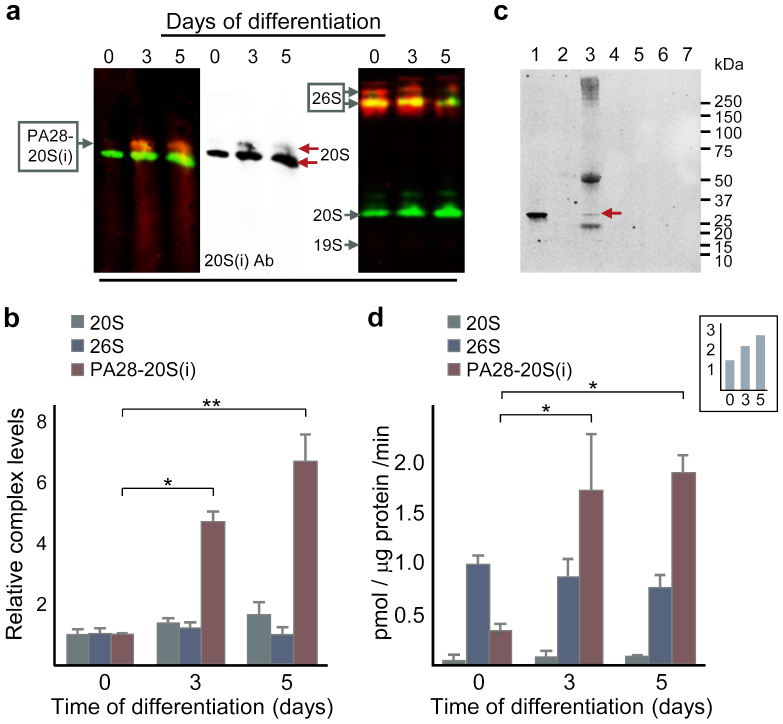
Using protocols optimizing for 20S interaction with either 19S or PA2817 and non-denaturing gel electrophoresis, we demonstrated that the PA28-20S complex is hardly detectable in undifferentiated cells, but increases dramatically upon differentiation (Fig. 3a and 3b). To confirm a physical interaction between PA28 and 20S, we immunoprecipitated 20S and found that PA28α co-precipitated with 20S in differentiated ES cells (Fig. 3c). The levels of the 26S proteasome displayed no major change upon differentiation (Fig. 3a and 3b; see also detection of 19S under PA28-20S favouring conditions in Supplementary figure S3). Moreover, as shown in figure 3d, the activity of the PA28-20S complex is induced 5-fold upon early differentiation (Supplementary figure S3 shows that 19S does not bind 20S during these conditions). In contrast, 26S activity does not increase during differentiation and the substrate cleavage activity of the PA28-20S complex in the cell extracts eventually surpasses that of the 26S (Fig. 3d). Thus, the increase in proteasome activity observed upon early differentiation (inset Fig. 3d) is predominantly due to elevated levels of the PA28-20S.
Figure 3. A functional proteasome activator PA28 is produced upon differentiation of ES cells.
(a) Levels of mature PA28-20S(i) (left; detected using antibodies against 20S α-subunits [green] and PA28α [red] on a blotted gel which had been run for 8.5 h; the middle blot displays the 20S channel alone) and 26S (right; detected using antibodies against the 19S subunit Rpn7 [red] and 20S α-subunits [green] on a blotted gel which had been run for 12 h) proteasomes. (b) Proteasome complexes PA28-20S(i) (red), 26S proteasome (blue), or 20S proteasome (grey) quantified from (a) and additional runs, error bars represent SD (n ≥ 2); p* = 0.013 and p** = 0.0040 (one-way ANOVA followed by Tukey's post hoc test, see Supplementary Fig. S8). (c) Immunochemical detection of PA28α before and after 20S immunoprecipitation (IP) of 3-day differentiated ES cells; lanes from the left: (1) untreated cell extract, (2) beads used to pre-clear extract, (3) 20S-IP beads (arrow indicates PA28α), followed by 4 lanes (4–7) of washes of the 20S-IP beads. (d) Proteasome activity measured under conditions optimizing for either PA28-20S(i) (red), 26S proteasome (blue), or 20S proteasome (grey) assembly and activity, error bars represent SD (n ≥ 2); p*day0-day3 = 0.037 and p*day0-day5 = 0.017 (one-way ANOVA followed by Tukey's post hoc test, see Supplementary Fig. S9). Inset shows the combined proteasome activity in pmol LLVY digested per μg protein and min, i.e. the sum of activity measurements favouring PA28-20S(i), 26S proteasome, and 20S proteasome after 0, 3, and 5 days of differentiation.
PA28 is required for the reduction in protein damage elicited by differentiation
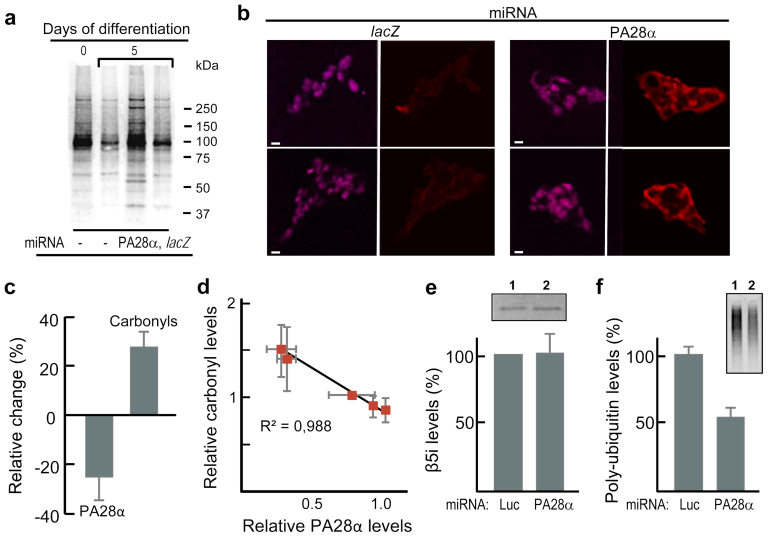
To test if the PA28 activator is required for the removal of carbonylated proteins triggered by differentiation, miRNA was used to counteract PA28α accumulation in differentiating cells. As shown in figure 4a, targeting PA28α abolished the reduction in protein carbonyls upon differentiation while the miRNA control (lacZ) did not. A similar increase in protein carbonyls upon reduced PA28α levels was observed in situ (Fig. 4b). Treating undifferentiated ES cells with miRNA targeting PA28α had no effect on protein carbonyl levels (Supplementary Fig. S4), consistent with the observation that these cells have almost no PA28α (Fig. 2b and 2c). We also used miRNA directed against luciferase (Luc) as a control and found that in these experiments, a 25.5% (±8.8) reduction in PA28α levels resulted in a 28.1% (±6.3) increase in the carbonyl load (Fig. 4c). Similarly, an alternative RNAi methodology (siRNA directed against PA28α mRNA transfected into differentiating ES cells with oligofectamine) generated a clear inverse correlation between PA28α levels and carbonyl content in these cells (R2 = 0.988; Fig. 4d).
Figure 4. PA28 is required for the clearance of protein carbonyls during differentiation of ES cells.
(a) Levels of carbonylated proteins in undifferentiated ES cells (0) and cells that have been differentiating for 5 days treated with miRNA as indicated; lacZ constitutes the miRNA non-targeting control. Lanes from the same blot image have been cut together. (b) Protein carbonyl detection (red) and nuclei (purple) in miRNA treated (as indicated) ES cells after 3 days of differentiation. (Scale bar, 25 μm.) (c) Levels of protein carbonyls and PA28α in cells that have been differentiating 5 days with miRNA treatment targeting PA28α, determined by SDS-PAGE followed by Western blotting and immunodetection, presented as percent difference compared to the miRNA non-targeting control (Luc). The mean value of control cells was set to 100%, error bars represent SEM (n = 3); pcarbonyls = 0.012 (unpaired two-tailed t-tests of Luc vs PA28α targeting miRNA). (d) Inverse correlation between PA28α levels and protein carbonyls in undifferentiated ES cells and cells that have been differentiating for 3 days while under siRNA treatment (non-targeting nucleotides and targeting PA28α) determined as described in c), error bars represents SD (n ≥ 2). (e) Levels of β5i and (f) poly-ubiquitin in cells that have been differentiating 5 days with miRNA treatment targeting luciferase (Luc) and PA28α determined as described in c). The mean value of control cells was set to 100%, error bars represent SEM (n = 3); ppoly-ub = 0.01019 (unpaired two-tailed t-tests of Luc vs PA28α targeting miRNA). Inset shows Western-blot β5i (e) and poly-ubiquitin patterns (f) of cells treated with miRNA targeting luciferase (1) and PA28α (2).
To examine whether the change in carbonyl content upon miRNA treatment against PA28α is specifically due to a loss of PA28 rather than 20Si or 26S, we analysed the effect of this treatment on β5i and poly-ubiquitin levels. As shown in figure 4e, miRNA targeting PA28α had no effect on β5i levels, excluding a concomitant down-regulation of this 20Si subunit as PA28α levels are lowered. A reduction in 19S (and thus 26S) would be expected to cause an increase in the levels of poly-ubiquitinated protein; however, we found that miRNA against PA28α lowered the levels of ubiquitinated proteins (Fig. 4f). This apparent increase in 26S activity might be due to competition between 19S and PA28 for 20S binding and in this scenario a reduction in PA28 levels would result in elevated levels of 26S. Taken together, the data support that the elevated carbonyl levels observed upon miRNA treatment against PA28α is the result of reduced PA28 levels.
Discussion
The proteasome activator PA28 is produced in many cells upon intensified immune responses, which often includes a replacement of the regular 20S proteasomes with 20Si immunoproteasomes6,10. Apart from being involved in antigen processing10, both PA28 and 20Si may have general protective functions related to oxidative stress. Specifically, recent data point to PA28 participating in the degradation of misfolded proteins and the management of protein aggregates18. Moreover, a link between PA28/20Si and protein quality control is evidenced by the fact that PA28 overexpression in cultured cardiomyocytes decreases protein carbonyl levels upon H2O2 stress19, that knockdown of either PA28 or 20Si limits cellular adaptation to H2O2 treatment20,21, and that 20Si is involved in the clearance of protein aggregates upon IFN-γ induced oxidative stress22. As demonstrated herein, PA28 appears to be required also for the clearance of carbonylated proteins upon the first signs of cell fate specification. The potential in vivo relevance of these results are underlined by the finding that PA28α, PA28β, and β5i are all expressed during early mouse embryogenesis23.
The 26S proteasome and the ubiquitin-proteasome system (UPS) have recently been implicated in the maintenance of the pluripotent state24,25,26. It has been demonstrated that proteasome inhibition induces differentiation24,25 but the quality of the resulting differentiated cells regarding protein damage upon such inhibition was not investigated. In addition, undifferentiated human ES cells (hESC) were found to exhibit higher 26S proteasome activity compared to differentiated cells24, consistent with our data showing a trend towards a decline in 26S activity after the onset of differentiation. Since the proteasome activity in the Vilchez et al., study24 was measured under conditions optimizing for the 26S proteasome (the MgCl2 present in the assay excludes all PA28-dependent proteasome activity17), it is not known if the reduced 26S activity in differentiated cells was preceded by a transient elevated activity of the PA28-20S proteasome. Thus, it would be interesting to test whether the onset also of hESC differentiation includes a boost in PA28-dependent proteasome activity.
PA28 knock-out mice are viable27 demonstrating that a failure to properly boost PA28-20S activity upon ES cell differentiation is not severely affecting the development or functionality of the embryo. However, it could be speculated that a diminished ability to clear out protein damage by the PA28-20S proteasome during early cell fate specification could accelerate deterioration of metabolically active tissues, such as neurons and muscles, in the adult and aging organism. Moreover, the involvement of the PA28-activated proteasome in purging ES cells from oxidative protein damage suggests that this may be accomplished without the need for ubiquitin tagging or ATP and is in accord with data demonstrating that cells with compromised ubiquitin-conjugating activity retain their ability to degrade oxidized intracellular proteins at near normal rates28. A question of interest is whether ectopic induction of the PA28 activator might counteract the accumulation of oxidatively damaged and/or aggregated proteins also in aging cells/tissues and whether this affects the progression or timing of age-related protein conformational disorders and the rate of aging itself.
Methods
ES cell line and culture conditions
The murine ES cell line R129,30 was used in all experiments, except for proteasome activity and Western analysis upon epoxomicin treatment and DHR FACS analysis which were done on E14.130,31. Several of the experiments were done on both cell lines with similar results. Cells were cultured on gelatin-coated plates in ES cell culture medium (Supplementary Table S1) as previously described9.
Immunocytochemistry
The undifferentiated status of the ES cells was confirmed by immunochemical staining of Oct-4, localized in the nucleus32, and SSEA-1, cell surface-localized33; as previously described9. Immunocytochemical detection of carbonyls was performed as described previously9. For detection of PA28α, the cells were fixed in 4% PFA (15 min), permeabilized in 0.5% Triton X-100 (5 min), blocked with 10% goat serum in PBS (40 min) at room temperature, and incubated with rabbit anti-PA28α (1:100; Abgent) overnight at 4°C, with goat anti-rabbit IgG A555 (1:250; Molecular probes, Invitrogen) for 60 min, and DAPI (1 μg/mL) for 4 min at room temperature.
ES cell differentiation and estimation of doubling time
Differentiation was induced as described9, by removal of LIF and omission of continuous passage for 1, 2, 3 and 5 days. Time point “0” of differentiation refers to undifferentiated cells on a plate of 80% confluency. This is when LIF is removed and passaging omitted to induce differentiation and thus occurs at day 0 of differentiation (start). Doubling time estimates of undifferentiated cells were based on the number of cells plated during passage, the time until next passage, and the number of cells after trypsin treatment at that time. For differentiated cells, the pellet weight at harvest was used as an approximation of the relative number of cells, so that the increase in pellet weight over time gave an estimation of the doubling rate.
Peroxide detection by DHR and DCFDA staining followed by FACS analysis
Trypsin treated ES cells were incubated in DHR (5 μM for 10 min) or DCFDA (20 μM for 30 min) and analysed using flow cytometry (FACS Aria, BD equipment for DHR and FACSCalibur, BectonDickinson for DCFDA) with FITC filter settings, gating on live cells.
Proteasome activity assays and proteasome inhibition
Cells were lysed in PA28-20S or 26S lysis buffer (Supplementary Table S1). Cell debris was removed by centrifugation and protein concentration was determined using the BCA Protein Assay kit (Pierce). The chemotryptic activity was assayed by hydrolysis of the fluorogenic peptide succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (suc-LLVY-AMC) (Bachem, Saffron Walden, U.K.). 20 μg total protein was incubated with 200 μM suc-LLVY-AMC in PA28-20S or 26S optimizing buffer, or 20S buffer (see Supplementary Table S1) in a total volume of 100 μl; fluorescence was read using 390 nm excitation and 460 nm emission filters with free AMC as standard (Bachem). Lactacystin (30 μM, Enzo Life Sciences) inhibited the suc-LLVY-AMC hydrolysis of the PA28-20S and 26S samples to 90.1% (±3.8). Coomassie stained SDS-PAGE gels were used to confirm equal protein levels in the assay. Proteasome inhibition during ES cell differentiation was done by including 20 nM Epoxomicin (Enzo Life Sciences) in the differentiation medium (without LIF).
Electrophoresis (SDS-PAGE and native PAGE) and western blot analysis
For protein separation by SDS-PAGE, cells were lysed with a modified RIPA buffer (Supplementary Table S1), cell debris was removed and protein concentration was determined using the BCA Protein Assay kit (Pierce). Native PAGE samples were extracted to optimize the 20S interaction to either PA28 or 19S as described above, mixed with native gel loading buffer (Supplementary Table S1), and run on non-denaturing PAGE (2.5% stacking and 4.5% separating gel)34. Samples were prepared for SDS-PAGE as described (for detection of protein carbonyls9 and absolute levels of specific proteins and ubiquitin35). Gel electrophoresis was followed by blotting the proteins onto a PVDF (polyvinylidene difluoride) membrane (Millipore). The following primary antibodies were used: rabbit pAb anti-2,4-dinitrophenyl (Chemicon Oxyblot kit, Millipore), mouse mAb anti-ubiquitin (Santa Cruz), mouse mAb anti-polyubiquitin (Abcam), rabbit pAb anti-β5, anti-Rpn7, anti-PA28α, anti-PA28β and mouse mAb anti-β5i, anti-α7, and anti 20S α subunits (a mixture of antibodies against α1, α2, α3, α5, α6 and α7) (Enzo Life Sciences). IRDye 800CW-labelled or 680CW-labelled goat anti-mouse or anti-rabbit IgG antibodies (LI-COR Biosciences) were used for detection and blots were analysed with the Odyssey infrared imaging system and software (LI-COR Biosciences).
20S immunoprecipitation
Cells were lysed in PA28-20S lysis buffer (Supplementary Table S1), cell debris was removed by centrifugation, and extracts were precleared with protein A agarose beads (GE Healthcare), incubated with rabbit pAb anti-20S subunits (Enzo Life sciences) and with beads. Beads were extensively washed with Lysis buffer, and subjected to SDS-PAGE followed by Western analysis.
RNA extraction and quantitative (qPCR) analysis
Total RNA was extracted using GenElute™ Mammalian Total RNA Miniprep Kit from Sigma-Aldrich, and DNase treated on-column with RNase-Free DNase Set (Qiagen) according to manufacturer's instructions. cDNA was synthesized on 1 μg total RNA in First strand buffer (Invitrogen), 50 nM random primers, 500 μM dNTP mixture and 5 mM DTT by 200 U Superscript™ III RT (Invitrogen) in 20 μl total volume; a reaction with no reverse transcriptase was included for each sample. 1/100 of total synthesized cDNA were analysed in triplicates by qPCR using iQ™ SYBR® Green Supermix and the iQ5 detection system from Bio-Rad. For primer sequences, see Supplementary Table S2.
RNAi Inhibition of PA28α expression
miRNA treatment against PA28α was performed using the BLOCK- iT™ Pol II miR RNAi expression vector kit with pre-designed BLOCK-iT™ miR RNAi Select hairpins directed towards PA28α, lacZ, and luciferase (the two latter being non-targeting controls) mRNA from Invitrogen (see Supplementary Table S3). miRNA was cloned into the pcDNA™6.2-GW/miR vector according to manufacturer's instructions and correct insertion was confirmed by sequencing. The vectors expressing miRNA were introduced into undifferentiated ES cells using a GenePulser (BioRad) with the following settings: 260 V, 500 μF, ∞ Ω, in a 4 mm cuvette. After 24 h recovery, selection for the miRNA vector (7.5 μg/mL blasticidin) was started in parallel to initiation of the differentiation protocol. Two miRNA variants targeting PA28α were combined to inhibit PA28α expression (see Supplementary Table S3). siRNA silencing was carried out as described36 using Oligofectamine (Invitrogen) as transfection reagent. siRNA duplexes (see Supplementary Table S4) were purchased from Dharmacon Research.
Statistical analysis
Comparisons between two groups were performed with unpaired t-tests assuming two-tailed distribution and equal variances and differences were considered significant at p < 0.05. Statistical calculations for figures 1a, 2d, 2e, 3b, and 3d were done in R-2.15.2 (www.r-project.org)37 by one-way ANOVA followed by Tukey's post hoc test to compare group means and the null hypothesis was rejected at the 0.05 level.
Author Contributions
M.H. and T.N. designed research; M.H., Å.F., M.L. and K.N. performed research; J.W. and H.S. contributed new analytical tools; M.H., T.N. and M.C. analysed the data; M.H. and T.N. wrote the paper.
Supplementary Material
Supplementary information
Acknowledgments
We thank Gabriella Brolén for valuable guidance in ES cultivation, Jennifer Rivett and Fiona Stratford for essential input regarding proteasome methodology, Julie Grantham for counsel on siRNA and IP protocols, Martin Kjerrulf for FACS analysis support, Anna-Lena Loyd for immunochemical assistance, the Centre for Cellular Imaging at the Sahlgrenska Academy (University of Gothenburg) for the use of imaging equipment and for the support from the staff, and past and present members of TN group for inspiring discussions and technical advice. miRNA treatments were performed at AstraZeneca Discovery Sciences. This work was supported by grants from the Swedish Natural Research Council (VR), Knut and Alice Wallenberg Foundation (Wallenberg Scholar), an Advanced ERC grant (QualiAge) and the EC (Acronym: Proteomage).
References
- Levine R. L. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radical Biology and Medicine 32, 790–796 (2002). [DOI] [PubMed] [Google Scholar]
- Requena J. R., Chao C.-C., Levine R. L. & Stadtman E. R. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci U S A 98, 69–74 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman E. Protein oxidation and aging. Science 257, 1220–1224 (1992). [DOI] [PubMed] [Google Scholar]
- Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J 24, 1311–1317 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farout L. & Friguet B. Proteasome Function in Aging and Oxidative Stress: Implications in Protein Maintenance Failure. Antioxidants & Redox Signaling 8, 205–216 (2006). [DOI] [PubMed] [Google Scholar]
- Jung T., Catalgol B. & Grune T. The proteasomal system. Molecular Aspects of Medicine 30, 191–296 (2009). [DOI] [PubMed] [Google Scholar]
- Aguilaniu H., Gustafsson L., Rigoulet M. & Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299, 1751–1753 (2003). [DOI] [PubMed] [Google Scholar]
- Goudeau J. & Aguilaniu H. Carbonylated proteins are eliminated during reproduction in C. elegans. Aging Cell 9, 991–1003 (2010). [DOI] [PubMed] [Google Scholar]
- Hernebring M., Brolèn G., Aguilaniu H., Semb H. & Nyström T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci U S A 103, 7700–7705 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl B. et al. Interferon-γ, the functional plasticity of the ubiquitin–proteasome system, and MHC class I antigen processing. Immunological Reviews 207, 19–30 (2005). [DOI] [PubMed] [Google Scholar]
- Stadtmueller B. M. & Hill C. P. Proteasome Activators. Molecular Cell 41, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaparthi K. & Elliott J. L. Non-neuronal induction of immunoproteasome subunits in an ALS model: Possible mediation by cytokines. Experimental Neurology 196, 441–451 (2005). [DOI] [PubMed] [Google Scholar]
- Moschonas A. et al. CD40 Induces Antigen Transporter and Immunoproteasome Gene Expression in Carcinomas via the Coordinated Action of NF-kB and of NF-kB-Mediated De Novo Synthesis of IRF-1. Mol. Cell. Biol. 28, 6208–6222 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Natoli G. & Ghosh G. Transcriptional regulation via the NF-kB signaling module. Oncogene 25, 6706–6716 (2006). [DOI] [PubMed] [Google Scholar]
- Schroder K., Hertzog P. J., Ravasi T. & Hume D. A. Interferon-γ: an overview of signals, mechanisms and functions. Journal of Leukocyte Biology 75, 163–189 (2004). [DOI] [PubMed] [Google Scholar]
- Heink S., Ludwig D., Kloetzel P.-M. & Krüger E. IFN-γ-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci U S A 102, 9241–9246 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivett A. J. et al. Assays of proteasome activity in relation to aging. Experimental Gerontology 37, 1217–1222 (2002). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. The Journal of Clinical Investigation 121, 3689–3700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Powell S. R. & Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. FASEB Journal 25, 883–893 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussong S. A., Kapphahn R. J., Phillips S. L., Maldonado M. & Ferrington D. A. Immunoproteasome deficiency alters retinal proteasome's response to stress. Journal of Neurochemistry 113, 1481–1490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering A. M. et al. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochemical Journal 432, 585–594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert U. et al. Immunoproteasomes Preserve Protein Homeostasis upon Interferon-Induced Oxidative Stress. Cell 142, 613–624 (2010). [DOI] [PubMed] [Google Scholar]
- Hamatani T., Carter M. G., Sharov A. A. & Ko M. S. H. Dynamics of Global Gene Expression Changes during Mouse Preimplantation Development. Developmental Cell 6, 117–131 (2004). [DOI] [PubMed] [Google Scholar]
- Vilchez D. et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489, 304–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley Shannon M. et al. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell Stem Cell 11, 783–798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S. et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics 10, 10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S. et al. Immunoproteasome assembly and antigen presentation in mice lacking both PA28α and PA28β. EMBO J 20, 5898–5907 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shringarpure R., Grune T., Mehlhase J. & Davies K. J. A. Ubiquitin Conjugation Is Not Required for the Degradation of Oxidized Proteins by Proteasome. Journal of Biological Chemistry 278, 311–318 (2003). [DOI] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W. & Roder J. C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A 90, 8424–8428 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T., Rodriguez I., Perry A. C. F., Yanagimachi R. & Mombaerts P. Mice cloned from embryonic stem cells. Proc Natl Acad Sci U S A 96, 14984–14989 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M., Hardy K., Handyside A., Hunter S. & Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326, 292–295 (1987). [DOI] [PubMed] [Google Scholar]
- Scholer H. R., Hatzopoulos A. K., Balling R., Suzuki N. & Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J 8, 2543–2550 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter D. & Knowles B. B. Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1). Proc Natl Acad Sci U S A 75, 5565–5569 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L., Pratt G. & Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. Journal of Biological Chemistry 267, 22362–22368 (1992). [PubMed] [Google Scholar]
- Ballesteros M., Fredriksson A., Henriksson J. & Nystrom T. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J 20, 5280–5289 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham J., Brackley K. I. & Willison K. R. Substantial CCT activity is required for cell cycle progression and cytoskeletal organization in mammalian cells. Experimental Cell Research 312, 2309–2324 (2006). [DOI] [PubMed] [Google Scholar]
- Team R. D. C. R: A language and environment for statistical computing <http://www.R-project.org/> (2011). [Google Scholar]
- Hendil K. B., Khan S. & Tanaka K. Simultaneous binding of PA28 and PA700 activators to 20S proteasomes. Biochem. J. 332, 749–754 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi N. et al. Hybrid Proteasomes. Journal of Biological Chemistry 275, 14336–14345 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information