Summary
Significant progress has been made in studies of the mechanisms by which RANKL induces terminal osteoclast differentiation. However, many crucial details in the RANKL-evoked signaling pathway for osteoclast differentiation remain to be defined. We characterized genes specifically expressed in osteoclasts by differential screening of a human osteoclastoma cDNA library, and found that the regulator of G-protein signaling 10A (RGS10A), but not the RGS10B isoform, was specifically expressed in human osteoclasts. The expression of RGS10A is also induced by RANKL in osteoclast precursors and is prominently expressed in mouse osteoclast-like cells. RGS10A silencing by RNA interference blocked intracellular [Ca2+]i oscillations, the expression of NFAT2, and osteoclast terminal differentiation in both bone marrow cells and osteoclast precursor cell lines. Reintroduction of RGS10A rescued the impaired osteoclast differentiation. RGS10A silencing also resulted in premature osteoclast apoptosis. RGS10A silencing affected the RANKL-[Ca2+]i oscillation-NFAT2 signaling pathway but not other RANKL-induced responses. Our data demonstrate that target components of RGS10A are distinct from those of RGS12 in the RANKL signaling mechanism. Our results thus show the specificity of RGS10A as a key component in the RANKL-evoked signaling pathway for osteoclast differentiation, which may present a promising target for therapeutic intervention.
Keywords: Differential screening, RGS10 RNA interference, [Ca2+]i oscillations, Osteoclast differentiation, RANKL signaling pathway
Introduction
Osteoclasts are bone-resorbing cells that exert a profound impact on skeletal metabolism. Disorders of skeletal insufficiency, such as osteoporosis, are characterized by enhanced osteoclastic bone resorption relative to bone formation. A complete understanding of the mechanisms by which osteoclasts differentiate from their precursors and degrade bone is therefore crucial in developing therapies for these often debilitating diseases.
The past several years have witnessed important new insights into the mechanisms of osteoclast formation and activation, particularly the role of RANKL (Alliston and Derynck, 2002; Li et al., 1999; Matsuo et al., 2000; Mocsai et al., 2004; Naito et al., 1999; Wagner and Karsenty, 2001). Binding of RANKL to its receptor RANK results in the recruitment of TRAF family proteins including TRAF6, which is linked to the NFκB and Jun N-terminal kinase (JNK) pathways. Both NFκB and Fos function downstream of the RANKL signal. There is genetic evidence that these factors exert an essential function in the development from monocyte/macrophage precursors to mature osteoclasts (Franzoso et al., 1997; Grigoriadis et al., 1994). However, the mechanism by which RANKL induces terminal differentiation of osteoclasts through specific transcriptional pathways remains unclear.
Recently, significant progress has been made by Takayanagi et al. who reported that RANKL evokes intracellular [Ca2+]i oscillations which lead to calcineurin-mediated activation of NFAT2, and trigger a sustained NFAT2-dependent transcriptional program during osteoclast differentiation (Takayanagi et al., 2002). They demonstrated that NFAT2-deficient embryonic stem cells fail to differentiate into osteoclasts in response to RANKL stimulation, and that overexpression of NFAT2 causes precursor cells to undergo efficient differentiation in the absence of RANKL signaling. Thus, NFAT2 might function downstream of RANKL as a master switch for regulating the terminal differentiation of osteoclasts. Koga et al. reported that, in osteoclast precursor cells, FcRγ and DAP12 associate with multiple immunoreceptors and activate Ca2+ signaling through phospholipase Cγ (PLCγ), indicating that phosphorylation of PLCγ is the crucial component involved in the RANKL-induced RANKL-Ca2+-NFAT2 pathway (Koga et al., 2004). Despite these new insights, the question remains as to how RANKL evokes the [Ca2+]i-oscillation–NFAT2 signaling pathway that triggers osteoclast-specific gene expression and differentiation.
Here, we found that the A isoform of RGS10 (NM_001005339) is prominently expressed in RANKL-stimulated osteoclast-like cells (OCLs) and human osteoclasts. Using gain-of-function and loss-of-function approaches, we demonstrate that RGS10A is a key regulator of [Ca2+]i oscillations and is an essential component in the RANKL–[Ca2+]i-oscillation–NFAT2 signaling pathway in the terminal differentiation of osteoclasts.
Results
To identify crucial components in the RANKL-evoked signaling pathway for osteoclast differentiation, we performed differential screening of a human osteoclastoma cDNA library. Approximately 1.2×104 clones from the pcDNAII osteoclastoma library were replicated and screened by differential hybridization, as described previously (Li et al., 1995). One clone contained a 0.5-kb insert that showed a positive hybridization signal with the cDNA probes derived from osteoclastoma mRNA but was negative with stromal cell cDNA probes. The DNA sequence of the clone was found to be 100% homologous to the 3′ sequence of the RGS10 human cDNA (data not shown). To clone the full length of the cDNA, we screened the whole human osteoclastoma cDNA library with 2×105 clones using the 0.5 kb 3′ sequence RGS10 human cDNA fragment as a probe. Eight clones that showed positive hybridization signals were obtained and one clone containing a 0.9-kb insert was found to be 100% homologous to the sequence of the human cDNA of RGS10A (NM_001005339; data not shown).
We performed northern blotting, RT-PCR and in situ hybridization to confirm the differential screening result. The northern blot data of mouse OCLs and various tissues indicated that RGS10 was prominently expressed in the OCLs derived from the monocytic osteoclast progenitor cell lines, MOCP-5 and RAW264.7 (Fig. 1A). Lower expression levels, about three times lower than in the cell lines, were observed in the brain. Expression was also observed – although to a far lesser extent – in the calvaria, long bones, spleen and lungs of mice. The expression of RGS10 was undetectable in the heart, liver, kidneys and skeletal muscles (Fig. 1A).
Fig. 1.
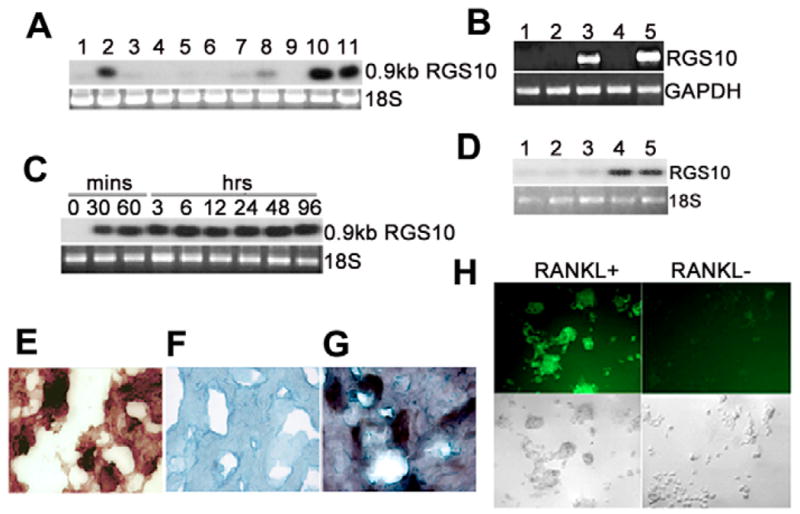
RGS10A is prominently expressed in osteoclasts and osteoclast precursors stimulated by RANKL and M-CSF. (A) Northern blot hybridization of mouse RGS10A cDNA to total RNA from mouse tissues and cell lines. Lane 1, calvaria; lane 2, brain; lane 3, long bone; lane 4, heart; lane 5, liver; lane 6, kidney; lane 7, spleen; lane 8, lung; lane 9, muscle; lane 10, RANKL-stimulated RAW264.7 cells; lane 11, RANKL-stimulated MOCP-5 cells. (B) RT-PCR analysis of isoforms RGS10A and RGS10B mRNA in human osteoclasts and brain, showing that hRGS10A is specifically expressed in osteoclasts. Lane 1, negtive control (H2O); lane 2, RNA of isoform A of hRGS10 from brain; lane 3, RNA of isoform B of hRGS10 from brain; lane 4, RNA of isoform B of hRGS10 from osteoclasts; lane 5, RNA of isoform A of hRGS10 from osteoclasts. The primer BOR10-F for RGS10A is located at exon 1 and the primer BOBRR10-R is located at exon 5. The distance between the two exons is 10.8 kb (Gene Access no: NC_000010), so the 669 bp size band is the true RT-PCR product. The RT-PCR fragments from lane 5 were cloned into pBluescript and sequenced. The RT-PCR cDNA fragment from human osteoclast mRNA was 100% homologous to the sequence of the human RGS10A cDNA. (C) Time course of RGS10A mRNA in RANKL-stimulated RAW264.7 cells by northern blotting. Cells were treated with RANKL and M-CSF for the indicated times. RGS10A mRNA was detectable at 30 minutes and continued to increase at 60 minutes, staying at high levels for 96 hours after stimulation with RANKL and M-CSF. (D) Northern blot analysis of RGS10A expression in osteoclasts and BMMs stimulated with M-CSF in the absence of RANKL. No RGS10A expression was detected in Lane 1–3, which are BMMs stimulated with M-CSF for 0, 12, and 24 hours. Lane 4–5, BMMs stimulated with RANKL and M-CSF for 12 and 24 hours. (E–G) In situ hybridization of RGS10A mRNA in a human osteoclastoma. Counterstain, Methyl Green. (E) TRAP+ human osteoclasts. (F) Sense probe as control. (G) Antisense probe. (H) Expression of RGS10A protein in RANKL-stimulated BMMs. Multinucleated osteoclasts were formed only in RANKL-stimulated BMMs, and strongly expressed RGS10A. RGS10A protein expression was absent in cells not stimulated with RANKL.
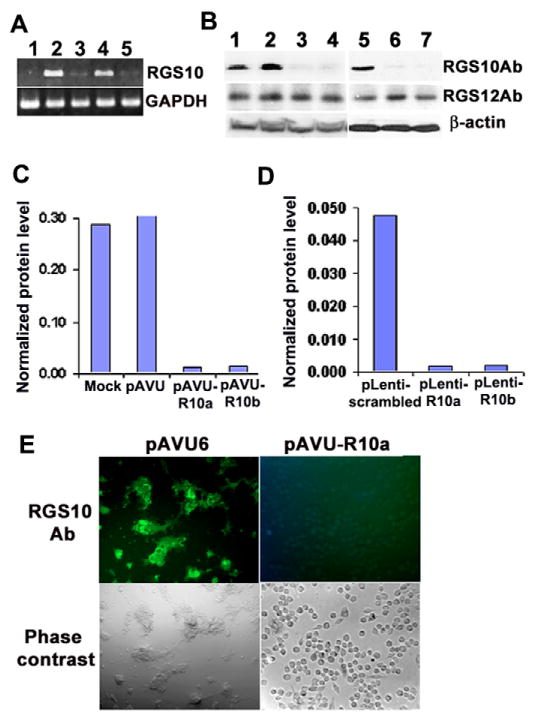
To define whether RGS10 expression in osteoclasts is isoform specific, we characterized isoform specificity by RT-PCR (Fig. 1B). We detected the expression of RGS10 isoform A (RGS10A) cDNA, but did not detect expression of RGS10 isoform B (RGS10B) in human osteoclast, and RGS10B but not RGS10A was detected in the human brain, indicating that only RGS10A was highly and fairly selectively expressed in osteoclasts. The primer BOR10-F for RGS10A is located at exon 1 and the primer BOBRR10-R is located at exon 5. The size between the two exons is 10.8 kb (Gene Access NO: NC_000010), so the 669 bp size band is the true RT-PCR product.
In order to determine when the cells are initiated to express RGS10A following the stimulation of RANKL, we studied the time course of RGS10A transcription in RANKL-stimulated bone marrow-derived monocytes (BMMs). We found that RGS10A mRNA could not be detected when RANKL was absent, but that transcription was induced as soon as 30 minutes after exposure to RANKL and M-CSF, and remained at high levels for up to 96 hours (Fig. 1C). We further observed that RGS10A proteins are not responsive to M-CSF in BMMs treated in the absence of RANKL with M-CSF alone (Fig. 1D). To evaluate whether RGS10A is prominently expressed in authentic osteoclasts, in situ hybridization was performed on a human osteoclastoma (giant cell tumor). Osteoclasts in human osteoclastomas are believed to be normal osteoclasts (Chambers et al., 1986; Flanagan et al., 1988). The result showed that RGS10A is highly expressed in osteoclasts but not in stromal cells (Fig. 1E–G). To analyze RGS10A expression in osteoclasts at the protein level, we examined the expression of RGS10A in RANKL-stimulated OCLs derived from BMMs using immunofluorescent cell staining. The results indicated that RGS10A is highly expressed in mouse OCLs, but is absent in BMMs without RANKL stimulation (Fig. 1H). Our results therefore demonstrate that RGS10A is prominently expressed in RANKL-stimulated mouse pre-osteoclasts, OCLs and human authentic osteoclasts, at both the mRNA and protein levels.
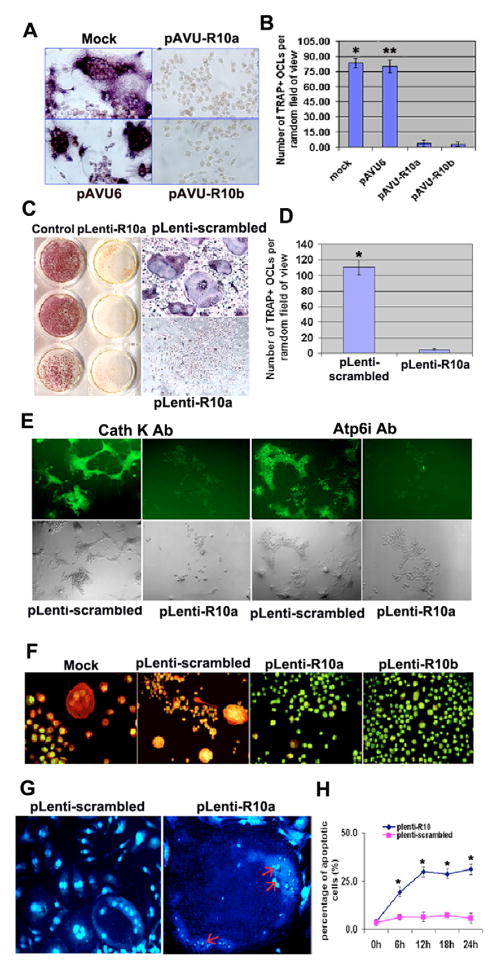
We used vector-based RNA interference (RNAi) gene silencing in RAW264.7 cells (Yu et al., 2002) to investigate the role of RGS10A in osteoclast differentiation. After stable transfection with constructs encoding small interfering RNA (siRNA) targeting RGS10A, we performed a quick analysis of short hairpin RNA (shRNA) expression with tartrate-resistant acid phosphatase (TRAP) staining and chose those colonies carrying the two constructs (pAVU-R10a and pAVU-R10b) for passage that had inserts located at 189–209 bp and 330–350 bp. To confirm the effect of RNAi, we performed RT-PCR (Fig. 2A), western blotting (Fig. 2B–D), and immunofluorescent staining (Fig. 2E) to assess silencing efficiency. As shown in Fig. 2A–E, RGS10A mRNA transcription and protein expression were silenced in stably transfected RGS10A-silenced cell lines after RANKL stimulation, as compared with control cells transfected with the pAVU-scrambled vector. Silencing of RGS10A did not affect the expression of RGS12 (Fig. 2B), indicating silencing specificity against RGS10A. The cells stably transfected with pAVU-scrambled, pAVU-R10a or pAVU-R10b were stimulated with RANKL and M-CSF for 96 hours. Approximately 80% of the cells differentiated into TRAP+ multinucleated cells (MNCs) when transfected with the control vector (Fig. 3A). By contrast, only 3–4% of the cells were stimulated to differentiate into TRAP+ MNCs with pAVU-R10a or pAVU-R10b transfection (Fig. 3A). The number of TRAP+ MNCs in RGS10A-silenced groups was 15 times lower than that in the control groups (Fig. 3B), indicating that RGS10A silencing almost completely blocks osteoclast differentiation from osteoclast precursor cells.
Fig. 2.
RGS10A silencing blocks RGS10A expression. RAW264.7 cells stably transfected with pAVU-scrambled, pAVU-R10a or pAVU-R10b constructs, and BMMs with pLenti-scrambled or pLenti-R10a were stimulated with RANKL and M-CSF for 96 hours. (A) RT-PCR analysis. Lane 1, negative control (H2O). The expression of RGS10A mRNA was weak or absent in RGS10A-silenced cells (lane 2, pAVU-R10a; lane 4, pAVU-R10b). (B) Western blotting of RGS10A protein and RGS12 protein in control and RGS10A-silenced RAW264.7 cells (lanes 1–4) and BMMs (lanes 5–7) stimulated with RANKL and M-CSF. For RGS10A protein, the signals were strong in control cells (lanes 1, 2, 5) and weak in RGS10A-silenced cells (lanes 3, 4, 6, 7). However, for RGS12 protein, there is no apparent difference in signals between control cells and RGS10A silenced cells. Lane 1, mock; lane 2, pAVU-scrambled shRNA; lane 3, pAVU-R10a; lane 4, pAVU-R10b; lane 5, pLenti-scrambled shRNA; lane 6, pLenti-R10a; lane 7, pLenti-R10b. Ab, antibody. (C,D) Quantification of RGS10A levels by western blotting as in B. RGS10A protein levels in transfected cells were normalized to the loading control β-actin. (E) Immunofluorescence staining. RGS10A expression was silenced in the cells transfected with pAVU-R10a. These are images of phase-contrast as an internal control.
Fig. 3.
RGS10A silencing blocks osteoclast differentiation. (A) TRAP+ MNCs were formed in RANKL-stimulated RAW264.7 cells transfected with mock shRNA or pAVU-scrambled shRNA, but not in the cells transfected with pAVU-R10a or pAVU-R10b. (B) Quantitative analysis of TRAP+ MNCs in (A). TRAP+ MNCs in control groups, mock (*) or pAVU-scrambled (**), are more than 15 times (P<0.05) that of RGS10A-silenced groups (pAVU-R10a or pAVU-R10b). (C) Formation of TRAP+ MNCs in RANKL-stimulated BMMs infected with pLenti-scrambled and the absence of TRAP+ MNCs in the cells infected with pAVU-R10a are shown. (D) Quantitative analysis of TRAP+ MNCs in (C), pLenti-R10a vs the RGS10A-silenced group (*P<0.05). (E) Immunofluorescent staining of cathepsin K and Atp6i in RGS10A-silenced BMMs stimulated with RANKL and M-CSF. RGS10A silencing blocks the expression of cathepsin K and Atp6i as showed in pLenti-R10a groups. (F) Acridine orange staining. Strong orange fluorescence indicates extracellular acidification in the mock or pLenti-scrambled, but not in pLenti-R10a or pLenti-R10b. (G,H) RGS10A silencing-mediated apoptosis in multinucleated osteoclasts with RANKL stimulation. (G) The nuclei of osteoclast cells are condensed and fragmented in the RGS10A-silenced cells as indicated by the arrows, compared with the control cells. (H) Time course of RGS10A silencing-mediated apoptosis. Apoptosis was quantified by counting multinucleated cells with condensed nuclei (each point *P<0.05 vs control).
We confirmed our findings in primary bone marrow cells, by using the same siRNA sequences to generate recombinant lentiviruses carrying RGS10A-scrambled shRNA, denoted as pLenti-scrambled, and RGS10A shRNA (pLenti-R10a and pLenti-R10b). The effect of silencing RGS10A expression was confirmed by western blotting (Fig. 2B, lanes 5–7). Similar to the results with the cell line, as many as 80% of the cells differentiated into TRAP+ MNCs when infected with control viruses, but only 5% differentiated with pLenti-R10a (Fig. 3C,D). To determine the effect of RGS10A RNAi on osteoclast gene expression, we also examined the expression of osteoclast marker genes cathepsin K and Atp6i using immunofluorescent staining. As shown in Fig. 3E, RGS10A silencing blocked both cathepsin K and Atp6i gene expression, two late markers of osteoclast differentiation.
To characterize the effects on osteoclast function, we studied whether extracellular acid compartments were formed beneath RGS10A-silenced cells. Acridine Orange staining, indicative of acid transport, showed strong orange fluorescence in the control cells, but not in the RGS10A-silenced cells (Fig. 3F). This result demonstrates that the RGS10A-silenced OCLs had lost their extracellular acidification function.
These results provided convincing evidence for the role of RGS10A in RANKL-mediated osteoclast differentiation. However, because RANKL is a survival factor for osteoclasts, the question remained whether RGS10A is also required for cell survival. Accordingly, OCLs derived from RANKL-stimulated BMMs were infected with pLenti-scrambled (control) or pLenti-R10a, because lentiviruses can infect non-dividing cells including mature osteoclasts, and were stimulated with RANKL for different time periods as indicated. Using Hoechst 33258 staining, we detected characteristic apoptotic changes in the nuclei. The nuclei of OCLs were condensed and fragmented in RGS10A-silenced groups (Fig. 3G, arrows). In control groups, the nuclei of OCLs showed no evidence of apoptosis (Fig. 3G). In the presence of RANKL and M-CSF, apoptosis caused by RGS10A silencing reached a maximum at 24 hours (Fig. 3H).
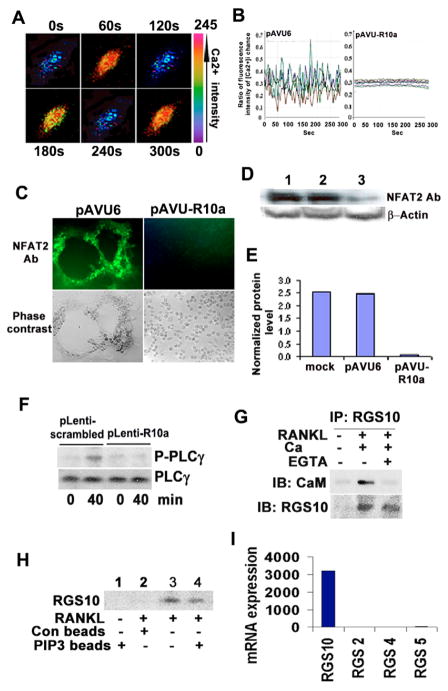
We also hypothesized that RGS10A is a specific regulator of [Ca2+]i oscillations during osteoclast differentiation. To test this proposition, we visualized fluctuations of [Ca2+]i by confocal microscopy. As shown in Fig. 4A,B, in control (pAVU6) cells, sustained oscillations occurred at a frequency of approximately 2-minute intervals. However, in the RGS10A-silenced cells, [Ca2+]i oscillations were completely blocked, indicating that RGS10A is an essential regulator of this function.
Fig. 4.
RGS10A signaling is essential for induction of [Ca2+]i oscillations by RANKL and NFAT2 expression. (A) [Ca2+]i oscillations were observed in RANKL-stimulated BMMs or RAW264.7 cells, 48–72 hours after RANKL stimulation. The picture shows an example of successive pseudocolored Ca2+ images of cells treated with RANKL for 48 hours. Oscillations were produced at 2-minute intervals. (B) Ca2+ changes were traced in single RGS10A-silenced or control cells treated with RANKL and M-CSF for 72 hours. Ca2+ changes were estimated as the ratio of fluorescence intensity of the Fluo-4 to Fura Red, plotted at 5-second intervals. Each color indicates a different cell in the same field. [Ca2+]i oscillations are blocked in RGS10A-silenced cells (pAVU-R10a). (C) Immunofluorescence revealed that the expression of NFAT2 was blocked in RGS10A-silenced cells. (D) There were weak signals of NFAT2 protein detected in RGS10A-silenced cells (lane 3) as compared to the controls (lane 1: mock; lane 2: pAVU6). (E) Quantification of the bands in D. NFAT2 signals in the mock or pAVU6, are 4–4.7 times stronger than in pAVU-R10a. (F) Western blot analysis of activation of PLCγ after stimulation with RANKL for 40 minutes. Phosphorylation of PLCγ was impaired in the RGS10A-silenced group (plenty-R10a) following a 40-minute stimulation with RANKL. (G) Co-immunoprecipitation of RGS10A and CaM. No interaction was observed in unstimulated cells. CaM bound to RGS10A in the presence of 1 mM CaCl2. This interaction was blocked by 0.5 mM EGTA. (H) PdIns(3,4,5)P3-bead binding assay. RGS10A was not detected after stimulation with M-CSF alone (lane 1) or with control beads without PdIns(3,4,5)P3 (negative control, lane 2). RGS10A was detected after stimulation with RANKL (positive control, lane 3). RGS10A was detected after stimulation with RANKL in a PdIns(3,4,5)P3 pull-down assay (lane 4). (I) mRNA expression of RGS 2, 4, 5, and 10A in RANKL-stimulated OCLs. RGS10A is predominantly expressed in OCLs, compared with RGS2, RGS4, and RGS5.
To determine whether RGS10A silencing affects NFAT2 expression, we examined the expression of NFAT2 protein in RANKL and M-CSF-stimulated RGS10A-silenced cells and control cells using immunofluorescent staining and western blotting. As shown in Fig. 4C, the expression of NFAT2 was significantly blocked in RGS10A-silenced cells, whereas it was highly expressed in the control cells. The western blot result showed that the NFAT2 level was 25 times lower in RGS10A-silenced cells (Fig. 4D,E), indicating that RGS10A also acts upstream of NFAT2 in the RANKL-[Ca2+]i oscillation-NFAT2 pathway during osteoclast differentiation.
To examine whether RGS10A silencing affects other signaling pathways evoked by RANKL and M-CSF, we characterized the activation of these pathways. We found that RANKL activation of NFκB, assessed by IκBα degradation, M-CSF-driven phosphorylation of Erk-1/2 and transcriptional induction of Fos, all of which are required for efficient osteoclastogenesis (Boyle et al., 2003; Faccio et al., 2003; Faccio et al., 2005), were normal in all RGS10A-silenced BMMs tested (supplementary material Fig. S1). This suggests that the absence of RGS10A did not affect these signaling pathways, and that RGS10A is specifically involved in the [Ca2+]i-oscillation–NFAT2 signaling pathway.
To gain further insight into the molecular mechanisms underlying RANKL-initiated [Ca2+]i oscillations, we analyzed the activation of PLCγ, which is a key participant in the PLCγ–[Ca2+]i-oscillation–NFAT2 pathway (Koga et al., 2004). BMMs were infected with pLenti-scrambled or pLenti-R10a viruses and then stimulated with RANKL and M-CSF for 40 minutes. Activation of PLCγ was detected using anti-PLCγ and anti-phospho-PLCγ antibodies. The results demonstrated that RANKL-induced phosphorylation of PLCγ was impaired in RGS10A-silenced cells (Fig. 4F). Furthermore, we examined the interaction between RGS10A and calmodulin (CaM) using co-immunoprecipitation of lysates from BMMs (Fig. 4G). When BMMs were stimulated with M-CSF alone, no binding between RGS10A and CaM was detected. In the presence of 1 mM Ca2+, CaM bound to RGS10A following stimulation with RANKL and M-CSF. To test whether CaM binds to RGS10A in a Ca2+-dependent manner, we added the Ca2+-chelator EGTA (0.5 mM) to remove free Ca2+. We found that in the absence of free Ca2+, CaM lost the ability to bind to RGS10A, indicating that Ca2+ is essential for the interaction between RGS10A and CaM.
To determine whether RGS10A binds phosphatidylinositol (3,4,5)-trisphosphate [PdIns(3,4,5)P3] – as shown previously for RGS4 (Ishii et al., 2005) – we performed a PdIns(3,4,5)P3-bead-binding assay (Fig. 4H). RGS10A was not detected with M-CSF stimulation alone or with control beads without PdIns(3,4,5)P3 following stimulation with RANKL and M-CSF. RGS10A was detected after PdIns(3,4,5)P3 pull-down, showing that PdIns(3,4,5)P3 binds to RGS10A. Our results showed that RGS10A provides biochemical control over RANKL-evoked [Ca2+]i oscillations through dual interaction with PdIns(3,4,5)P3 and CaM in a Ca2+-dependent manner.
Our previous studies have shown that RGS12 regulates [Ca2+]i oscillations by controlling Ca2+ influx through interacting with N-type Ca2+ channels (Cav2.2) (Yang and Li, 2007). To further test whether RGS10A has the same function as RGS12, we performed co-immunoprecipitation assays. We found that RGS10A does not act on N-type Ca2+ channels, as shown in supplementary material Fig. S2, indicating that RGS12 and RGS10A plays different roles in regulating [Ca2+]i oscillations. RGS proteins modulate the Ca2+-sensing receptor (CaR) through inhibition of trimetric G protein Giα signaling. Mentaverri et al. found that stimulation of CaR may play a pivotal role in the control of the signaling pathway involving activation of CaR for osteoclast differentiation and activation of PLC for osteoclast apoptosis (Mentaverri et al., 2006), but is not specifically linked to [Ca2+]i oscillations, which are in a different pathway. We found that CaR is expressed in pre-osteoclast and osteoclasts, and that RGS12 binds to CaR; however, we have not found any evidence that RGS10A modulates CaR (supplementary material Fig. S3). After finding that RGS10A interacts with CaM in a Ca2+-dependent manner (Fig. 4G) we determined whether RGS12 also interacts with CaM. We found that RGS12 does not interact with CaM, whereas RGS10A does (supplementary material Fig. S4). This indicates the specificity of RGS10A in this particular role in the regulation of [Ca2+]i oscillation, whereas RGS12 plays a different role. There are reports in which RGS2, RGS4 and RGS5 have been implicated in [Ca2+]i oscillations in other cells (Ishii et al., 2005; Wang et al., 2004; Zhou et al., 2001). To test whether they are indeed expressed in RANKL-stimulated OCLs, we performed microarrays. Our results showed that RGS10A is predominantly expressed in OCLs, and expression of RGS2, RGS4, and RGS5 is much lower than RGS10A (Fig. 4I).
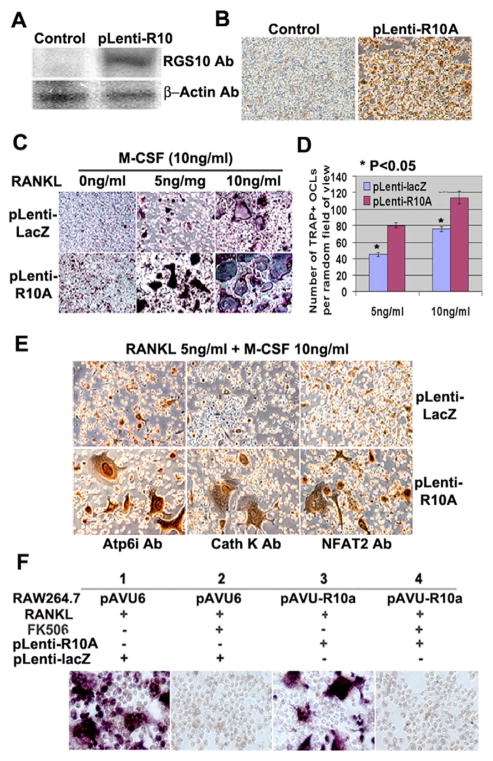
Our data demonstrated that silencing of RGS10A results in a block of [Ca2+]i oscillations and the expression of NFAT2. However, it remained unclear whether RGS10A directly causes [Ca2+]i oscillations, without stimulation of RANKL. To address this question, RGS10A was ectopically expressed in BMM cells. We first confirmed that RGS10A is highly expressed in cells stably infected with pLenti-R10A (Fig. 5A,B). Interestingly, as many as 8% of the pLenti-R10A-infected cells spontaneously differentiated to TRAP+ mononuclear cells in the absence of RANKL (Fig. 5C). When the cells were treated with 5 or 10 ng/ml of RANKL in the presence of 10 ng/ml of M-CSF for 4 days and stained for TRAP+ cells, numbers of TRAP+ MNCs were increased 1.8-fold or 1.5-fold, respectively, compared with pLenti-LacZ-infected cells (Fig. 5C,D). We further investigated the expression of osteoclast marker genes Atp6i, cathepsin K, and NFAT2 in pLenti-R10A infected cells stimulated with lower concentrations of 5 ng/ml RANKL and 10 ng/ml M-CSF. As expected (and shown in Fig. 5E), pLenti-R10A infected cells expressed Atp6i, cathepsin K, and NFAT2 at higher levels than the control cells. These results demonstrated that overexpression of RGS10A increased the sensitivity of osteoclast differentiation to RANKL signaling. However, RGS10A overexpression in pLenti-R10A infected cells was unable to evoke [Ca2+]i oscillations in the absence of RANKL (data not shown), indicating that RGS10A is necessary but not sufficient for [Ca2+]i oscillations.
Fig. 5.
Overexpression of RGS10A significantly increases the sensitivity to RANKL signaling during osteoclast differentiation. BMMs were infected with pLenti-LacZ or pLenti-R10A. (A) Western blotting. Without RANKL stimulation, the signals of RGS10A were only detectable in cells infected with pLenti-R10A but not in the pLenti-LacZ-transfected cells (control). (B) Immunostaining. 80–90% of the cells express RGS10A in pLenti-R10A; however, there are no stained cells in pLenti-LacZ- infected cells. (C) TRAP+ staining. The cells were treated with 0, 5 and 10 ng/ml of RANKL in the presence of 10 ng/ml of M-CSF for 4 days. Without RANKL stimulation, 8% of precursor cells differentiated into mononuclear TRAP+ cells in pLenti-R10A (left lower panel). In pLenti-R10A cells, there are more TRAP+ cells and mature multinuclear cells compared with the control group in the presence of 5 or 10 ng/ml RANKL together with 10 ng/ml M-CSF (middle and right lower panels). (D) Quantitative analysis of TRAP+ MNCs in (C). The number of TRAP+ MNCs in pLenti-R10A is 1.5 times higher than that of the control group (P<0.05 vs pLenti-LacZ). (E) Immunostaining. With 5 ng/ml RANKL stimulation, pLenti-R10A-infected cells expressed Atp6i, cathepsin K, and NFAT2 at higher levels than the control cells. (F) The effect of FK506 on osteoclastogenesis induced by RGS10A overexpression in RGS10A-silenced RAW264.7 or control cells. FK506 (1 μg/ml) inhibited osteoclast differentiation from RANKL-stimulated RAW264.7 cells infected by pLenti-LacZ (panel 2) as compared with the culture without FK506 (panel 1). FK506 inhibited the rescue effect of RGS10A reintroduction as shown in panel 3 in RGS10A-silenced RAW264.7 cells infected with pLenti-RGS10a (panel 4).
FK506 is a specific inhibitor of calcineurin (Liu et al., 1991; Sun et al., 2006), which also inhibits RANKL-induced osteoclast differentiation from osteoclast precursor cells in a dose-dependent manner (Takayanagi et al., 2002). To test whether RGS10A acts upstream of calcineurin in the RANKL–[Ca2+]i-oscillation–calcineurin–NFAT2 pathway, we analyzed whether FK506 could block the rescue of the defective osteoclastogenesis of RGS10A-silenced RAW264.7 cells (pAVU-R10a) by the reintroduction of RGS10A expression. We found that FK506 inhibited the rescue effect (Fig. 5F, panel 4), indicating that FK506 can block RGS10A signaling. Our data showed that RGS10A acts upstream of calcineurin in the RANKL–[Ca2+]i-oscillation–calcineurin– NFAT2 pathway. Our results also confirmed that RANKL evokes [Ca2+]i oscillations that lead to calcineurin-mediated activation of NFAT2 and, therefore, triggers a sustained NFAT2-dependent transcriptional program during osteoclast differentiation (Takayanagi et al., 2002).
Discussion
Here, we demonstrate that RGS10A expression is specific and prominent in RANKL-stimulated osteoclasts and osteoclast precursors, that RGS10A silencing can block [Ca2+]i oscillations and indirectly blocks NFAT2 expression and osteoclast-specific gene expression. Although the regulation of RGS proteins in [Ca2+]i oscillations has primarily been studied in the immune (Kehrl, 1998), neural (Sinnarajah et al., 2001) and cardiovascular systems (Ishii et al., 2002), this is the first report to demonstrate that RGS10A is involved in the RANKL-evoked signaling pathway as a crucial regulator of [Ca2+]i oscillations and is an essential factor for the terminal differentiation of osteoclasts stimulated with RANKL.
Takayanagi et al. have reported that Ca2+ oscillations are initiated as late as 24 hours after RANKL stimulation (Takayanagi et al., 2002), and supernatants from RANKL-stimulated BMMs have a stimulatory effect on osteoclastogenesis and Ca2+ signaling, an effect that was not suppressed by OPG. This result suggests that RANKL activates molecule(s) that can indirectly trigger [Ca2+]i oscillations. We also found that [Ca2+]i oscillations appeared between 24 and 72 hours after RANKL stimulation. We further noted that RGS10A mRNA was detectable 30 minutes after RANKL stimulation and persisted at a high level through 96 hours. Our loss-of-function experiments showed that RGS10A silencing blocked [Ca2+]i oscillations and osteoclast differentiation, suggesting that, before the beginning of [Ca2+]i oscillations, at least one signaling protein, i.e. RGS10A, is activated by RANKL. In addition, it has been reported that Ca2+ signals regulate calcineurin, which in turn de-phosphorylates and induces the nuclear localization of the cytoplasmic components of NFAT transcription complexes (Crabtree and Olson, 2002; Timmerman et al., 1996). In the nucleus, NFAT transcription complexes assemble on DNA to activate specific gene expression in different tissues. These results provide the explanation that impaired NFAT2 expression may result from loss of the regulation of [Ca2+]i oscillations and calcinuerin caused by silencing of RGS10A. Collectively, these findings provide evidence that RGS10A, as a signaling protein, plays an essential role in the RANKL-evoked [Ca2+]i-oscillation–NFAT2 pathway for osteoclast differentiation.
To address the question of whether or not RGS10A directly causes the [Ca2+]i oscillations and whether it is sufficient for osteoclast differentiation, we ectopically expressed RGS10A in osteoclast precursor cells. The results showed that overexpression of RGS10A increased the sensitivity of osteoclast differentiation to RANKL signaling after RANKL stimulation. However, without RANKL, overexpression of RGS10A was unable to evoke [Ca2+]i oscillations and induce osteoclast differentiation, indicating that RGS10A cooperates with other factor(s) activated by RANKL to regulate [Ca2+]i oscillations and terminal differentiation. This is perhaps not surprising because, thus far, only NFAT2 has been demonstrated, by ectopic NFAT2 expression, to act as a master switch for regulating the terminal differentiation of osteoclasts in a RANKL-independent manner. By contrast, overexpression of many osteoclast differentiation regulators (e.g. Fos) could not induce osteoclast precursor differentiation, with only 7% and 1% of cells becoming TRAP+ cells by Fos and Jun overexpression, respectively (Takayanagi et al., 2002).
Our results indicated that NFAT2 expression in RANKL-stimulated RGS10A-silenced cells was very weak when compared with that in RANKL-stimulated control cells. The normalized protein level of NFAT2 in RGS10A silenced cells was 18-fold lower than that in control cells. It was suggested that auto-amplification of NFAT2 is regulated by calcineurin, which was activated by RANKL-induced [Ca2+]i oscillations during osteoclastogenesis (Takayanagi et al., 2002). Our results show that the calcineurin-specific inhibitor FK506 inhibited the rescue effect of RGS10A reintroduction in RANKL-stimulated RGS10A-silenced cells and the formation of TRAP+ cells from the control cells in the absence of RANKL, which indicates that calcineurin acts downstream of RGS10A signaling.
An advantage to studies using lentiviruses is that, since lentiviruses can infect non-dividing cells including mature osteoclasts, we can study all stages of osteoclast development. In our study of RGS10A function in mature osteoclasts we found that RGS10A-silencing induced apoptosis. This finding implies that RGS10A not only functions in the [Ca2+]i oscillation-NFAT2 signaling pathway for osteoclast differentiation, but also in osteoclast survival. It is logical that, because RGS10A is downstream of RANKL in the RANKL–[Ca2+]i-oscillation–NFAT2 pathway, and because RANKL inhibits osteoclast apoptosis through regulation of [Ca2+]i and CaM (El Hajj Dibb et al., 2006; Seales et al., 2006), silencing of RGS10A would induce apoptosis. The mechanism of this function of RGS10A is currently unknown. However, this could be a secondary consequence due to the lack of oscillations, which broadly affects cell signaling and proliferation, differentiation and survival.
Koga et al. have found that PLCγ is an essential component of the [Ca2+]i-oscillation–NFAT2 pathway for osteoclast differentiation (Koga et al., 2004), and Maffucci and Falasca reported that PdIns(3,4,5)P3 is a crucial player in PLCγ activation (Maffucci and Falasca, 2007). Ishii et al. found that RGS4 binds PdIns(3,4,5)P3 and that the Ca2+/CaM complex competes with PdIns(3,4,5)P3 for binding to RGS4 (Ishii et al., 2005). In our studies, based on these reports, we found that RGS10A-silencing inhibits PLCγ activation. We also found that RGS10A binds CaM as a complex of Ca2+/CaM-RGS10A and that RGS10A binds PdIns(3,4,5)P3. Based on our data and those of others, we propose an RGS10A working model: RANKL mediates phosphorylation of both FcRγ and DAP12, the membrane adaptor molecules that contain an ITAM motif, which then activate PLCγ. Once activated, PLCγ can hydrolyze phosphatidylinositol (4,5)-trisphosphate [PdIns(4,5)P2] to generate inositol (1,4,5)-trisphosphate [Ins(1,4,5)P3], which then triggers a transient initial release of Ca2+ from intracellular stores. Intracellular Ca2+ release allows an increase in [Ca2+]i to reach peak concentration and leads to formation of the Ca2+/CaM complex. The Ca2+/CaM complex competes for the PdIns(3,4,5)P3-binding site on RGS10A and frees the bound PdIns(3,4,5)P3. Once the [Ca2+]i reaches its peak, it begins to reload into the endoplasmic reticulum (ER) without further PLCγ activation, and the combination of reloading the ER with Ca2+ and Ca2+ binding to CaM results in a decrease of [Ca2+]i. The Ca2+/CaM complex dissociates from RGS10A at low Ca2+ concentration. Free PdIns(3,4,5)P3 activates PLCγ and then binds RGS10A again without competition by the Ca2+/CaM complex. PLCγ activation triggers a release of Ca2+ from intracellular stores by generating Ins(1,4,5)P3 to cause a second peak. This cycle continues, causing [Ca2+]i oscillations. In this way, RGS10A mediates PLCγ activation and triggers [Ca2+]i oscillations through its [Ca2+]i-dependent dual interaction with Ca2+/CaM and PdIns(3,4,5)P3. The RGS10A-mediated [Ca2+]i oscillations activate calcineurin and NFAT2 expression for osteoclast terminal differentiation.
To demonstrate the speficity of RGS10A in regulating [Ca2+]i oscillation in osteoclasts with the described target components, we tested the interaction of RGS10A and RGS12 with CaM, and with the target components described in our previous study (Yang and Li, 2007), the membrane N-type Ca2+ channel and the CaR. We found that RGS12, but not RGS10A, interacts with the N-type Ca2+ channel and the CaR, whereas RGS10A, and not RGS12, interacts with CaM, indicating that these two RGS proteins are specific to two different pathways in the regulation of [Ca2+]i oscillations. The specificity of RGS10A and RGS12 to these functions is not surprising owing to the differences in their protein structure. RGS12 has a molecular mass of 174 kDa, whereas that of RGS10A is 20 kDa. In addition, RGS12 contains several interaction domains, such as RBD, PDZ, and PTB domains, and the GoLoco motif, which have not been explored, whereas RGS10A only contains the RGS domain. Even the RGS domain of RGS12 only shows 51% similarity with the RGS domain of RGS10A. This shows that they are structurally different and are likely to have different target components, which probably contributes to the specificity of their roles in the regulation of [Ca2+]i oscillation. Our results also demonstrate that, even though RGS10A and RGS12 can be silenced (causing lack of [Ca2+]i oscillations) RGS12 regulates membrane channels, including the Ca2+ channel and the CaR (Yang and Li, 2007). RGS10A, however, functions internally, interacting with signaling proteins in the cytoplasm as described in the working model. Owing to the number of interaction domains in RGS12 we cannot exclude other possible functions of RGS12 in osteoclast differentiation. To further demonstrate the specificity of RGS10A, we found that RGS10A is predominantly expressed in OCLs, in contrast to the other RGS family members RGS2, RGS4 and RGS5.
As a key intermediate in the RANKL–[Ca2+]i-oscillation–NFAT2 signaling pathway, RGS10A may provide a therapeutic target in osteolytic bone diseases. Further studies will be required to identify other molecules that might interact with RGS10A and might be involved in the RANKL–[Ca2+]I-oscillation–NFAT2 signaling pathway.
Materials and Methods
Cells and cell culture
MOCP-5 and RAW264.7 are monocytic osteoclast progenitor cell lines. MOCP-5 was generated in our laboratory (Chen and Li, 1998). RAW264.7 was obtained from the American Type Culture Collection. Human osteoclastoma tumors were obtained from Andrew Rosenberg (Chambers et al., 1986; Flanagan et al., 1988). Stromal cells from tumors were obtained as described (Li et al., 1995).
Differential screening of a human osteoclastoma cDNA library
Two human osteoclastoma cDNA libraries were prepared in pcDNAII (Invitrogen) and the Lambda-ZAP system (Stratagene) (Li et al., 1995). Differential screening was performed as described (Li et al., 1995).
Northern blotting analysis
Total RNA from mouse tissues and OCLs derived from the osteoclast progenitor cell lines MOCP-5 and RAW264.7, and from BMMs was isolated using Trizol reagent. A 0.9 kb mouse full-length RGS10A cDNA was used as a probe. Hybridization was performed as described previously (Yang et al., 2003).
RT-PCR cDNA cloning and sequencing analysis
Total RNA from human osteoclasts, human brain and RANKL-stimulated BMMs was isolated using Trizol reagent. One step RT-PCR was performed using the Access RT-PCR system (Promega). RT-PCR analysis of RGS10A, RGS10B or FOS mRNA was performed in RANKL-stimulated OCLs. The specific intron-spanning primers for isoform A of hRGS10 were BOR10-F 5′-ATGTTCAACCGCGCCGTGA and BOBRR10-R:. 5′-CATCCCATTGAAGGGTTTTG-3′. For isoform B of hRGS10 primers were BRR10-F 5′-ATGGAACACATCCACGACA-3′ and BOBRR10-R 5′-CATCCCATTGAAGGGTTTTG-3′. The specific intron-spanning primer sequences for the mouse Fos gene were Fos-S 5′-CTGGTGCAGCCCACTCTGGTC-3′ and Fos-AS 5′-CTTTCAGCAGATTGGCAATCTC-3′.
In situ hybridization
In situ hybridization with RGS10A mRNA probes for RGS10A in human osteoclasts was performed as described (Li et al., 1995).
Immunostaining
The cells were stimulated with RANKL and M-CSF for the indicated times, plated on sterile polylysine-coated coverslips and incubated overnight. The cells were sequentially fixed with 100% methanol for 10 minutes, blocked with 10% normal donkey, rabbit or goat serum in PBS for 60 minutes, then correspondingly incubated in goat anti-RGS10 antibody (1 μg/ml, Santa Cruz Biotechnology), rabbit anti-Apt6i specific polyclonal antibody, rabbit anti-cathepsin K polyclonal antibody (from our lab), and mouse anti-NFAT2 antibody (1 μg/ml, Santa Cruz Biotechnology) in PBS containing 1.5% normal rabbit, donkey, or goat serum, respectively, for 60 minutes. Cells were then incubated in FITC- or HRP-conjugated anti-goat, rabbit, or mouse IgG (1 μg/ml, Santa Cruz Biotechnology) with 1.5% normal rabbit, donkey, or goat serum, respectively, for 60 minutes. For cells with FITC-conjugated antibody, coverslips were washed three times in PBS, mounted on glass slides and examined with microscopes. For cells with HRP-conjugated antibody, coverslips were further stained with VECTASTAIN Elite ABC kit (Vector Laboratories).
Co-immunoprecipitation
RAW264.7 cells were stimulated with RANKL and M-CSF for 96 hours and then washed with PBS and lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 1% Nonidet P-40, and phosphatase and protease inhibitors (Yang et al., 2003). Cell lysates were incubated at 4°C for 2 hours with anti-RGS10 (Santa Cruz Biotechnology), anti-RGS12 (GenWay), anti-CaM (Santa Cruz Biotechnology) or anti-CaR (Santa Cruz Biotechnology) antibodies and then incubate with protein G-Sepharose beads (Amersham Biosciences) for overnight. The precipitates were separated by 4–15% SDS-PAGE, followed by immunoblotting with anti-CaM antibody (Santa Cruz Biotechnology).
RGS10A silencing using plasmid-based siRNA expression in RAW264.7 cells
Oligonucleotides encoding shRNA directed against the RGS10A gene (Accession No. NM_026418) were designed using Insert Design Tool for the Vectors (Ambion) according to the manufacturer’s instructions, and ensured by BLAST search that they did not have significant sequence homology with other genes including other members of the RGS family. The oligonucleotides were then cloned into the SalI/XbaI site of the pAVU6+27 siRNA expression vector (gift by Paul D. Good, University of Michigan, Ann Arbor, MI) driven by the RNA polymerase III promoter of U6 small nuclear RNA gene. The inserts contain the 9-nucleotide spacer and five thymine residues. The target sites were located at 189–209 bp and 330–350 bp. The sequences of oligonucleotides were RGS10A-siR1S, 5′-TCGACCC-GTCTTCTGGAAGACCCAGAATTCAAGAGATTCTGGGTCTTCCAGAAGAT-TTTTGGAAA-3′; RGS10A-siR1AS, 5′-CTAGTTTCCAAAAATCTTCTGGAA-GACCCAGAATCTCTTGAATTCTGGGTCTTCCAGAAGACGGG-3′; RGS10A-siR2S, 5′-TCGACCCGGAGATCTACATGACCTTCTTCAAGAGAGAAGGTCA-TGTAGATCTCCTTTTTGGAAA-3′; RGS10A-siR2AS, 5′-CTAGTTTCCAAA-AAGGAGATCTACATGACCTTCTCTCTTGAAGAAGGTCATGTAGATCTCCG-GG-3′. RAW264.7 cells were plated on 35-mm dishes at a density of 2.5×105 cells per cm2. After 24 hours, cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Each transfection contained 0.5 μg of the expression vector. The cells were cultured in growth medium for 24 hours and then switched to the medium containing 400 μg/ml G418, to select stable colonies.
By stable transfection with these constructs into RAW264.7 cells, we performed a quick cell clone screening analysis of shRNA expression using TRAP staining and selected the cell colonies carrying the two constructs for further study, pAVU-R10a and pAVU-R10b. The insert sequences of the two constructs were 5′-AATCTTCTGGAAGACCCAGAA-3′ and 5′-AAGGAGATCTACATGACCTTC-3′, located at 189–209 bp and 330–350 bp, respectively, of RGS10A mRNA. Cells carrying pAVU-R10a or pAVU-R10b and control construct (pAVU-scrambled) were seeded at 5×104 cells per well in a 24-well plate and cultured in α-MEM (GIBCO-BRL) with 10% FBS (GIBCO-BRL) containing M-CSF (10 ng/ml). After 24 hours, RGS10A-silenced cells were further cultured and stimulated with 10 ng/ml soluble RANKL (Peprotech) and M-CSF (10 ng/ml) to generate osteoclasts. For suppressive effects of a calcineurin inhibitior, RGS10A-stably silenced RAW264.7 cells and RAW264.7 cells were infected with pLenti-LacZ or pLenti-RGS10A, then 1 μg/ml FK506 was added to the medium to generate mature osteoclasts. The identity of osteoclasts was confirmed by TRAP staining.
RGS10A silencing by expressing lentivirus siRNA in mouse bone marrow cells
Oligonucleotides encoding the same shRNAs directed against the RGS10A gene (Accession No. NM_026418) were designed using BLOCK-iT™ RNAi Designer (Invitrogen) according to the manufacturer’s instructions, and then cloned into pENTR/H1/TO vector (Invitrogen). The LR recombination reaction was completed between the pENTR/H1/TO-RGS10A vector and the pLenti6/BLOCK-iT™-DEST construct to generate pLenti6/BLOCK-iT™ expression constructs according to the manufacturer’s instructions. Sequences of the oligonucleotides were lenR10-top1, 5′-CACCTCTTCTGGAAGACCCAGAACGAATTCTGGGTCTTCCAGAAGA-3′; lenR10-bottom1, 5′-AAAATCTTCTGGAAGACCCAGAATTCGTTCTGGGTCT-TCCAGAAGA-3′; lenR10-top2, 5′-CACCGGAGATCTACATGACCTTCCGAA-GAAGGTCATGTAGATCTCC-3′; lenR10-bottom2, 5′-AAAAGGAGATCTACAT-GACCTTCTTCGGAAGGTCATGTAGATCTCC-3′. Recombinant lentiviral production was carried out according to the manufacturer’s instructions from the BLOCK-iT™ Lentiviral RNAi system (Invitrogen). Briefly, the 293FT producer cell line was co-transfected with the expression constructs (pLenti-scrambled shRNA, pLenti-R10a, or pLenti-R10b) and packaging mixture. The viral supernatant was harvested after 48–72 hours and titers were determined by infecting HeLa cells with serial dilutions of concentrated lentivirus. BMMs was obtained as described (Kelly et al., 1998). The BMMs were infected with pLenti-R10a or pLenti-R10b viral supernatant plus 6 μg/ml polybrene (Sigma) for 24 hours, and then treated with 10 ng/ml soluble RANKL (Peprotech) and M-CSF (10 ng/ml). Ninety-six hours later, the cells were fixed for determination of differentiation.
TRAP+ staining
TRAP was used as a marker for osteoclasts. Pre-osteoclasts and OCLs derived from BMMs, cell lines stably expressing RGS10 siRNA, and cell lines overexpressing RGS10 were fixed and stained for TRAP activity using a commercial kit (Sigma) according to the manufacturer’s instructions. Multinucleated (>3 nuclei) TRAP+ cells appeared as dark purple cells and were counted by light-microscopy. All data are expressed as the mean ± s.d.
Western blotting
The cells were incubated with RANKL and M-CSF as above for 40 minutes or 96 hours. Western blots were performed as described (Yang et al., 2003), visualized, and quantified using a Fluor-S MultiImager and Multi-Analyst software (Bio-Rad). Activation of PLCγ was detected using anti-PLCγ 1 and anti-phospho-PLCγ 1 antibodies (Santa Cruz Biotechnology). Expression of RGS10, NFAT2 or RGS12 was detected using anti-RGS10 and NFAT2 antibodies (Santa Cruz Biotechnology) and anti-RGS12 antibody (GenWay). The expression of IκBα and phosphorylated (P)-Erk was detected using anti-IκBα and anti-P-Erk antibodies from Cell Signaling Technology.
Apoptosis assay
Apoptosis was measured by Hoechst 33258 staining of condensed chromatin (Kameda et al., 1996; Wu et al., 2003). The osteoclasts derived from RANKL-stimulated BMMs were infected with recombinant lentivirus carrying pLenti-scrambled or pLenti-R10 constructs, and were continuously treated with RANKL for 0, 6, 12, 18 and 24 hours. Next, cells were fixed with 2% glutaraldehyde solution (Wako) for 10 minutes and stained with 0.2 mM Hoechst 33258 to visualize DNA localization. Cells were examined under a fluorescence microscope for determination of osteoclasts with chromatin condensation and/or nuclear fragmentation.
Ca2+ measurements
Ca2+ measurements were performed as described (Takayanagi et al., 2002). The cells were incubated with M-CSF or RANKL and M-CSF for 24, 48 or 72 hours and then with 5 μM fluo-4 AM, 5 μM Fura Red AM, and 0.05% pluronic F127 for 30 minutes. The cells were post-incubated in DMEM medium with 10 ng/ml M-CSF for 20 minutes and mounted on a confocal microscope (Leica). To estimate [Ca2+]i concentration in single cells, the ratio of the fluorescence intensity of Fluo-4 to Fura Red was calculated.
Overexpression of RGS10A in osteoclast precursor cell line and BMMs
A 0.6 kb full-length mouse RGS10A cDNA was amplified by RT-PCR and cloned into pDONR221 vector (Invitrogen) according to the manufacturer’s instructions. The LR recombination reaction was completed between the pDONR-RGS10 vector and pLenti6/V5-Dest vector to generate pLenti-R10A, which is engineered to express RGS10A. pLenti-LacZ was a control. Lentivirus packaging was performed according to the manufacturer’s instructions by co-transfection of these vectors and packaging mixtures (Invitrogen) into 293T cells. The viral supernatant was harvested after 48–72 hours, and titers were determined. BMMs or RGS10A stably silenced RAW264.7 cells were infected with the pLenti-R10A or control virus pLenti-LacZ. Protein expression of RGS10A was confirmed by immunoblotting and immunostaining in part of pLenti-RGS10-transfected BMMs. The rest of the cells were further cultured with M-CSF in the presence or absence of RANKL for the osteoclast formation assay.
Ca2+/CaM-binding assay
BMMs were stimulated with RANKL and M-CSF for 96 hours and then washed with PBS and lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 1% Nonidet P-40, and phosphatase and protease inhibitor cocktail (Sigma) (Yang et al., 2003). Prior to incubation, 0.5 mM EGTA or 1-mM aliquots of CaCl2 were added to those lysates and incubated at 4°C for 1 hour (Edlich et al., 2005; Erickson-Viitanen and DeGrado, 1987), followed by incubation at 4°C for 2 hours with anti-RGS10 antibody (Santa Cruz Biotechnology) and protein A/G-Sepharose beads (Amersham Biosciences). Ca2+ was added to the lysis buffer to mimic the cytoplasmic environment at high levels of Ca2+, and EGTA was added to the lysis buffer to mimic the cytoplasmic environment at levels of low or no Ca2+. The precipitates were separated by 4–15% SDS-PAGE, followed by immunoblotting with anti-CaM antibody (Santa Cruz Biotechnology).
PdIns(3,4,5)P3 binding assay
BMMs were treated with M-CSF for 24 hours or RANKL and M-CSF for 96 hours, and suspended in 0.5 ml of binding assay buffer (100 mM KCl, 2 mM MgCl2, 0.5% Lubrol, and 20 mM Tris-HCl, pH 7.5) in the presence of 1% protease inhibitor cocktail (Sigma). After brief sonication, cell lysates were centrifuged at 1500 g for 5 minutes. The supernatant was collected and incubated with control beads or PdIns(3,4,5)P3 beads (50 μl slurry; Echelon Research Laboratories) at 4°C for 12 hours. Then the mixture was centrifuged at 1500 g for 5 minutes. The collected beads were washed three times with 1 ml of binding assay buffer, and the bound proteins were subjected to western blot analysis (Lee et al., 2003; Tseng et al., 2004).
Statistical analysis
Where indicated, experimental data are reported as mean ± s.d. of triplicate independent samples. Studies using multiple groups were analyzed using ANOVA followed by Tukey-Kramer multiple-comparisons test to determine statistically significant differences between groups. Statistical significance for two groups was assessed using an unpaired t test.
Supplementary Material
Acknowledgments
We thank Carrie Soltanoff for excellent assistance with the manuscript. This work was supported by NIH grant AR-44741 (Y.P.L.), AR-48133-01 (Y.P.L.).
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/120/19/3362/DC1
References
- Alliston T, Derynck R. Medicine: interfering with bone remodelling. Nature. 2002;416:686–687. doi: 10.1038/416686a. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation 1. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Fuller K, Darby JA, Pringle JA, Horton MA. Monoclonal antibodies against osteoclasts inhibit bone resorption in vitro. Bone Miner. 1986;1:127–135. [PubMed] [Google Scholar]
- Chen W, Li YP. Generation of mouse osteoclastogenic cell lines immortalized with SV40 large T antigen. J Bone Miner Res. 1998;13:1112–1123. doi: 10.1359/jbmr.1998.13.7.1112. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Edlich F, Weiwad M, Erdmann F, Fanghanel J, Jarczowski F, Rahfeld JU, Fischer G. Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin 1. EMBO J. 2005;24:2688–2699. doi: 10.1038/sj.emboj.7600739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj Dibb I, Gallet M, Mentaverri R, Sevenet N, Brazier M, Kamel S. Imatinib mesylate (Gleevec) enhances mature osteoclast apoptosis and suppresses osteoclast bone resorbing activity. Eur J Pharmacol. 2006;551:27–33. doi: 10.1016/j.ejphar.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Erickson-Viitanen S, DeGrado WF. Recognition and characterization of calmodulin-binding sequences in peptides and proteins 1. Meth Enzymol. 1987;139:455–478. doi: 10.1016/0076-6879(87)39106-2. [DOI] [PubMed] [Google Scholar]
- Faccio R, Takeshita S, Zallone A, Ross FP, Teitelbaum SL. c-Fms and the alphavbeta3 integrin collaborate during osteoclast differentiation 2. J Clin Invest. 2003;111:749–758. doi: 10.1172/JCI16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, Ross FP, Swat W. Vav3 regulates osteoclast function and bone mass 4. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- Flanagan AM, Nui B, Tinkler SM, Horton MA, Williams DM, Chambers TJ. The multinucleate cells in giant cell granulomas of the jaw are osteoclasts. Cancer. 1988;62:1139–1145. doi: 10.1002/1097-0142(19880915)62:6<1139::aid-cncr2820620617>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Xing L, Poljak L, Shores EW, Brown KD, Leonardi A, Tran T, Boyce BF, Siebenlist U. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- Ishii M, Inanobe A, Kurachi Y. PIP3 inhibition of RGS protein and its reversal by Ca2+/calmodulin mediate voltage-dependent control of the G protein cycle in a cardiac K+ channel. Proc Natl Acad Sci USA. 2002;99:4325–4330. doi: 10.1073/pnas.072073399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action 35. Biochem J. 2005;385:65–73. doi: 10.1042/BJ20040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda T, Miyazawa K, Mori Y, Yuasa T, Shiokawa M, Nakamaru Y, Mano H, Hakeda Y, Kameda A, Kumegawa M. Vitamin K2 inhibits osteoclastic bone resorption by inducing osteoclast apoptosis. Biochem Biophys Res Commun. 1996;220:515–519. doi: 10.1006/bbrc.1996.0436. [DOI] [PubMed] [Google Scholar]
- Kehrl JH. Heterotrimeric G protein signaling: roles in immune function and fine-tuning by RGS proteins. Immunity. 1998;8:1–10. doi: 10.1016/s1074-7613(00)80453-7. [DOI] [PubMed] [Google Scholar]
- Kelly KA, Tanaka S, Baron R, Gimble JM. Murine bone marrow stromally derived BMS2 adipocytes support differentiation and function of osteoclast-like cells in vitro. Endocrinology. 1998;139:2092–2101. doi: 10.1210/endo.139.4.5915. [DOI] [PubMed] [Google Scholar]
- Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, Iwata T, Ohnishi H, Matozaki T, Kodama T, et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Xu H, Kang LW, Amzel LM, Montell C. Light adaptation through phosphoinositide-regulated translocation of Drosophila visual arrestin 6. Neuron. 2003;39:121–132. doi: 10.1016/s0896-6273(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Li YP, Alexander M, Wucherpfennig AL, Yelick P, Chen W, Stashenko P. Cloning and complete coding sequence of a novel human cathepsin expressed in giant cells of osteoclastomas. J Bone Miner Res. 1995;10:1197–1202. doi: 10.1002/jbmr.5650100809. [DOI] [PubMed] [Google Scholar]
- Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes 3. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Maffucci T, Falasca M. Phosphoinositide 3-kinase-dependent regulation of phospholipase Cgamma. Biochem Soc Trans. 2007;35:229–230. doi: 10.1042/BST0350229. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet. 2000;24:184–187. doi: 10.1038/72855. [DOI] [PubMed] [Google Scholar]
- Mentaverri R, Yano S, Chattopadhyay N, Petit L, Kifor O, Kamel S, Terwilliger EF, Brazier M, Brown EM. The calcium sensing receptor is directly involved in both osteoclast differentiation and apoptosis. FASEB J. 2006;20:2562–2564. doi: 10.1096/fj.06-6304fje. [DOI] [PubMed] [Google Scholar]
- Mocsai A, Humphrey MB, Van Ziffle JA, Hu Y, Burghardt A, Spusta SC, Majumdar S, Lanier LL, Lowell CA, Nakamura MC. The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRgamma) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci USA. 2004;101:6158–6163. doi: 10.1073/pnas.0401602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4:353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Seales EC, Micoli KJ, McDonald JM. Calmodulin is a critical regulator of osteoclastic differentiation, function, and survival. J Cell Biochem. 2006;97:45–55. doi: 10.1002/jcb.20659. [DOI] [PubMed] [Google Scholar]
- Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S, Dennis JC, Morrison EE, Vodyanoy V, Kehrl JH. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- Sun L, Peng Y, Zaidi N, Zhu LL, Iqbal J, Yamoah K, Wang X, Liu P, Abe E, Moonga BS, et al. Evidence that calcineurin is required for the genesis of bone resorbing osteoclasts 1. Am J Physiol Renal Physiol. 2006;292:F285–F291. doi: 10.1152/ajprenal.00415.2005. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Clipstone NA, Ho SN, Northrop JP, Crabtree GR. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- Tseng PH, Lin HP, Hu H, Wang C, Zhu MX, Chen CS. The canonical transient receptor potential 6 channel as a putative phosphatidylinositol 3,4,5-trisphosphate-sensitive calcium entry system 1. Biochemistry. 2004;43:11701–11708. doi: 10.1021/bi049349f. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Karsenty G. Genetic control of skeletal development. Curr Opin Genet Dev. 2001;11:527–532. doi: 10.1016/s0959-437x(00)00228-8. [DOI] [PubMed] [Google Scholar]
- Wang X, Huang G, Luo X, Penninger JM, Muallem S. Role of regulator of G protein signaling 2 (RGS2) in Ca(2+) oscillations and adaptation of Ca(2+) signaling to reduce excitability of RGS2−/− cells. J Biol Chem. 2004;279:41642–41649. doi: 10.1074/jbc.M406450200. [DOI] [PubMed] [Google Scholar]
- Wu X, McKenna MA, Feng X, Nagy TR, McDonald JM. Osteoclast apoptosis: the role of Fas in vivo and in vitro. Endocrinology. 2003;144:5545–5555. doi: 10.1210/en.2003-0296. [DOI] [PubMed] [Google Scholar]
- Yang S, Li YP. RGS12 is essential for RANKL-evoked signaling for terminal differentiation of osteoclasts in vitro 1. J Bone Miner Res. 2007;22:45–54. doi: 10.1359/jbmr.061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wei D, Wang D, Phimphilai M, Krebsbach PH, Franceschi RT. In vitro and in vivo synergistic interactions between the Runx2/Cbfa1 transcription factor and bone morphogenetic protein-2 in stimulating osteoblast differentiation. J Bone Miner Res. 2003;18:705–715. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Moroi K, Nishiyama M, Usui H, Seki N, Ishida J, Fukamizu A, Kimura S. Characterization of RGS5 in regulation of G protein-coupled receptor signaling 2. Life Sci. 2001;68:1457–1469. doi: 10.1016/s0024-3205(01)00939-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.