Abstract
We previously analyzed the differential localization patterns of five septins (AspAÊ), including a filamentous fungal-specific septin, AspE, in the human pathogen Aspergillus fumigatus. Here we utilized the A. fumigatus strain expressing an AspE-EGFP fusion protein and show that this novel septin with a tubular localization pattern in hyphae is phosphorylated in vivo and interacts with the other septins, AspA, AspB, AspC and AspD. The other major proteins interacting with AspE included the cytoskeletal proteins, actin and tubulin, which may be involved in the organization and transport of the septins. This is the first report analyzing the phosphorylation of AspE and localizing the sites of phosphorylation, and opens opportunities for further analysis on the role of post-translational modifications in the assembly and organization of A. fumigatus septins. This study also describes the previously unknown interaction of AspE with the actin-microtubule network. Furthermore, the novel GFP-Trap® affinity purification method used here complements widely-used GFP localization studies in fungal systems.
Keywords: Aspergillus, Septin, AspE, Phosphorylation, GFP-Trap® affinity purification, Mass spectrometry
1. Introduction
Septins are a highly conserved family of GTP-binding proteins distributed in all eukaryotes except plants and function by forming heteropolymeric structures [1,2]. In contrast to mammalian septins that are encoded by a multi-gene family comprising 12 genes (SEPT1-12) [1,3], the model yeast Sacchromyces cerevisiae contains 7 septin genes (CDC3, CDC10, CDC11, CDC12, SHS1, SPR3 and SPR28) that are differentially expressed during vegetative growth and sporulation [4,5,6]. However, the filamentous group of fungi, including Aspergillus species, consists of 4 septins (AspA~D) that are orthologous to S. cerevisiae septins and a fifth septin (AspE) unique to filamentous fungi [7]. Although septins have been proposed to regulate a wide variety of functions in mammalian cells [1,2,3,8] and the yeasts [1,4,5,6,9], knowledge of their roles in filamentous fungi is limited to morphogenetic events involving hyphal branching, septation, and conidiophore development [10,11,12]. Earlier reports implicated a role for septins in tissue invasion and virulence of the pathogenic yeast Candida albicans [13], and recent data from the plant pathogenic filamentous fungus Magnaporthe grisea revealed the importance of septins for plant cell invasion [14,15]. Therefore, studies directed towards understanding septin organization and their roles in the opportunistic human pathogen A. fumigatus could help decipher invasive pathogenesis and lead to identification of better molecular targets to combat invasive aspergillosis in patients.
Cellular mechanisms involved in the formation of higher order septin structures and the dynamics of septin assembly still remain unknown. In mammals and the yeasts, septin organization and dynamics have been linked to post-translational modifications involving phosphorylation [3,16,17,18]. Three kinases, Elm1, Cla4 and Gin4, control septin organization in S. cerevisiae [19,20,21]. After Tachikawa et al [22] reported that a Gip1p-Glc7p phosphatase complex is required for proper septin organization and initiation of spore wall formation in S. cerevisiae, the Shs1p septin, which serves as the outer component of the septin complex, was shown to bind to the Gin4 kinase, indicating the possibility of Shs1 undergoing Gin4-mediated phosphorylation [23]. This was further strengthened by another report which indicated that the association of the septins Shs1p and Cdc11p involves some yeast-specific factor or post-translational modification [24]. Phosphorylation of the septin Cdc3p by the Cdc28 kinase was shown to be essential for proper disassembly of the septin ring [25]. Septin collar formation and septin filament assembly was also shown to require both GTP binding and Cla4-mediated phosphorylation of septins [24]. Furthermore, Dobbelaere et al [17] showed that RTS1 (the regulatory subunit of protein phosphatase-2A) mediated dephosphorylation of septins and is required for the disassembly process, indicating that activation of septin dynamics by phosphorylation events controls the rigidity of the septin ring.
Although these results indicated that phosphorylation and dephosphorylation regulate septin architecture in the yeasts, there is limited information on the analysis of septin interactions and phosphorylation in filamentous fungi. Only recently in Ashbya gossypii, a filamentous fungus belonging to the saccharomycotina subphyla and closely related to yeast, the assembly of a subset of septin rings was shown to be associated with two kinases, Elm1p and Gin4p [26]. Multiple phosphorylation sites were identified in the A. gossypii septin, Shs1p [27], and also the other septins [28]. Although mutation of Shs1p phosphorylable sites led to decreased septin dynamics, phosphomimetic mutations were lethal [28], revealing a dynamic regulation of septin organization by phosphorylation /dephosphorylation mechanisms. While A. gossypii belongs to the filamentous group of fungi, it lacks the ortholog of AspE which is present in pezizomycota, the largest subphyla of filamentous fungi.
We previously reported the differential localization patterns of all the 5 septins in the human pathogenic fungus A. fumigatus, and showed that the filamentous fungal-specific septin, AspE, localized as long tubular or filament-like structures which were disrupted in presence of actin and microtubule inhibitors [29]. In order to further understand the mechanism of AspE assembly in the septin complex, here we purified the AspE-GFP fusion protein to identify its interactants and also examine its phosphorylation status in vivo. We show that AspE interacted with all of the other 4 septins (AspA, AspB, AspC and AspD) as well as the other major binding proteins, actin and tubulin. In addition, we also show for the first time that AspE is highly phosphorylated at the N-terminus, within the G-domain, and at its C-terminal region, indicating the possible role for phosphorylation in the assembly of A. fumigatus septins.
2. Materials and methods
2. 1. Organism and culturing, Protein extraction and AspE-GFP purification
The A. fumigatus strain expressing the aspE-egfp fusion construct under the control of the otef promoter was grown in glucose minimal media (GMM) liquid medium as a shaking culture for 24 h at 37°C. Total cell lysate was extracted by homogenizing the fungal tissue (1.5~2 mg wet weight) using liquid nitrogen and suspended in 5 ml lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.01% Triton X-100, 1mM DTT, 1mM PMSF, 1:100 Protease Inhibitory Cocktail) and centrifuged at 5000 rpm for 10 min at 4°C to remove cell debris. The crude supernatant was clarified by centrifugation at 7000 rpm for 15 min at 4°C. Total protein in the crude extract was quantified by Bradford method and normalized to contain ~10 mg protein in the sample before GFP-Trap® affinity purification (Chromotek). GFP-Trap® resin (35 μl) was equilibrated by washing three times in 500 μl ice-cold dilution buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1mM PMSF, 1:100 Protease Inhibitory Cocktail) according to the manufacturer instructions and finally resuspended in 100 μl ice cold dilution buffer. The GFP-Trap® resin suspension was then mixed with total crude cell lysate containing ~10 mg total protein and incubated at 4°C by gentle agitation for 2 h. The suspension was centrifuged at 2000 rpm for 10 min at 4°C and the pelleted GFP-Trap® resin was washed once in 500 μl of ice-cold dilution buffer and then twice with 500 μl of wash buffer (10 mM Tris-HCl pH 7.5, 350 mM NaCl, 0.5 mM EDTA, 1mM PMSF, 1:100 Protease Inhibitory Cocktail).
2.3. Sample Preparation and Nano-Flow Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-MS/MS) Analysis
Protein bound GFP-Trap® resins were washed three times with 50 mM ammonium bicarbonate, pH 8.0, and then suspended in 30 μl 50 mM ammonium bicarbonate, pH 8.0, supplemented with 0.1% Rapigest SF surfactant (Waters Corp). Samples were reduced with 5 mM dithiolthreitol for 30 min at 70°C and free sulfhydryls were alkylated with 10 mM iodoacetamide for 45 min at room temperature. Proteolytic digestion was accomplished by the addition of 500 ng sequencing grade trypsin (Promega) directly to the resin with incubation at 37°C for 18 hours. Supernatants were collected following a 2 min centrifugation at 1,000 rpm, acidified to pH 2.5 with TFA, and incubated at 60°C for 1 h to hydrolyze the remaining Rapigest surfactant. Insoluble hydrolyzed surfactant was cleared by centrifugation at 15,000 rpm for 5 min. Ninety percent (by volume) of the sample was then removed for subsequent phosphopeptide analysis and the remaining ten percent (by volume) was subjected to an unbiased protein interaction analysis. For the phosphopeptide analysis, samples were dried using vacuum centrifugation and resuspended in 100 μl 80% acetonitrile, 1% TFA, 50 mg/mL MassPrep Enhancer, pH 2.5 (Waters Corp). Peptides were subjected to phosphopeptide enrichment using a 200 μL TiO2 Protea Tip (Protea Bio) and subsequently washed with 200 μl 80% acetonitrile, 1% TFA, 50 mg/ml MassPrep Enhancer followed by 200 μl 80% acetonitrile, 1% TFA. Peptides were eluted in 50 μl 20% acetonitrile, 5% aqueous ammonia, pH 10.5 and then acidified to pH 2.5 with formic acid prior to drying using vacuum centrifugation.
Samples were resuspended in 10 μl 2% acetonitrile, 0.1% formic acid and subjected to chromatographic separation on a Waters NanoAquity UPLC equipped with a 1.7 μm BEH130 C18 75 μm I.D. × 250 mm reversed-phase column. Phosphopeptide enriched samples were additionally supplemented with 10 mM citric acid. The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. Following a 5 μl injection, peptides were trapped for 5 min on a 5 μm Symmetry C18 180 μm I.D. × 20 mm column at 20μl/min in 99.9% A. The analytical column was held at 5% B for 5 min then switched in-line and a linear elution gradient of 5% B to 40% B was performed over 90 min at 300 nl/min. The analytical column was connected to a fused silica PicoTip emitter (New Objective, Cambridge, MA) with a 10 μm tip orifice. Phosphopeptide enriched samples were analyzed on an LTQ-Orbitrap XL mass spectrometer with a precursor MS scan in the Orbitrap from m/z 400-2000 with r = 60,000 at m/z 400 and a target AGC setting of 1e6 ions. In a data-dependent mode of acquisition, MS/MS spectra of the three most abundant precursor ions were acquired in the linear ion-trap with a target AGC setting of 1e3 ions. Max fill times were set to 1000 ms for full MS scans and 250 ms for MS/MS scans with minimum MS/MS triggering thresholds of 5000 counts. For all experiments, fragmentation occurred in the LTQ linear ion trap with a CID energy setting of 35% and a dynamic exclusion of 60 s was employed for previously fragmented precursor ions. For unbiased protein interaction studies, data were acquired on a Waters Synapt G2 mass spectrometer with precursor MS scans from m/z 50-2000 operating in a data-dependent mode of acquisition. The top three most abundant ions were selected for MS/MS using a charge state dependent CID energy setting with a 60s dynamic exclusion list employed.
2.4. Data analysis
Raw LC-MS/MS data files were processed in Mascot distiller (Matrix Science) and then submitted to independent Mascot database searches (Matrix Science) against a NCBI fungus entry. database (download date; Feb 18, 2012) appended with the reverse sequence of each forward entry. Search tolerances were 10 ppm for precursor ions and either 0.8 Da or 0.04 Da for product ions for LTQ-Orbitrap XL or Synapt G2 data, respectively, using trypsin specificity with up to two missed cleavages. Carbamidomethylation (+57.0214 Da on C) was set as a fixed modification, whereas oxidation (+15.9949 Da on M) and phosphorylation (+79.9663 Da on S, T, and Y) were considered a variable modifications. All searched spectra were imported into Scaffold (v 3.6.2, Proteome Software) and confidence thresholds were set using a Bayesian statistical algorithm based on the PeptideProphet and ProteinProphet algorithms which yielded a peptide and protein false discovery rate of 0% [30,31]. Only those proteins with at least two unique peptides to match were considered as being correct. Phosphorylation site localization was assessed by exporting peak lists directly from Scaffold into the online AScore algorithm (http://ascore.med.harvard.edu/ascore.html) [32]. A scaffold file containing the raw data is available for download here: https://discovery.genome.duke.edu/express/resources/3339/AspE_Phosphorylation_Interation.sf3
3. Results and Discussion
3.1. AspE interacts with actin-microtubule network and rest of the septin complex
We previously noted that, in contrast to AspA, AspB, AspC and AspD (the core septin homologs of cdc11p, cdc3p, cdc12p and cdc10p), the filamentous fungal-specific septin AspE showed filament-like tubular localization pattern along the A. fumigatus hyphae. Drugs that inhibit actin polymerization and microtubule stability mislocalized AspE from filaments to dot-like structures, which were presumed to be Microtubule Organizing Centers (MTOCs) [29]. Previous studies with mammalian septins have revealed the importance of actin and microtubules in the organization of septins [2]. In order to investigate if AspE interacted with the cytoskeletal proteins and the whole septin complex in vivo, we utilized the A. fumigatus strain expressing AspE-EGFP fusion protein to purify the AspE protein-complex by exploiting the EGFP-tag that bound to the GFP-Trap® agarose resin; bound proteins were directly subjected to LC-MS after proteolytic digestion. As shown in Table 1, a total of 48 proteins were identified with at least 2 unique peptides to match. The top identified protein was AspE with 40 unique peptides to match and with 63% sequence coverage. All of the other septins (AspB, AspD, AspA and AspC) were identified as AspE-binding partners along with other major proteins, including actin, tubulin (beta chain, alpha-1 subunit, beta subunit), Hsp70 chaperone, and translational elongation factor subunits (EF-1 and EF-2). Previously, the interaction of the mammalian septin, Sept6p, with non-septin binding partners, including Hsp70 and Hsp70-like proteins, was shown but significance of these interactions was not studied further [33]. Apart from ribosomal proteins, we also identified some metabolic-related enzymes such as phosphoglycerate kinase, isocitrate dehydrogenase, succinyl-CoA synthetase and calmodulin-dependent protein kinase as AspE binding proteins.
Table 1.
| Protein | NCBI accession number |
Molecular weight (kDa) |
Identified peptides |
|---|---|---|---|
| Septin Proteins | |||
| AspE | gi|71000419 | 62 | 40 |
| AspB | gi|70986973 | 59 | 13 |
| AspD | gi|70994952 | 39 | 10 |
| AspA | gi|119479605 | 43 | 8 |
| AspC | gi|70985058 | 44 | 7 |
| Proteins related to cytoskeleton organization | |||
| Actin | gi|115386236 | 40 | 11 |
| Tubulin beta chain | gi|119495802 | 50 | 8 |
| Tubulin alpha-1 subunit | gi|70990312 | 50 | 9 |
| Tubulin beta subunit | gi|70982752 | 50 | 4 |
| Tubulin TubB subunit | gi|71002302 | 50 | 3 |
| Proteins related to heat shock | |||
| Hsp70 chaperone (HscA) | gi|70983346 | 67 | 10 |
| Heat shock 70 kDa protein | gi|115385867 | 70 | 5 |
| Mitochondrial Hsp70 chaperone (Ssc70) | gi|159129408 | 72 | 3 |
| Heat shock protein 60, mitochondrial precursor | gi|115443330 | 62 | 2 |
| Proteins related to protein synthesis and folding | |||
| Translation elongation factor EF-1 alpha subunit | gi|146322501 | 54 | 8 |
| Translation elongation factor EF-2 subunit | gi|71002010 | 93 | 8 |
| 40S Ribosomal protein S3 | gi|70990926 | 29 | 5 |
| 40S Ribosomal protein S14 | gi|115384692 | 16 | 4 |
| Ribosomal protein S5 | gi|119488785 | 28 | 4 |
| T-complex protein 1, theta subunit | gi|119500564 | 61 | 4 |
| Eukaryotic Translation initiation factor 4 | gi|71000162 | 46 | 4 |
| T-complex protein 1, gamma subunit | gi|115391547 | 57 | 3 |
| Ribosomal protein S13 | gi|154280018 | 17 | 3 |
| 40S Ribosomal protein S7 | gi|115396282 | 23 | 2 |
| 40S Ribosomal protein S27 | gi|115402529 | 9 | 2 |
| 40S Ribosomal protein S5-A | gi|119186515 | 24 | 2 |
| Ribosomal protein S13p/S18e | gi|119473095 | 18 | 2 |
| 40S Ribosomal protein S16 | gi|115384680 | 16 | 2 |
| 60S Ribosomal protein L12 | gi|119497097 | 18 | 2 |
| 40S Ribosomal protein S17 | gi|116194718 | 17 | 2 |
| 40S Ribosomal protein S20 | gi|115398580 | 15 | 2 |
| 40S Ribosomal protein S3aE | gi|115438592 | 29 | 2 |
| 60S Ribosomal protein L4 | gi|119498945 | 41 | 2 |
| T-complex protein 1, zeta subunit | gi|119195049 | 59 | 2 |
| Proteins related to metabolism | |||
| Alcohol dehydrogenase | gi|159126256 | 37 | 11 |
| Bifunctional pyrimidine biosynthesis protein | gi|70992333 | 248 | 6 |
| Phosphoglycerate kinase (pgkA) | gi|119495924 | 45 | 6 |
| Putative flavohaemoglobin | gi|50788082 | 46 | 5 |
| Isocitrate dehydrogenase LysB | gi|119468338 | 38 | 4 |
| Isocitrate dehydrogenase, NAD-dependent | gi|119495397 | 42 | 4 |
| Succinyl–CoA synthetase beta subunit | gi|119486728 | 48 | 3 |
| ATP synthase alpha chain (mitochondrial) | gi|340521314 | 59 | 3 |
| ATP synthase beta chain (mitochondrial) | gi|115401284 | 54 | 3 |
| S-adenosylmethionine synthetase | gi|119495861 | 42 | 2 |
| 5-oxo-L-prolinase | gi|159130239 | 139 | 2 |
| Homocitrate synthase (mitochondrial) | gi|115492729 | 51 | 2 |
| Isocitrate dehydrogenase subunit 1 (mitochondrial) | gi|115400882 | 42 | 2 |
| Calmodulin dependent protein kinase | gi|119481599 | 46 | 2 |
3.2. Phosphorylation of AspE occurs in the polybasic region, G-domain and the C terminal end
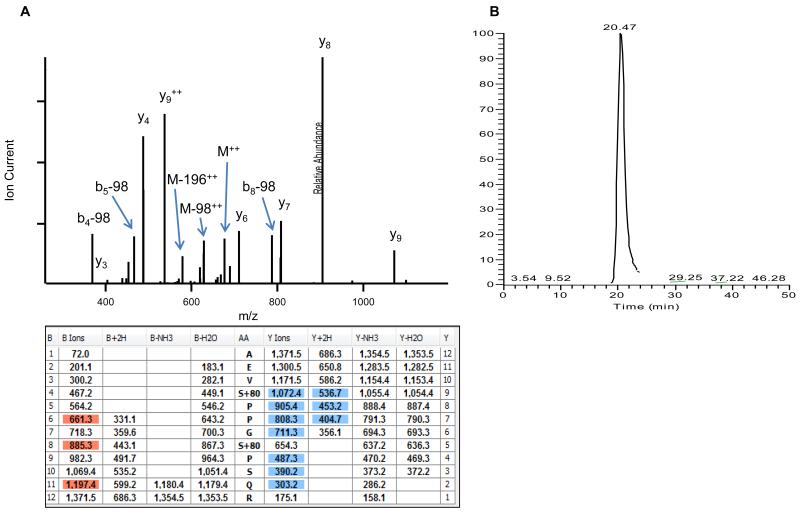
Earlier reports in S. cerevisiae have shown that phosphorylation of cdc3p regulates the ratio between septin rings and unbound monomeric septin [25], and phosphorylation of cdc10p and shs1p regulate septin collar formation and the fluidity of the septin ring at the bud neck, respectively [9,17,23,24]. A recent report analyzing septin phosphorylations and mutation of the phosphorylatable residues from A. gossypii also showed that phosphorylation plays a key role in septin organization [28]. Because there are no data available on phosphorylation of septins in any Aspergillus species [16], coupled with the probability of phosphorylation resulting in the formation of higher ordered septin structures, we were interested in examining the phosphorylation status of the A. fumigatus septins. To accomplish this, the AspE-GFP purified protein complex was subjected to phosphopeptide enrichment and the sample was subjected to LC-MS/MS to identify the specific phosphorylation sites on AspE. As shown in Table 2, a total of 6 phosphorylation sites were detected in 5 unique phosphorylated peptides. All of the phosphorylated residues in AspE were serines, located at positions Ser48, Ser209, Ser283, Ser430, Ser434 and Ser533. An example annotated MS/MS spectrum of the doubly phosphorylated peptide AEVS*PPGS*PSQR is shown in Figure 1A with the corresponding fragment ion coverage table. Selected ion chromatogram traces of the intact precursor ion (Figure 1B) illustrate a single peak at retention time ~20.5 min. As localization of the phosphorylation modification cannot be directly assessed by Mascot ion scores alone, each of these peptides were subjected to AScore scoring, a site localization probability algorithm publically available at http://ascore.med.harvard.edu/ascore.html [32]. The results indicated high confidence localizations (>99%) for all 6 of the phosphorylated residues (Table 2). As shown in Fig. 2, while Ser48 is present in the N-terminal polybasic domain and the Ser533 is at the C-terminus just after the septin unique element (SUE), 2 phosphorylated residues (Ser209 and Ser283) were present between the G1 and G3 domains. Two other sites, Ser430 and Ser434, were close to each other and resided between G4 domain and the SUE.
Table 2.
| Unique Phosphopeptide |
Phosphorylated Residue |
Mascot Ion Score |
Ascore1 | Localized2 |
|---|---|---|---|---|
| AKS*TEPLSER | S48 | 40.5 | 24.4 | Yes |
| S*PEEVEAYER | S209 | 31.0 | 65.5 | Yes |
| VIRS*PGFR | S283 | 28.4 | Unambiguous | Yes |
| AEVS*PPGS*PSQR | S430, S434 | 62.9 | 24.8, 38.5 | Yes, Yes |
| NGISAS*PMQK | S533 | 31.3 | 51.6 | Yes |
Unique phosphorylated residues within AspE identified by TiO2-based phosphoenrichment followed by LC-MS/MS analysis.
AScore phosphorylation localization score reported from http://ascore.med.harvard.edu/.
Ascore values >20 indicate greater than 99% chance of correct localization versus other phosphorylatable residues.
Fig. 1.
(A) Annotated MS/MS spectrum of the doubly phosphorylated peptide AEVS*PPGS*PSQR identified within AspE following TiO2 enrichment and high-resolution LC-MS/MS analysis. (B) Extracted ion chromatogram of m/z 686.2700 +/−10ppm, corresponding to the doubly charged precursor ion of AEVS*PPGS*PSQR, resulted in a single chromatographic elution profile at 27.5 min.
Fig. 2.
Septin AspE sequence showing various domains and sites of phosphorylations. Aspergillus fumigatus AspE amino acid sequence showing the 3 major domains including the N-terminus, the G-domain (in orange) and the C-terminus (in grey). The respective G-motifs (G1, G3 and G4) are underlined and in bold red color. The phosphorylated serine residues are boxed in yellow. Arrows indicate the phosphorylated residues within the G-domain and arrow heads indicate the phosphorylated residues in the N and C-termini. The septin specific motifs (S1, S2, S3 and S4) are double underlined. The other conserved single amino acid residues are indicated in green and underlined. The Septin Unique Element at the C-terminus is boxed. All the motifs were assigned based on Pan et al [7].
3.3. Conservation of the AspE phosphorylated residues among Aspergillus species
Although AspE from different Aspergillus species is highly conserved in the G domain, the N and C-terminal regions are variable (Fig. 3). Among the 6 phosphorylation sites detected, 4 sites were conserved at positions Ser48, Ser283, Ser434 and Ser533. The polybasic region in the N-terminus is thought to be involved in membrane interaction of septin filaments through phosphatidylinositol-4,5-bisphosphate (PIP2) binding and the presence of PIP2 promoted filament assembly and organization [34]. It is therefore possible that phosphorylation of the conserved Ser48 residue within the polybasic region is involved in membrane association of AspE. Moreover, the phosphorylation we observed at the conserved Ser533 site in the divergent C-terminus may also be very significant for AspE filamentation because phosphorylations occurring in the N and C termini of the core septins (cdc3p, cdc10p, cdc11p and cdc12p) are known to regulate the assembly and disassembly of the septin ring [16]. As shown in Fig. 2, only one (Ser283) of the two phosphorylation sites (Ser209 and Ser283) within the G1 and G3 motifs is conserved. Between the G4 motif and S4 motif, phosphorylation occurred at a short serine-proline rich region on the Ser430 and Ser434, but only Ser434 was conserved among different Aspergilli (Fig. 3). While further experiments are required to understand the significance of these phosphorylation sites in AspE, it is possible that more than one kinase is responsible for the phosphorylation of the 6 serine residues we identified. Ser430 and Ser434 are present in a short proline-rich domain that could be a target of proline-directed kinases. The sequence 45RAKSTEPLS53 in which Ser48 was phosphorylated resembles the consensus site (RXXS/T) of phosphorylation by a calmodulin dependent protein kinase [35], and it is interesting to note that calmodulin dependent protein kinase was identified as an AspE interacting protein (Table 1). Further mutational analyses of these individual residues to either non-phosphorylable or phosphomimetic residues would reveal their specific roles in the localization of AspE to filaments, and as well as their role in the organization or assembly of the whole septin complex. Additionally, analyzing the phosphorylation status of the other septins in A. fumigatus would also be helpful because specific alterations in individual septin phosphorylation sites can result in severe cell defects [28]. Further studies directed towards deletion of the A. fumigatus septin genes and understanding the importance of post-translational modifications for their organization will be useful for future antifungal drug design and targeting.
Fig. 3.
Clustal alignment of AspE homologs from other Aspergillus species showing conservation of phosphorylated residues. AspE homologs from other Aspergillus species were aligned using the ClustalW program. The sites of conserved phosphorylated residues are boxed and indicated in red color. The non-conserved phosphorylated residues are indicated by arrowheads. The short serine-proline rich region is boxed in red. The N terminal (in white) and C terminal domains (in grey) and the G-domain (in orange) are shown.
Highlights.
In vivo interactions of the novel septin AspE were identified by GFP-Trap® affinity purification
Septins AspA, AspB, AspC and AspD interacted with AspE in vivo
Actin and tubulin interacted with AspE in vivo
AspE is phosphorylated at 6 serine residue in vivo
Acknowledgements
This work was supported by 1R21AI097541-01A1 from the NIH/NIAID to WJS.
Abbreviations
- GFP
green fluorescent protein
- Asp
Aspergillus septin
- LC-MS/MS
Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Cao L, Yu W, Wu Y, Yu L. The evolution, complex structures and function of septin proteins. Cellular and Molecular Life Sciences. 2009;66:3309–3323. doi: 10.1007/s00018-009-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- [3].Kinoshita M. Assembly of Mammalian Septins. Journal of Biochemistry. 2003;134:491–496. doi: 10.1093/jb/mvg182. [DOI] [PubMed] [Google Scholar]
- [4].Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends in Cell Biology. 2011;21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. The Journal of Cell Biology. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Onishi M, Koga T, Hirata A, Nakamura T, Asakawa H, Shimoda C, Bähler J, Wu J-Q, Takegawa K, Tachikawa H, Pringle JR, Fukui Y. Role of Septins in the Orientation of Forespore Membrane Extension during Sporulation in Fission Yeast. Molecular and Cellular Biology. 2010;30:2057–2074. doi: 10.1128/MCB.01529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pan F, Malmberg R, Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evolutionary Biology. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Saarikangas J, Barral Y. The emerging functions of septins in metazoans. EMBO Rep. 2011;12:1118–1126. doi: 10.1038/embor.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Garcia G, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. Subunit-dependent modulation of septin assembly: Budding yeast septin Shs1 promotes ring and gauze formation. The Journal of Cell Biology. 2011;195:993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gladfelter AS. Guides to the final frontier of the cytoskeleton: septins in filamentous fungi. Current Opinion in Microbiology. 2010;13:720–726. doi: 10.1016/j.mib.2010.09.012. [DOI] [PubMed] [Google Scholar]
- [11].Hernández-Rodríguez Y, Hastings S, Momany M. The Septin AspB in Aspergillus nidulans Forms Bars and Filaments and Plays Roles in Growth Emergence and Conidiation. Eukaryotic Cell. 2012;11:311–323. doi: 10.1128/EC.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lindsey R, Cowden S, Hernández-Rodríguez Y, Momany M. Septins AspA and AspC Are Important for Normal Development and Limit the Emergence of New Growth Foci in the Multicellular Fungus Aspergillus nidulans. Eukaryotic Cell. 2010;9:155–163. doi: 10.1128/EC.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].L L, González-Novo A, Jiménez A, Sánchez-Pérez M, Jiménez J. Role of the septin Cdc10 in the virulence of Candida albicans. Microbiol Immunol. 2006;50:499–511. doi: 10.1111/j.1348-0421.2006.tb03820.x. [DOI] [PubMed] [Google Scholar]
- [14].Dagdas YF, Yoshino K, Dagdas G, Ryder LS, Bielska E, Steinberg G, Talbot NJ. Septin-Mediated Plant Cell Invasion by the Rice Blast Fungus, Magnaporthe oryzae. Science. 2012;336:1590–1595. doi: 10.1126/science.1222934. [DOI] [PubMed] [Google Scholar]
- [15].Saunders DGO, Dagdas YF, Talbot NJ. Spatial Uncoupling of Mitosis and Cytokinesis during Appressorium-Mediated Plant Infection by the Rice Blast Fungus Magnaporthe oryzae. The Plant Cell Online. 2010;22:2417–2428. doi: 10.1105/tpc.110.074492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hernández-Rodríguez Y, Momany M. Posttranslational modifications and assembly of septin heteropolymers and higher-order structures. Current Opinion in Microbiology. 2012;15:660–668. doi: 10.1016/j.mib.2012.09.007. [DOI] [PubMed] [Google Scholar]
- [17].Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-Dependent Regulation of Septin Dynamics during the Cell Cycle. Developmental Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- [18].Sinha I, Wang Y-M, Philp R, Li C-R, Yap WH, Wang Y. Cyclin-Dependent Kinases Control Septin Phosphorylation in Candida albicans Hyphal Development. Developmental Cell. 2007;13:421–432. doi: 10.1016/j.devcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- [19].Cvrcková F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes & Development. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- [20].Longtine MS, Fares H, Pringle JR. Role of the Yeast Gin4p Protein Kinase in Septin Assembly and the Relationship between Septin Assembly and Septin Function. The Journal of Cell Biology. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bouquin N, Barral Y, Courbeyrette R, Blondel M, Snyder M, Mann C. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. Journal of Cell Science. 2000;113:1435–1445. doi: 10.1242/jcs.113.8.1435. [DOI] [PubMed] [Google Scholar]
- [22].Tachikawa H, Bloecher A, Tatchell K, Neiman AM. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. The Journal of Cell Biology. 2001;155:797–808. doi: 10.1083/jcb.200107008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mortensen EM, McDonald H, Yates J, Kellogg DR. Cell Cycle-dependent Assembly of a Gin4-Septin Complex. Molecular Biology of the Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. The Journal of Cell Biology. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tang CS RS. Phosphorylation of the Septin Cdc3 in G1 by the Cdc28 Kinase Is Essential for Efficient Septin Ring Disassembly. Cell Cycle. 2002;1:38–45. [PubMed] [Google Scholar]
- [26].DeMay BS, Meseroll RA, Occhipinti P, Gladfelter AS. Regulation of Distinct Septin Rings in a Single Cell by Elm1p and Gin4p Kinases. Molecular Biology of the Cell. 2009;20:2311–2326. doi: 10.1091/mbc.E08-12-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meseroll RA, Howard L, Gladfelter AS. Septin ring size scaling and dynamics require the coiled-coil region of Shs1p. Molecular Biology of the Cell. 2012;23:3391–3406. doi: 10.1091/mbc.E12-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Meseroll RA, Occhipinti P, Gladfelter AS. Septin phosphorylation and coiled-coil domains function in cell and septin ring morphology in the filamentous fungus Ashbya gossypii. Eukaryotic Cell. 2012 doi: 10.1128/EC.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Juvvadi PR, Fortwendel JR, Rogg LE, Steinbach WJ. Differential localization patterns of septins during growth of the human fungal pathogen Aspergillus fumigatus reveal novel functions. Biochemical and Biophysical Research Communications. 2011;405:238–243. doi: 10.1016/j.bbrc.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [31].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Analytical Chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- [32].Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotech. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- [33].Kremer BE, Haystead T, Macara IG. Mammalian Septins Regulate Microtubule Stability through Interaction with the Microtubule-binding Protein MAP4. Molecular Biology of the Cell. 2005;16:4648–4659. doi: 10.1091/mbc.E05-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bertin A, McMurray MA, Thai L, Garcia G, Iii, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate Promotes Budding Yeast Septin Filament Assembly and Organization. Journal of Molecular Biology. 2010;404:711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Braun AP, Schulman H. The Multifunctional Calcium/Calmodulin-Dependent Protein Kinase: From Form to Function. Annual Review of Physiology. 1995;57:417–445. doi: 10.1146/annurev.ph.57.030195.002221. [DOI] [PubMed] [Google Scholar]